Abstract
Reduced host cell reactivation (HCR) of a reporter gene containing 8-oxoguanine (8-oxoG) lesions in Cockayne syndrome (CS) fibroblasts has previously been attributed to increased 8-oxoG-mediated inhibition of transcription resulting from a deficiency in repair. This interpretation has been challenged by a report suggesting reduced expression from an 8-oxoG containing reporter gene occurs in all cells by a mechanism involving gene inactivation by 8-oxoG DNA glycosylase and this inactivation is strongly enhanced in the absence of the CS group B (CSB) protein. The observation of reduced gene expression in the absence of CSB protein led to speculation that decreased HCR in CS cells results from enhanced gene inactivation rather than reduced gene reactivation. Using an adenovirus-based β-galactosidase (β-gal) reporter gene assay, we have examined the effect of methylene blue plus visible light (MB + VL)-induced 8-oxoG lesions on the time course of gene expression in normal and CSA and CSB mutant human SV40-transformed fibroblasts, repair proficient and CSB mutant Chinese hamster ovary (CHO) cells and normal mouse embryo fibroblasts. We demonstrate that MB + VL treatment of the reporter leads to reduced expression of the damaged β-gal reporter relative to control at early time points following infection in all cells, consistent with in vivo inhibition of RNA polII-mediated transcription. In addition, we have demonstrated HCR of reporter gene expression occurs in all cell types examined. A significant reduction in the rate of gene reactivation in human SV40-transformed cells lacking functional CSA or CSB compared to normal cells was found. Similarly, a significant reduction in the rate of reactivation in CHO cells lacking functional CSB (CHO-UV61) was observed compared to the wild-type parental counterpart (CHO-AA8). The data presented demonstrate that expression of an oxidatively damaged reporter gene is reactivated over time and that CSA and CSB are required for normal reactivation.
Introduction
The correct chemical structure of DNA bases allows the specific pairing of adenine with thymine and guanine with cytosine, forming the basis by which accurate transmission of hereditary information through DNA replication and functional information through transcription occurs. The chemical properties of the bases leave them susceptible to alteration by a number of factors including reactive oxygen species (ROS) generated endogenously by metabolic processes of the cell, from environmental sources including ultraviolet (UV) radiation, ionising radiation and dietary sources. Due to its low redox potential, guanine is particularly susceptible to oxidation by ROS and can form a large number of oxidised products (1). Of the many oxidised guanine products formed, 7,8-dihydro-8-oxoguanine (8-oxoguanine; 8-oxoG) is one of the most frequent forms of oxidative DNA lesions generated in living cells (2). 8-oxoG can functionally mimic T, allowing it to easily base pair with either A or C during replication and transcription. Failure to recognise and repair an improper 8-oxoG:A following replication results in G to T transversions during replication and mutant transcripts during transcription of nascent mRNAs (1). The base excision repair (BER) pathway is responsible for removing oxidised bases, including 8-oxoG from chromosomal DNA and relies on DNA glycosylases to recognise specific lesions and initiate the repair process (3). 8-oxoguanine DNA glycosylase (hOGG1 in humans; OGG1 in mice) recognises 8-oxoG lesions base-paired with C and catalyses excision of the damaged base, initiating its repair (for a review of BER, see ref. 3). In addition, the Cockayne syndrome group B protein (CSB) has been shown to be involved in processing of the lesion (4–6). Among the many functions of CSB, the best studied to date is the role it plays in conjunction with the Cockayne syndrome group A (CSA) protein in coupling transcription by RNA polymerase II (polII) to repair of nucleotide excision repair (NER) substrates in actively transcribed genes via the transcription-coupled repair (TCR) subpathway of NER (7). Mutations in CSA and CSB in humans normally causes Cockayne syndrome, a segmental progeroid syndrome characterised by premature ageing and neurodegeneration beginning in early childhood (8). Using a plasmid-based β-galactosidase (β-gal) reporter gene treated with the photosensitiser methylene blue (MB) activated by visible light (VL) to generate 8-oxoG lesions, Spivak and Hanawalt (9) showed a reduced level of expression in SV40-transformed human fibroblasts deficient for CSA and CSB compared to normal. This observation was interpreted as defective host cell reactivation (HCR) of the reporter gene in CSA and CSB cells resulting from impaired removal of 8-oxoG, with the persistent 8-oxoG lesions hindering reporter gene transcription by RNA polII. The interpretation of this observation as an inability of cells lacking CSA and CSB to properly process methylene blue plus visible light (MB + VL)-induced 8-oxoG and reactivate gene expression has recently been challenged. Using a plasmid-based reporter construct, Khobta et al. (10) examined the expression of a MB + VL-treated green fluorescent protein (GFP) reporter gene in spontaneously transformed mouse embryo fibroblasts (MEFs) between 8 and 48 h after transfection. Compared to the non-damaged control vector, the 8-oxoG containing reporter demonstrated the same level of transcription at 8 h with ∼50% reduction in transcription over a subsequent 40-h period (10). In addition to wild-type (WT) MEFs, reporter gene expression was also examined in Csbm/m, Ogg1−/− and Csbm/mOgg1−/−-immortalised MEFs. The authors suggest that Ogg1 mediates inactivation of the reporter gene and that the effect is considerably enhanced by the absence of functional CSB (10). Based on their finding of no difference in reporter gene expression from damaged and undamaged plasmids at 8 h after infection, Khobta et al. (10) also conclude that 8-oxoG does not impede RNA polII. In addition, they conclude that the decreased expression levels of the oxidatively damaged reporter gene in CSB-deficient cells compared to normal cells (9,11) are not due to the lack of CSB-mediated repair/reactivation of the reporter construct, but rather Ogg1/hOGG1 mediated inactivation that is enhanced in the absence of functional CSB. Recently presented data by the same group (12) demonstrated decreased acetylation of histone H4 in oxidatively damaged plasmid DNA introduced into HeLa cells, suggesting gene silencing may be mediated by chromatin alterations in the 8- to 48-h period following transfection of the plasmid.
To further study the effect of 8-oxoG on gene expression, we have examined expression of a recombinant adenovirus-encoded β-gal reporter gene damaged by MB + VL in normal and CSA and CSB mutant SV40-transformed human fibroblasts; WT MEFs and repair-proficient and repair-deficient CHO cells with a mutation in the hamster CSB homologue. We demonstrate that MB + VL treatment of the reporter gene leads to reduced expression relative to control at early time points following infection in all cells examined and that HCR of reporter gene expression occurs in all cell types examined by a mechanism requiring CSA and CSB.
Materials and methods
Cells, virus and culture conditions
The SV40-transformed fibroblasts GM637F (normal), CSA-SV40 (CS3BE.S3.G1) and CSB-SV40 (CS1AN.S3.G2) were obtained from NIGMS (Camden, NJ, USA). The spontaneously transformed MEF line WT MEF (ERCC1+/+; referred to as WT MEF hereafter) (13) was obtained from Dr X-D Zhu McMaster University, Hamilton, Ontario, Canada. The SV40-transformed mouse embryo fibroblast cell line BC1-6 was from Dr S.E. Andrew University of Alberta, Edmonton, Alberta and has been previously described (14). The Chinese hamster ovary (CHO) cell lines CHO-AA8 (repair proficient parental) and CHO-UV61 (repair deficient) were provided by Dr Larry Thompson, Lawrence Livermore National Laboratory, Livermore, CA with the help of Dr Gordon Whitmore, Physics Division, Ontario Cancer Institute, Toronto, Ontario. Cell cultures were grown at 37°C in a humidified incubator in 5% CO2 and cultured in Eagle's α-minimal essential media (α-MEM) supplemented with 10% foetal bovine serum and antimycotic antibiotic (100 μg/ml penicillin, 100 μg/ml streptomycin and 250 ng/ml amphotericin B). The recombinant adenoviruses Ad5HCMVlacZ (AdCA17) and Ad5MCMVlacZ (AdCA35) were obtained from The Robert E. Fitzhenry Vector Laboratory, McMaster University, Hamilton, Ontario. The viruses were propagated, collected and titered as described previously (15).
Treatment of the virus with MB + VL
Preparation of MB was carried out under minimal ambient light conditions as described previously (16). Treatment of the virus was performed under minimal ambient light conditions as described previously (17). Briefly, a 4 ml volume of 20 μg/ml (53.5 μM) MB solution was prepared by diluting the MB stock in cold phosphate-buffered saline (PBS) (4°C) in a 35-mm Petri dish and subsequently shielded from any ambient light. An appropriate volume of stock Ad5MCMVlacZ was added to the Petri dish containing the MB solution in order to obtain the desired multiplicity of infection (MOI) upon infecting cells. Prior to exposing the virus to VL, an aliquot was removed for use as the undamaged control. The solution was then exposed to VL using a 1000 W bulb (General Electric, GE R1000) at a distance of 82 cm from the source for a defined period of time while being kept on ice with continuous stirring. After each defined VL exposure, aliquots were removed for use in infecting cells. The aliquots were diluted in unsupplemented α-MEM and used to infect cells.
Treatment of the virus with UVC
UV irradiation of the virus was carried out as previously described (18). Briefly, the virus was resuspended in a 35-mm Petri dish in cold PBS at the appropriate dilution to achieve an MOI of 100 pfu/ml upon infection of cells. Using a General Electric germicidal lamp (model G8T5) emitting predominately at 254 nm, the virus was irradiated with stirring on ice with an incident fluence rate of 2 J/m2/sec. After each UV exposure, 200 μl aliquots were removed from the viral preparation and appropriately diluted using unsupplemented α-MEM.
β-Galactosidase reporter gene expression assay (HCR)
We have previously reported a HCR assay for examining BER of MB + VL-induced 8-oxoG lesions in a number of different cell strains (16,17,19). The HCR assay utilises a recombinant non-replicating adenovirus (Ad) expressing the β-gal reporter gene under control of (16,17,19) the murine cytomegalovirus immediate early promoter (20) to examine the ability of different cell types to remove damage and reactivate reporter gene expression. Upon exposure to VL, MB leads to the formation of 8-oxoG lesions with a small number of other single-base oxidative lesions occurring (21,22). Cells were seeded for confluence (SV40-transformed cells at 3.5 × 104 cells per well; MEFs and CHO cells at 4 × 104 cells per well) in 96-well plates (Falcon, Franklin Lakes, NJ, USA). After seeding, cells were incubated for 18–24 h and subsequently infected with 40 μl of untreated or MB + VL-treated virus for 90 min at a MOI of 100 pfu/cell. Following the 90-min viral absorption period, the infection medium was aspirated and cells were overlaid with 200 ml of complete α-MEM and incubated for a further 1, 2, 3, 6, 12, 24 or 44 h before harvesting for measurement of β-gal activity. For the time course experiments, cells from the same pool were seeded into separate 96-well plates for each time point and infected with virus from the same preparation. A single HCR experiment consisted of triplicate wells for each treatment of the virus and triplicate wells of non-infected cells were used to obtain background levels of β-gal activity. β-Gal activity was scored as previously described (23).
Graphing and statistical analysis
All curves for inactivation of β-gal activity following MB + VL treatment of the virus were plotted using Origin Laboratory software. Each point on the graphs represents an arithmetic mean ± standard error of triplicate determinations of the β-gal activity at each VL exposure to the virus relative to the untreated control for single representative experiments. D37 values for each cell line were obtained by extrapolation from the HCR survival curves from each independent experiment. D37 values for each cell type were then calculated relative to the indicated repair-proficient cell line used within the same experiment and used as a measure of relative HCR capacity. For individual time course experiments, the D37 values for each cell line at each time point were calculated relative to the indicated repair proficient normal at 12 h after infection. The data calculated in this manner were pooled from independent experiments to construct the D37 reactivation time course curves. Statistical analysis of differences between relative D37 values was carried out using a one-sample two-tailed t-test with a confidence interval of 0.05. The χ2 goodness of fit test was used to quantify how well the D37 reactivation time course curves for repair-deficient cell lines matched the curves for the appropriate repair-proficient control cell line over the range of time points examined (24). For comparison of two cell types, χ2 values were obtained for each time point and summed over the indicated ranges to obtain P-values representing the goodness of fit between the two D37 reactivation curves.
Results
Normal HCR of the MB + VL-damaged reporter gene in SV40-transformed human skin fibroblasts requires CSA and CSB
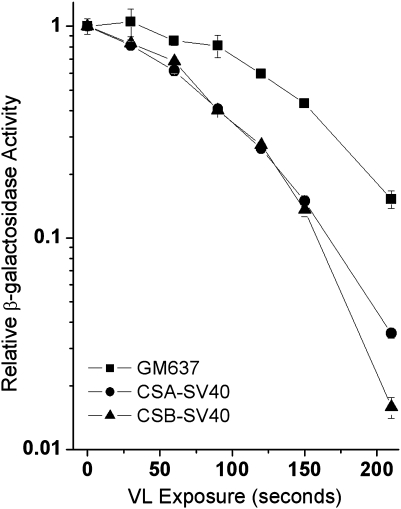
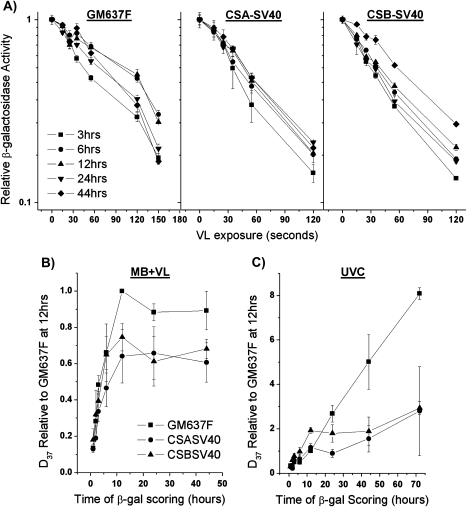
In the present work, we first examined expression from a MB + VL-damaged reporter gene using our recombinant adenovirus-based β-gal gene. In agreement with the results of Spivak and Hanawalt (9) using a plasmid-based reporter gene, we find a significant deficiency in reporter gene expression from CSA-SV40 and CSB-SV40 fibroblasts compared to the normal GM637F fibroblasts 44 h following infection with the MB + VL-treated adenovirus (Figure 1). Similar results were obtained at 24 h following infection with the MB + VL-treated adenovirus (data not shown). To investigate whether reactivation of reporter gene expression was taking place, we examined the expression of the MB + VL-treated reporter gene at various times after infection with the MB + VL-treated adenovirus. To facilitate examining expression at very early time points following infection, we used the AdCA35 virus, which drives higher levels of transgene expression than AdCA17 (20). The expression of the MB + VL-treated reporter gene was reduced compared to the non-treated reporter at early times after infection and expression of the damaged reporter gene increased when β-gal expression was assayed at later times (Figure 2A). The D37 time course curve (Figure 2B) was plotted using D37 values obtained from HCR plots. Expression of the β-gal reporter is reactivated by the host cell reaching a maximum level by 12 h in both normal and CS-deficient SV40-transformed fibroblasts. The rate of reactivation was significantly reduced in CSA-SV40 and CSB-SV40 compared to GM637F as measured by the χ2 goodness of fit test (P < 0.05).
Fig. 1.
Reduced HCR of the MB + VL-treated reporter in SV40-transformed CSA and CSB fibroblasts compared to the SV40-transformed normal fibroblast GM637F. CSA-SV40 and CSB-SV40 fibroblasts demonstrate a reduced capacity to reactivate β-gal expression from the MB + VL-treated reporter gene (AdCA17) 44 h after infection compared to the normal SV40-transformed fibroblast GM637F. The above β-gal survival curve is from a representative experiment done in triplicate. Each point represents β-gal expression for the given VL dose ± standard error. The average relative D37 values compared to G637F for CSA-SV40 and CSB-SV40 at 44 h (0.70 and 0.71, respectively) for three independent experiments were significantly decreased by one-sample two-tailed t-test (P < 0.05). A similar significant decrease in relative D37 was observed for CSA-SV40 and CSB-SV40 compared to GM637F at 24 h after infection (data not shown).
Fig. 2.
Time course of gene reactivation for MB + VL or UVC-damaged AdCA35 in SV40-transformed normal and CS fibroblasts. (A) HCR curves for β-gal expression from MB + VL-treated AdCA35 over time (3–44 h) in SV40-transformed human skin fibroblasts. Cells were infected with the MB + VL-treated (or mock treated) AdCA35 at an MOI of 100 pfu/cell and subsequently harvested for β-gal expression at 3, 6, 12, 24 and 44 h following infection. Representative survival curves for β-gal expression in SV40-transformed cells are shown. Each point is an average ± standard error (SE) of triplicate determinations. (B) Time course of gene reactivation (relative D37 ± SE) for MB + VL-damaged AdCA35 in SV40-transformed normal and CS-deficient fibroblasts shows reduced reactivation of reporter gene expression in CSA- and CSB-deficient fibroblasts. The increase in gene reactivation is significantly reduced in CSA-SV40 (6–44 h, P = 0.04 and 12–44 h, P = 0.02) and CSB-SV40 (3–44 h, P = 0.03; 6–44 h, P = 0.03; 12–44 h, P = 0.01; 24–44 h, P = 0.009) compared to GM637F as determined by the χ2 goodness of fit test. (C) Time course of gene reactivation (relative D37 ± SE) of UV damaged AdCA35 in SV40-transformed normal and CS-deficient fibroblasts. The increase in gene reactivation is significantly reduced in CSA-SV40 (6–72 h, P = 0.03; 12–72 h, P = 0.01; 24–72 h, P = 0.006 and 44–72 h, P = 0.02) and CSB-SV40 (for all time ranges: 3–72 h, 6–72 h, 12–72 h, 24–72 h, 44–72 h, P < 0.0001) compared to GM637F as determined by the χ2 goodness of fit test. For both (B and C), each point on the curves is the arithmetic average of at least three independent experiments done in triplicate determinations ± SE.
It has been reported that repair of UV-induced cyclobutane pyrimidine dimers in plasmids results in a time dependant recovery of transcription from damaged genes resulting from the gradual removal of UV-induced transcription blocking lesion (9,10,25,26). In contrast to the levelling off of reactivation by 12 h after infection for the MB + VL-treated reporter gene, infection of cells with a UVC-treated reporter gene resulted in a continual increase in reporter gene expression from 1 to 72 h after infection of GM637F cells (Figure 2C). Expression from the UVC-damaged virus increased at a significantly slower rate in CSA-SV40 and CSB-SV40 (P < 0.05 by χ2 goodness of fit test), consistent with defective TCR–NER and proficient global genome repair–NER in these cells.
Reactivation of expression from a MB + VL-damaged β-gal reporter gene in rodent cells
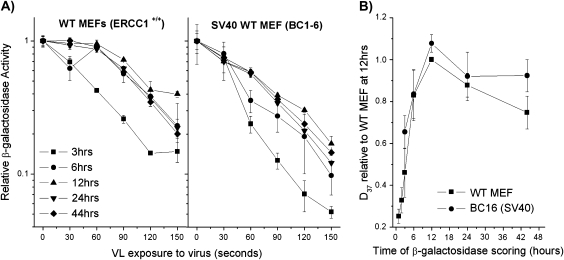
The experiments showing inactivation of a reporter gene containing oxidative DNA damage were carried out using normal and CSB-deficient MEFs (10). To address the possibility of a difference between human and rodent cells, we next examined expression of the MB + VL-damaged adenovirus-based reporter gene in MEFs and CHO cells. Expression of the MB + VL-treated reporter gene was measured in two different MEF lines. It can be seen that expression of the MB + VL-treated reporter gene increased over time reaching a maximum at 12 h in both WT MEF and BC1-6 (Figure 3A and B). Reporter gene expression was found to be significantly increased between 3 and 12 h for both WT MEF (P = 0.02) and BC1-6 (P = 0.009) by a two-sample t-test. At time points from 12 to 44 h after infection, we observed a slight decrease in the relative D37 value consistent with results published for the expression of a plasmid-based reporter gene during this time period (10,12). However, in the current work, the difference in the relative D37 value at 24 or 44 h compared to 12 h was not significant.
Fig. 3.
Time course of gene reactivation for MB + VL-damaged AdCA35 in WT MEFs (A) HCR curves for β-gal expression over time in MEFs. The spontaneously transformed MEFs WT MEF and the SV40-transformed MEFs BC1-6 were infected with MB + VL-damaged (or mock treated) AdCA35 at an MOI of 100 pfu/cell and subsequently harvested for β-gal expression at 1, 2, 3, 6, 12, 24 and 44 h after infection. Representative survival curves for β-gal expression for both MEF cell lines examined 3–44 h after infection are shown. Each point is an average ± standard error (SE) of triplicate determinations. (B) Change in the D37 value for each cell line. D37 values for each cell line at each time point were normalised to the D37 value obtained for WT MEF at 12 h for at least three independent experiments each done in triplicate. Each point is the average of the pooled results ± SE.
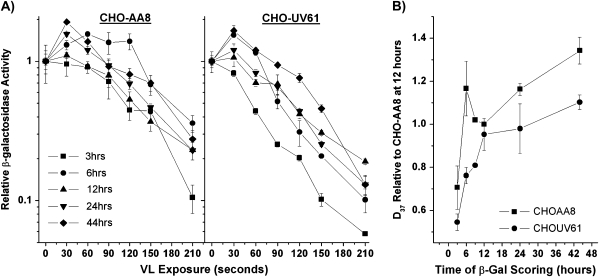
We also examined the time course of β-gal expression in repair-proficient and repair-deficient CHO cells to determine if the observed reactivation was specific to mice or if it occurred in other rodent species and to examine the role of CSB in gene reactivation. Figure 4A shows typical inactivation curves for β-gal activity in the repair-proficient parental line CHO-AA8 and the repair-deficient line CHO-UV61 carrying a mutation in the hamster homologue of CSB (27) and Figure 4B shows the time course of gene reactivation. These results demonstrate that the expression of the β-gal reporter was reactivated by the host cell in both CHO-AA8 and CHO-UV61 cells. This reactivation was significant by two-sample t-test from 3 to 6 h for CHO-AA8 (P = 0.009) and 3 to 12 h in CHO-UV61 (P = 0.001). In addition, the increase in reactivation over time (3–44 h) was significantly reduced in the CSB mutant CHO-UV61 compared to CHO-AA8 as determined by the χ2 goodness of fit test (P = 0.0439).
Fig. 4.
Time course of gene reactivation for MB + VL-damaged AdCA35 in repair-proficient and repair-deficient CHO cells. (A) HCR curves for β-gal expression over time in CHO cells. CHO-AA8 and the CHO-UV61 cells were infected with MB + VL-damaged (or mock treated) AdCA35 at an MOI of 100 pfu/cell and subsequently harvested for β-gal expression at 3, 6, 9, 12, 24 and 44 h after infection. Representative survival curves for β-gal expression for both CHO cell lines examined 3–44 h after infection are shown. Each point is an average ± standard error (SE) of triplicate determinations. (B) Change in the D37 value for each cell line. D37 values for each cell line at each time point were normalised to the D37 value obtained for CHO-AA8 at 12 h for at least three independent experiments each done in triplicate (n = 1 for 9-h time point, n = 2 for 24 and 44 h). Each point is the average of the pooled results ± SE. The increase in gene reactivation is significantly reduced in CHO-UV61 compared to CHO-AA8 cells of the time points examined (3–44 h, P = 0.0439; 6–44 h, P = 0.025; 12–44 h, P = 0.0301; 24–44 h, P = 0.0085) as determined by the χ2 goodness of fit test.
Discussion
The data presented here demonstrate that expression of a reporter gene containing oxidative DNA lesions is reactivated over time (1–12 h) by host cell mechanisms and that a normal level of reactivation in human fibroblasts requires the CSA and CSB gene products. The previous report by Khobta et al. examined the time course of expression from a plasmid-based reporter containing low levels of 8-oxoG in MEFs beginning 8–12 h after introduction through to 48 h. A reduction in gene expression with time was observed in repair-proficient WT MEFs, repair-deficient Ogg1−/− MEFs and to a greater extent in Csbm/m MEFs expressing a mutated copy of CSB mimicking the mutation in the human patient CS1AN (10,28). Based on their results, they suggested that oxidative DNA damage led to Ogg1/CSB-mediated reporter gene inactivation (10).
In the current work, we show that the change in relative D37 for expression of the MB + VL-treated adenovirus-encoded reporter gene slows and levels off from 12 to 44 h; however, no significant differences were observed between pooled relative D37 values over that time period. More importantly, compared to early times after infection (1–3 h), expression from the MB + VL-treated reporter was significantly increased at the later time points consistent with gene reactivation.
While the ability of 8-oxoG to block RNA polII transcription has been disputed (29–32), the observation that CSB can improve RNA polII elongation through 8-oxoG lesions suggests that at the very least, 8-oxoG leads to transient stalling of the RNA polII transcription complex (29). We demonstrate here that at early times after infection (1–3 h) with the reporter gene containing MB + VL-induced oxidative damage, gene expression is inhibited compared to the undamaged control in all cell types examined. The decrease in expression from the MB + VL-damaged adenovirus reporter compared to the undamaged control for all VL doses is consistent with an in vivo decrease of RNA polII transcription due to 8-oxoG lesions on the template strand. Reactivation of gene expression is consistent with BER and/or bypass of 8-oxoG lesions.
It has been shown that repair of 8-oxoG in MEF genomic DNA, as measured by loss of formamidopyrimidine-DNA glycosylase-sensitive sites (FSS) is ∼50% complete 4 h after induction by a photosensitiser (Ro 19-8022) and VL, >80% complete by 8 hrs and nearly 100% complete at 16 h (33). In the current work, expression of the MB + VL-damaged reporter gene in MEFs, CHO cells and human fibroblasts was found to be significantly increased between 1 and 12 h, but levelled off by 12 h after infection. This suggests that reactivation of the reporter gene is near completion by 12 h after infection, consistent with the time course for removal of 8-oxoG lesions from cellular DNA. In contrast, reactivation of the UVC-damaged reporter gene in the GM637F normal human fibroblasts continued to increase from 1 to 72 h after infection, consistent with the longer time course for removal of UVC-induced DNA damage in human cellular DNA (34,35).
Similar to human fibroblasts and MEFs, we observed a significant increase in reporter gene expression relative to control in WT CHO-AA8 cells and mutant CHO-UV61. The results presented are consistent with a role for the CSB homologue in hamsters in repair and/or bypass of MB + VL-induced 8-oxoG. The rate of change in relative D37 over time in CHO cells slows between 12 and 24 h; however, the continuous increase in gene expression over the time points examined suggests reactivation of MB + VL-induced 8-oxoG continues over a longer period of time in CHO cells compared to MEFs. Hamster cells do indeed repair 8-oxoG, as measured by loss of FSS at a slower rate than mouse cells with only 56% of FSS removed at 8 h (36) compared to >80% in mouse cells at the same time point (33). The hamster homologue of CSB, which is involved in repair of UV-induced damage (27) and oxidative damage (36), shows no significant involvement in RNA polII-mediated transcription (27). This suggests that CSB-stimulated lesion bypass and/or coupling of repair to transcription may be absent in hamster cells, possibly accounting for slower repair and subsequent gene reactivation.
It is possible that the decreased level of gene expression seen in MEFs lacking functional CSB compared to WT MEFs, as previously reported (10), could result from a failure of cells lacking CSB to reactivate gene expression from a damaged reporter to WT levels prior to 12 h. In the current work, we were unable to detect a significant change in expression of the MB + VL-treated reporter between 12 and 44 h after infection, whereas Khobta et al. (10) demonstrated a significant decrease in reporter gene expression over this time period. This difference could result from differences in the experimental conditions and the reporter gene employed.
Both studies employed MB + VL to generate 8-oxoG lesions in the reporter gene. MB + VL induces high yields of 8-oxoG, while oxidative pyrimidine modifications, sites of base loss and single-strand breaks (SSBs) are rare (37). The generation of SSBs by MB + VL has been shown to occur at a rate of ∼0.1 modifications per 10 000 bp, while 8-oxoG occurs at a rate of 2.7 modifications per 10 000 bp in supercoiled plasmid DNA (37). A similar ratio of SSBs to 8-oxoG lesions was observed in DNA isolated from treated bacteria (37) suggesting the mechanism and profile of DNA damage is independent of the system in which it is treated. Khobta et al. (10) employed MB + VL treatment of covalently closed plasmids of 0.8 μM MB + VL exposures resulting in the induction of an average of three FSS per plasmid, corresponding to an average of ∼0.5 FSS per transcribed strand of their GFP-encoding reporter gene. In the present work, we employed 53.5 μM MB plus from 10 to 150 sec VL treatment to the recombinant adenovirus-encoding lacZ reporter gene. Previous reports have shown that treatment of adenovirus with MB + VL has negligible effects on the protein capsid and subsequent infectivity of viral particles (38). Preliminary results indicate that VL exposures to the virus of 10–150 sec would result in an average of about one to six FSS per transcribed strand of the lacZ reporter gene (data not shown). This indicates that the 8-oxoG lesion frequency from VL exposures to the virus of 10–150 sec used in the HCR experiments reported here were somewhat greater those used in the study by Khobta et al., although of a similar order of magnitude.
In the work presented here, we utilised an adenovirus-based reporter system with confluent cultures, whereas Khobta et al. used a plasmid-based system with exponentially growing cultures (10,39). Regardless of the level of DNA damage, p53 upregulation, transcriptional activation and p53-dependant apoptosis are attenuated in confluent cultures compared to sparse growing cultures lacking cell–cell contacts (40). The higher cell density of the confluent cultures used in the current work may more closely reproduce the microenvironment in a living organism and account for the observed difference in gene inactivation. Differences in the level of expression and stability of the adenovirus encoded compared to the plasmid encoded reporter gene may account for the difference in the level of gene inactivation observed. Differences in the degree of chromatin association/modification between the two systems may also contribute to the difference (12).
Funding
National Cancer Institute of Canada with funds from the Canadian Cancer Society (16066).
Acknowledgments
We thank Robert W. Cowan, Faculty of Health Sciences, McMaster University and Shaqil Kassam, Faculty of Science, McMaster University, for performing initial experiments showing reduced HCR of the MB + VL-treated adenovirus-encoded reporter gene in CHO-UV61 compared to CHO-AA8 cells and in CSB-SV40 fibroblasts compared to GM637F, respectively.
Conflict of interest statement: None declared.
References
- 1.Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 2.Park EM, Shigenaga MK, Degan P, Korn TS, Kitzler JW, Wehr CM, Kolachana P, Ames BN. Assay of excised oxidative DNA lesions: isolation of 8-oxoguanine and its nucleoside derivatives from biological fluids with a monoclonal antibody column. Proc. Natl. Acad. Sci. U.S.A. 1992;89:3375–3379. doi: 10.1073/pnas.89.8.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dianov G, Bischoff C, Sunesen M, Bohr VA. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 1999;27:1365–1368. doi: 10.1093/nar/27.5.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osterod M, Larsen E, Le Page F, Hengstler JG, Van Der Horst GT, Boiteux S, Klungland A, Epe B. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene. 2002;21:8232–8239. doi: 10.1038/sj.onc.1206027. [DOI] [PubMed] [Google Scholar]
- 6.Tuo J, Jaruga P, Rodriguez H, Bohr VA, Dizdaroglu M. Primary fibroblasts of Cockayne syndrome patients are defective in cellular repair of 8-hydroxyguanine and 8-hydroxyadenine resulting from oxidative stress. FASEB J. 2003;17:668–674. doi: 10.1096/fj.02-0851com. [DOI] [PubMed] [Google Scholar]
- 7.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat. Rev. Genet. 2009;10:756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 8.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 9.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but normal in UV-sensitive syndrome fibroblasts. DNA repair. 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 10.Khobta A, Kitsera N, Speckmann B, Epe B. 8-Oxoguanine DNA glycosylase (Ogg1) causes a transcriptional inactivation of damaged DNA in the absence of functional Cockayne syndrome B (Csb) protein. DNA repair. 2009;8:309–317. doi: 10.1016/j.dnarep.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Pastoriza-Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb(-)/Ogg1(-) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 12.Khobta A, Anderhub S, Kitsera N, Epe B. Gene silencing induced by oxidative DNA base damage: association with local decrease of histone H4 acetylation in the promoter region. Nucleic Acids Res. 2010;38:4285–4295. doi: 10.1093/nar/gkq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 14.Fritzell JA, Narayanan L, Baker SM, et al. Role of DNA mismatch repair in the cytotoxicity of ionizing radiation. Cancer Res. 1997;57:5143–5147. [PubMed] [Google Scholar]
- 15.Graham FL, Prevec L. Methods in Molecular Biology. Clifton, NJ, USA: Humana Press; 1991. [Google Scholar]
- 16.Pitsikas P, Lee D, Rainbow AJ. Reduced host cell reactivation of oxidative DNA damage in human cells deficient in the mismatch repair gene hMSH2. Mutagenesis. 2007;22:235–243. doi: 10.1093/mutage/gem008. [DOI] [PubMed] [Google Scholar]
- 17.Kassam SN, Rainbow AJ. Deficient base excision repair of oxidative DNA damage induced by methylene blue plus visible light in xeroderma pigmentosum group C fibroblasts. Biochem. Biophys. Res. Commun. 2007;359:1004–1009. doi: 10.1016/j.bbrc.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Bennett CB, Rainbow AJ. Enhanced reactivation and mutagenesis of UV-irradiated adenovirus in normal human fibroblasts. Mutagenesis. 1988;3:157–164. doi: 10.1093/mutage/3.2.157. [DOI] [PubMed] [Google Scholar]
- 19.Kassam SN, Rainbow AJ. UV-inducible base excision repair of oxidative damaged DNA in human cells. Mutagenesis. 2009;24:75–83. doi: 10.1093/mutage/gen054. [DOI] [PubMed] [Google Scholar]
- 20.Addison CL, Hitt M, Kunsken D, Graham FL. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 1997;78:1653–1661. doi: 10.1099/0022-1317-78-7-1653. [DOI] [PubMed] [Google Scholar]
- 21.Floyd RA, West MS, Eneff KL, Schneider JE. Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch. Biochem. Biophys. 1989;273:106–111. doi: 10.1016/0003-9861(89)90167-7. [DOI] [PubMed] [Google Scholar]
- 22.Tuite EM, Kelly JM. Photochemical interactions of methylene blue and analogues with DNA and other biological substrates. J. Photochem. Photobiol. 1993;21:103–124. doi: 10.1016/1011-1344(93)80173-7. [DOI] [PubMed] [Google Scholar]
- 23.Pitsikas P, Francis MA, Rainbow AJ. Enhanced host cell reactivation of a UV-damaged reporter gene in pre-UV-treated cells is delayed in Cockayne syndrome cells. J. Photochem. Photobiol. 2005;81:89–97. doi: 10.1016/j.jphotobiol.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Bevington PR, Robinson DK. Data Reduction and Error Analysis for the Physical Sciences. Boston, MA, USA: McGraw-Hill; 2003. [Google Scholar]
- 25.Ganesan AK, Hunt J, Hanawalt PC. Expression and nucleotide excision repair of a UV-irradiated reporter gene in unirradiated human cells. Mutat. Res. 1999;433:117–126. doi: 10.1016/s0921-8777(98)00070-6. [DOI] [PubMed] [Google Scholar]
- 26.Johnson JM, Latimer JJ. Analysis of DNA repair using transfection-based host cell reactivation. Methods Mol. Biol. 2005;291:321–335. doi: 10.1385/1-59259-840-4:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orren DK, Dianov GL, Bohr VA. The human CSB (ERCC6) gene corrects the transcription-coupled repair defect in the CHO cell mutant UV61. Nucleic Acids Res. 1996;24:3317–3322. doi: 10.1093/nar/24.17.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Horst GT, van Steeg H, Berg RJ, et al. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell. 1997;89:425–435. doi: 10.1016/s0092-8674(00)80223-8. [DOI] [PubMed] [Google Scholar]
- 29.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kathe SD, Shen GP, Wallace SS. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 31.Kuraoka I, Endou M, Yamaguchi Y, Wada T, Handa H, Tanaka K. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 2003;278:7294–7299. doi: 10.1074/jbc.M208102200. [DOI] [PubMed] [Google Scholar]
- 32.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA repair. 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 33.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kantor GJ, Setlow RB. Rate and extent of DNA repair in nondividing human diploid fibroblasts. Cancer Res. 1981;41:819–825. [PubMed] [Google Scholar]
- 35.Konze-Thomas B, Levinson JW, Maher VM, McCormick JJ. Correlation among the rates of dimer excision, DNA repair replication, and recovery of human cells from potentially lethal damage induced by ultraviolet radiation. Biophys. J. 1979;28:315–325. doi: 10.1016/S0006-3495(79)85179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunesen M, Stevnsner T, Brosh RM, Jr., Dianov GL, Bohr VA. Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene. 2002;21:3571–3578. doi: 10.1038/sj.onc.1205443. [DOI] [PubMed] [Google Scholar]
- 37.Epe B, Pflaum M, Boiteux S. DNA damage induced by photosensitizers in cellular and cell-free systems. Mutat. Res. 1993;299:135–145. doi: 10.1016/0165-1218(93)90091-q. [DOI] [PubMed] [Google Scholar]
- 38.Schagen FH, Moor AC, Cheong SC, Cramer SJ, van Ormondt H, van der Eb AJ, Dubbelman TM, Hoeben RC. Photodynamic treatment of adenoviral vectors with visible light: an easy and convenient method for viral inactivation. Gene Ther. 1999;6:873–881. doi: 10.1038/sj.gt.3300897. [DOI] [PubMed] [Google Scholar]
- 39.Kitsera N, Khobta A, Epe B. Destabilized green fluorescent protein detects rapid removal of transcription blocks after genotoxic exposure. Biotechniques. 2007;43:222–227. doi: 10.2144/000112479. [DOI] [PubMed] [Google Scholar]
- 40.Bar J, Cohen-Noyman E, Geiger B, Oren M. Attenuation of the p53 response to DNA damage by high cell density. Oncogene. 2004;23:2128–2137. doi: 10.1038/sj.onc.1207325. [DOI] [PubMed] [Google Scholar]