Abstract
Trichloroethylene (TCE) is a widely used industrial chemical and a common environmental contaminant. It is a well-known carcinogen in rodents and a probable carcinogen in humans. Studies utilizing panels of mouse inbred strains afford a unique opportunity to understand both metabolic and genetic basis for differences in responses to TCE. We tested the hypothesis that strain- and liver-specific toxic effects of TCE are genetically controlled and that the mechanisms of toxicity and susceptibility can be uncovered by exploring responses to TCE using a diverse panel of inbred mouse strains. TCE (2100 mg/kg) or corn oil vehicle was administered by gavage to 6- to 8-week-old male mice of 15 mouse strains. Serum and liver were collected at 2, 8, and 24 h postdosing and were analyzed for TCE metabolites, hepatocellular injury, and gene expression of liver. TCE metabolism, as evident from the levels of individual oxidative and conjugative metabolites, varied considerably between strains. TCE treatment-specific effect on the liver transcriptome was strongly dependent on genetic background. Peroxisome proliferator–activated receptor–mediated molecular networks, consisting of the metabolism genes known to be induced by TCE, represent some of the most pronounced molecular effects of TCE treatment in mouse liver that are dependent on genetic background. Conversely, cell death, liver necrosis, and immune-mediated response pathways, which are altered by TCE treatment in liver, are largely genetic background independent. These studies provide better understanding of the mechanisms of TCE-induced toxicity anchored on metabolism and genotype-phenotype correlations that may define susceptibility or resistance.
Keywords: trichloroethylene, metabolism, genetics, peroxisome proliferator-activated receptor alpha, acute exposure, gene expression
Trichloroethylene (TCE), a ubiquitous environmental contaminant, continues to be a major recurrent challenge in mechanistic and regulatory toxicology (NRC, 2006). TCE is widely used as an industrial chlorinated organic solvent, and the potential for human exposure is substantial from a variety of sources such as indoor and ambient air, soil, and groundwater. TCE is thought to cause adverse health effects through multiple organ toxicities (IARC, 1995). Acute exposure results primarily in the adverse effects on the nervous system, whereas long-term exposures can result in liver- (Bull, 2000), kidney- (Lash et al., 2000b), and neurotoxicity (Barton and Clewell, 2000), as well as autoimmune disorders (Cooper et al., 2009). Adverse reproductive and developmental effects of TCE have also been reported (Pastino et al., 2000). Although TCE is classified as “reasonably anticipated to be a human carcinogen” (U.S. DHHS/PHS/NTP, 2005), it has been difficult to achieve consensus on its risks to humans (NRC, 2006). Despite the fact that a large body of information exists on adverse health effects of TCE, significant gaps remain because of the complexity of TCE metabolism and apparent existence of many modes of action.
The metabolic transformation of TCE involves formation of many metabolites that vary in their toxicity and organ-specific effects (Lash et al., 2000a). Cytochrome P450–dependent oxidation and glutathione conjugation are two primary pathways for metabolism in both humans and animals, albeit important species differences exist in the relative contribution of each pathway to liver and kidney toxicity that ensues (NRC, 2006). Although the precise mechanisms of TCE action and the reasons for species-specific differences in toxicity are yet to be fully understood, it is well accepted that knowledge of TCE metabolism is critical for determining susceptibility, target organ specificity, and gender and species differences (NRC, 2006). Two recent National Research Council reports addressing the challenges in chlorinated solvent toxicity (NRC, 2006, 2010) posit that the development of sensitive analytical methodologies for assessment of TCE metabolism and quantitative assessment of the extent of interindividual variability in TCE metabolism are among the critical needs in TCE assessment.
The development of a sensitive analytical method for key oxidative and glutathione conjugation TCE metabolites dichloroacetic acid (DCA), trichloroacetic acid (TCA), S-(1,2-dichlorovinyl)-L-cysteine (DCVC), and S-(1,2-dichlorovinyl) glutathione (DCVG) allows for an increase in throughput and a decrease in the number of animals required for mechanistic studies on TCE in rodents (Kim et al., 2009b). This method was used to investigate the time-concentration profiles of DCA, TCA, DCVG, and DCVG formation and elimination in the mouse (Kim et al., 2009a). In this study, we aimed to quantify the extent of interstrain differences, analogous to interindividual differences in humans (Rusyn et al., 2010), in TCE metabolism through oxidative and glutathione conjugation pathways in a panel of genetically diverse mouse strains. We found that TCE metabolism varies considerably between strains, and this knowledge may not only be used to determine the extent of within-species variability but could also serve as the scientific basis for selection of strains for assessment of organ-specific toxic effects in longer term studies. In addition, to advance the understanding of the mode of action of TCE, including interindividual differences in toxicity, we performed pathway-based assessment of the liver gene expression data. The mouse population experimental approach revealed that TCE-induced liver toxicity is a predominant genetic background–independent mode of action, whereas peroxisomal proliferation is a key genotype-dependent pathway altered by TCE.
MATERIALS AND METHODS
Animals and treatments.
Male mice (aged 7–9 weeks) were obtained from the Jackson Laboratory (Bar Harbor, ME) and housed in polycarbonate cages on Sani-Chips irradiated hardwood bedding (P. J. Murphy Forest Products Corp., Montville, NJ). Animals were fed NTP-2000 wafer diet (Zeigler Brothers, Inc., Gardners, PA) and water ad libitum on a 12-h light-dark cycle. Mice utilized in this study comprise 14 inbred strains that have been densely genotyped (Frazer et al., 2007): 129S1/SvImJ, A/J, BALB/cByJ, BTBR+ tf/J, CAST/EiJ, C3H/HeJ, C57BL/6J, DBA/2J, FVB/NJ, MOLF/EiJ, NOD/LtJ, NZW/LacJ, PWD/PhJ, and KK/HlJ. An F1 hybrid mouse strain, B6C3F1/J, was also used. On the day of treatment (at 9 a.m.), fed mice were treated orally by gavage with TCE (2100 mg/kg) in corn oil (vehicle, 10 ml/kg). Mice were sacrificed at 2, 8, and 24 h after treatment. The experimental design was selected based on the previous pharmacokinetic analysis of TCE metabolism in male B6C3F1 mice (Kim et al., 2009b). Liver and kidney sections were placed in neutral buffered formalin, and the remainder of the tissue was frozen in liquid nitrogen and stored at −80°C. Hematoxylin/eosin-stained sections of liver and kidney were evaluated using light microscopy. Blood was collected from vena cava, and serum was prepared by centrifugation using Z-gel tubes (Sarstedt, Germany) according to the manufacturer’s instructions and stored at −80°C. Serum tissue damage biomarkers (blood urea nitrogen, BUN; alanine aminotransferase, ALT; and aspartate aminotransferase, AST) were analyzed using standard protocols. All studies were conducted with approval of the Institutional Animal Care and Use Committee.
Determination of TCE metabolites in serum.
Concentrations of DCA, TCA, DCVG, and DCVC in mouse serum were determined as detailed elsewhere (Kim et al., 2009a). DCA and TCA were measured by high performance liquid chromatography-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) with a Finnigan Surveyor autosampler and pump coupled to a Finnigan TSQ Quantum triple-quadrupole mass spectrometer (Thermo Finnigan, San Jose, CA). Determination of DCVC and DCVG was performed by HPLC-ESI-MS/MS with an Aquity UPLC (Waters, Milford, MA) system coupled to a TSQ Quantum Ultra triple-quadrupole mass analyzer (Thermo Finnigan) using a heat-assisted electrospray ionization source in positive ion mode. Quantification was based on peak areas relative to the stable isotope-labeled internal standards, and the calibration curve was constructed in each batch. Limits of detection were determined by a signal-to-noise ratio of 3:1 and were as follows: TCA, 0.4 nmol/ml; DCA, 0.01 nmol/ml; DCVG and DCVC, 0.001 nmol/ml. ANOVA was used to treat metabolite differences as a function of strain. To evaluate whether there was a significant strain effect on the concentrations of TCE metabolites in serum, a one-way ANOVA model (concentration = strain + error) was applied for each of TCA, DCA, DCVC, and DCVG at each time point (2, 8, and 24 h). For each model, the ANOVA F statistic was used to assess the significance of the strain effect.
Microarray analysis of liver gene expression.
Total RNA was isolated from frozen liver samples (three vehicle and three TCE-treated animals per strain, 24-h time point) using the RNeasy (Qiagen, Valencia, CA) kit according to the manufacturer's instructions. RNA concentration and quality were assessed using ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), respectively. All RNA samples were hybridized to arrays individually; none were pooled. RNA amplifications and labeling were performed using Low RNA Input Linear Amplification kits (Agilent Technologies). Following the recommendations of the Toxicogenomics Research Consortium's standardization experiment (Bammler et al., 2005), a two-color array design was used. Specifically, 750 ng of total RNA from each mouse liver was amplified and labeled with fluorescent dye Cy5, and 750 ng of a common mouse reference RNA (Icoria Inc., RTP, NC) was labeled with the fluorescent dye Cy3. Labeled complementary RNA was then processed and hybridized to Agilent Mouse Arrays (catalog# G4122F; ∼44,000 features) according to the manufacturer's protocol. Raw data from scanned arrays were processed using Agilent's Feature Extraction software (v 7.5) according to the manufacturer's instructions. Quality control reports provided by the software were used to judge the quality of each array. No arrays failed in this experiment. The output data were uploaded into the UNC microarray database for storage and processing. Cy5:Cy3 ratios were generated for each spot, and these values were normalized for intensity and spatially dependent systematic variation by regional lowess smoothing (Cui et al., 2003). Using the normalized data, the log2 of the Cy5:Cy3 ratios were obtained for a given gene and used in subsequent analyses. The data are available from Gene Expression Omnibus (GSE24278).
Data processing and analysis.
Transcripts with over 30% missing values across all samples were excluded from further analysis, reducing the data to 25,315 transcripts. Remaining missing values were imputed by a K-nearest neighbor algorithm with K = 2 neighbors. Samples were randomly assigned into six separate batches for microarray experiments; no mice within the same strain and treatment group were analyzed in the same batch. ANOVA revealed a significant batch effect in 7299 transcripts (α = 0.01; Benjamini-Hochberg step-up false discovery rate [FDR] correction; Benjamini and Hochberg, 1995). Batch effects were adjusted using the ComBat method (Johnson et al., 2007).
Principal components analysis (PCA) was used for exploratory analysis of the transcript data (Joliffe, 2002). Distance weighted discrimination (DWD) was used to examine global differences between TCE-treated and vehicle samples (Marron et al., 2007). DWD finds a single direction providing good separation between two sets of samples. DiProPerm, a randomization test based on the DWD direction, was used to identify a significant overall difference between the treated and the control groups. Randomization testing was used to identify transcripts that contributed significantly to the discriminating direction. PCA and DWD analyses were performed in MATLAB (Mathworks, Natick, MA) using code available at www.unc.edu/∼marron/marron_software.html.
ANOVA models were used to assess the significance of individual transcripts. An ANOVA model with main effects for strain and treatment and an interaction between strain and treatment were applied for each transcript. Pearson correlation (transcript level and serum TCA at 24 h after treatment) coefficients were calculated. ANOVA effects and correlations were concluded to be significant if the p value (for the F statistic or t statistic, respectively) was below a threshold determined by the step-up FDR procedure (Benjamini and Hochberg, 1995) for multiple comparisons (α = 0.01). The R programming environment for statistical computing and graphics (2.10.0, R Development Core Team, Vienna, Austria) was used for the ANOVA, correlation analysis, and FDR adjustments. Heatmaps were generated using the Cluster (v 2.11) and TreeView (v 1.60) applications (Eisen et al., 1998).
Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) software was used for functional analysis of the significant genes. Significance of the enriched canonical pathways was determined by a right-tailed Fisher's exact test, with the step-up FDR correction for multiple comparisons. Gene networks were constructed based on predefined molecular interactions in the Ingenuity database.
Gene expression analysis by real-time PCR.
Total RNA (2 μg) was reverse transcribed using random primers and the high capacity complementary DNA archive kit (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The following gene expression assays (Applied Biosystems) were used for quantitative real-time PCR: cytochrome P450, family 4, subfamily a, polypeptide 10 (Cyp4a10, Mm01188913_g1); peroxisome proliferator–activated receptor alpha (Pparα, Mm00440939_m1); acyl-CoA thioesterase I, (Acot1, polypeptide A1, Mm01622471_s1); and beta glucuronidase (Gusb, Mm00446953_m1). Reactions were performed in a 96-well assay format. Each plate contained one experimental gene and a housekeeping gene, and all samples were plated in duplicate. Reactions were processed using Roche 480 instrument (Roche Applied Science, Indianapolis, IN). The cycle threshold (Ct) for each sample was determined from the linear region of the amplification plot. The ΔCt values for all genes relative to the control gene Gusb were determined. The ΔΔCt were calculated using treated group means relative to strain-matched control group means. Fold change data were calculated from the ΔΔCt values.
Protein level and activity measurements.
Liver tissue lysates were prepared by homogenization of 30–40 mg of tissue in 500 μl of lysis buffer (50mM Tris-HCl, pH 7.4; 1% nonyl phenosylpolyethoxylethanol 40; 1% SDS; 0.25% sodium deoxycholate; 150mM NaCl; 1mM EDTA; 1mM PMSF; 1 μg/ml each aprotinin, leupeptin, and pepstatin; 1mM Na3VO4; and 1mM NaF), sonication, and incubation at 4°C for 30 min followed by centrifugation at 10,000 × g at 4°C for 20 min. Extracts containing equal quantities of proteins were separated by SDS-PAGE on 10% polyacrylamide gels and transferred to polyvinylidene fluoride membranes. Membranes were blocked in 5% nonfat dry milk in PBS-T (0.1% Tween 20 in PBS), probed with 1:1000 anti-Cyp2E1 (Biomol, Plymouth Meeting, PA) and 1:2000 c-myc (Bethyl Laboratories Inc., Montgomery, TX), and incubated overnight at 4°C. Blots were then washed in tris buffered saline with tween incubated with the appropriate secondary antibody, and detected using chemiluminescence Western blotting analysis kit (Amersham, Pittsburgh, PA). Equal protein loading was confirmed by immunostaining against β-actin (1:4000; Sigma-Aldrich Corporation, St Louis, MO). The signal intensity was analyzed by ImageQuant software (Molecular Dynamics, Sunnyvale, CA) and normalized to β-actin. Cyp2e1 activity was measured by the rate of hydroxylation of p-nitrophenol to p-nitrocatechol by isolated hepatic microsomes prepared as detailed in Koop (1986).
RESULTS
TCE (2100 mg/kg) or corn oil vehicle was administered by gavage to 6- to 8-week-old male mice of 15 inbred strains. A single dose of TCE (2100 mg/kg, i.g. in corn oil vehicle) did not lead to adverse effects in liver or kidney as assessed by histopathology (data not shown). As expected (Yuan et al., 2009), the activity of serum AST and ALT, as well as levels of BUN, varied between strains (Supplementary table 1). TCE had no effect at either of the time points examined (2, 8, and 24 h) with regard to AST or BUN. A marginal, yet significant, increase in serum ALT (at 24 h) was observed only in 129S1/SvlmJ and CAST/EiJ strains. Body weight, liver weight, and liver:body weight ratios were also recorded (Supplementary table 1), and no significant (p < 0.05) effect of TCE treatment was observed at all time points.
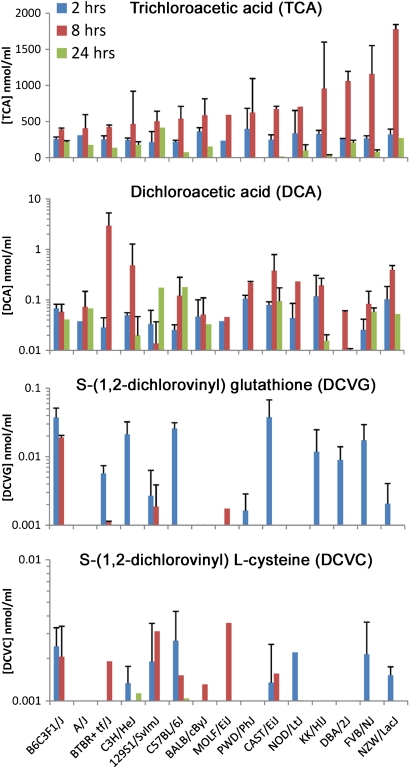
Serum concentration-time profiles of four oxidative and glutathione conjugation metabolites of TCE were examined (Fig. 1, Supplementary table 2). A more than fourfold difference in peak serum concentrations of TCA (at 8 h), the main product of TCE metabolism in mice (Lash et al., 2000a), was found across the individual strains. An ANOVA was used to evaluate the metabolite differences as a function of strain at each time point. For TCA, a significant (p < 0.01) strain effect was observed at 8 and 24 h (R2 of 0.73 and 0.87 for the fraction of variation due to strain at each time point, respectively). The amount of TCA formed over a 24-h period after single dosing (estimated by using the area under the curve from the concentration-time profiles) across strains was significantly correlated (r2 = 0.499, p < 0.05) with the basal liver levels of Cyp2e1 protein (data not shown).
FIG. 1.
Interstrain variability in TCE metabolism in the mouse. Serum levels of TCA, DCA, DCVG, and DCVC were assessed 2, 8, and 24 h following an acute dose (2100 mg/kg, intragastric in corn oil vehicle) of TCE administered to mice of 14 inbred and 1 hybrid strains and were measured by liquid chromatography coupled with tandem mass spectrometry. See Supplementary table 2 for strain average values, SDs, number of animals in each group, and the coefficients of variability of the measurements.
Although the levels of DCA in serum were approximately 1000-fold lower than those of TCA, similar to observations reported earlier (Kim et al., 2009a; Sano et al., 2009), considerable differences in concentration-time profiles and peak levels of DCA also exist between strains. Serum levels of DCVG and DCVC were the lowest of the four TCE metabolites assessed in this study and were below the limit of quantification in some strains, especially at 8- and 24-h time points. A significant strain effect from an ANOVA model was observed for DCA at 8 and 24 h (R2 of 0.69, p < 0.01, and 0.86, p < 0.05, respectively) and DCVG at 2 h (R2 = 0.67, p < 0.05). Serum levels of DCA, DCVG, or DCVC did not correlate with Cyp2e1, as expected (Kim et al., 2009a).
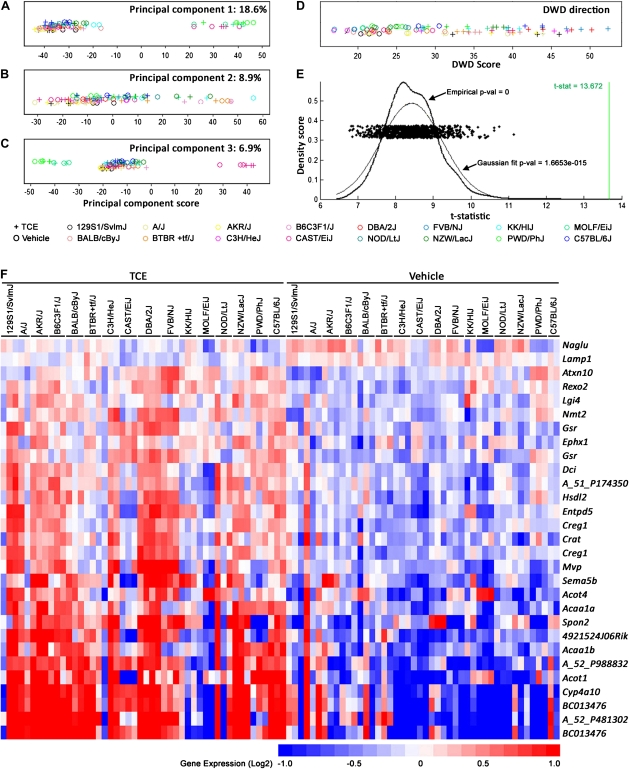
The regulatory pathways that control gene expression levels in liver and other tissues are known to be influenced by genetic factors, which results in considerable differences in messenger RNA (mRNA) levels between individuals even in the absence of any exogenous stimuli (Chesler et al., 2003; Gatti et al., 2007). In order to evaluate liver effects of TCE in the individual strains, microarray-based analysis was performed on tissues collected at 24 h after dosing. The PCA of the liver transcriptome data collected in this study revealed a strong strain effect, which concealed the TCE treatment effect (Figs. 2A–C). The wild-derived strains CAST/EiJ, MOLF/EiJ, and PWD/PhJ were most divergent with regards to liver transcriptome (principal component 1, Fig. 2A), an observation reported previously (Gatti et al., 2009), as they are most genetically distinct from other strains.
FIG. 2.
Whole-liver gene expression data (24 h after TCE treatment) were used for the principal component (panels A–C) and DWD (panels D and E) analyses (strains are identified by colors; “o” vehicle treatment; “+” TCE treatment). (E) A distribution of the t statistic values (each was derived from DWD analysis) of 2000 random permutations of the data is shown in comparison to the t statistic (green vertical line) derived from the DWD analysis (panel D). (F) A heatmap of genes identified as significant (p = 0.0005 under 2000 permutations, FDR = 0.041) TCE-specific response using DWD analysis. The color bar identifies up- and downregulated genes. See Supplementary table 3 for a complete list of genes and gene expression values.
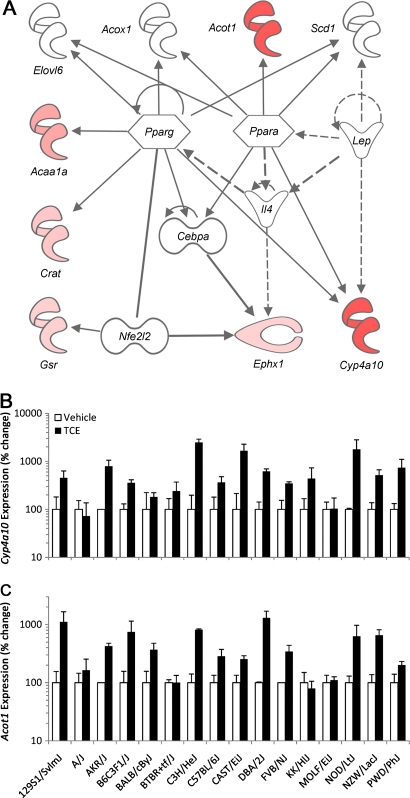
Despite these major differences, a highly significant (p < 0.001) effect of TCE could be observed across the entire population using DWD analysis, which considers a global distinction between TCE- and vehicle-treated animals (Figs. 2D and 2E). A DWD direction-derived signature consisting of 29 transcripts that were significantly discriminating between TCE and vehicle groups (α = 0.01; step-up FDR correction) was established (Fig. 2F, Supplementary table 3). The signature was enriched in genes that were strongly associated with lipid metabolism, small molecule biochemistry, and nucleic acid metabolism pathway (Ingenuity score 9) centered on PPARα and PPARγ (Fig. 3A). Interestingly, two well-known PPARα-regulated genes, Cyp4a10 (Barclay et al., 1999) and Acot1 (Dongol et al., 2007), were among the most discriminating genes with regards to TCE effect in the population of strains. Whereas expression of Pparα, a transcription factor, did not change in response to this short-term treatment (data not shown), mRNA induction of Pparα-target genes Cyp4a10 and Acot1 were confirmed using real time-PCR and shown to be elevated in most but not all strains (Figs. 3B–C).
FIG. 3.
(A) Network representation of the genes identified as significantly discriminating among vehicle- and TCE-treated groups (see Fig. 2E). Ingenuity analysis software was used to visualize the molecular interactions between the genes in lipid metabolism, small molecule biochemistry, and nucleic acid metabolism pathways. Expression differences for Cyp4a10 (B) and Acot1 (C) genes were confirmed using PCR.
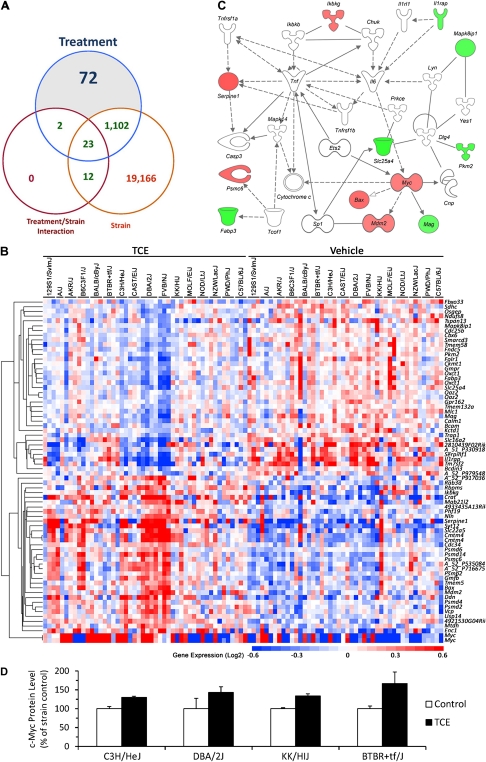
To further evaluate the changes in gene expression, an ANOVA model was fit for each transcript with main effects for strain (i.e., genotype), treatment (vehicle or TCE), or an interaction effect between the two because of the observed genotype-specific effects on TCE metabolism (Fig. 1). The number of transcripts significantly altered by each of these experimental factors is depicted in a Venn diagram (Fig. 4A). Of the 25,315 transcripts used for the analysis (see “Materials and Methods” section), 20,303 had a significant strain effect, 1199 a significant treatment effect, and 37 were significant for treatment × strain interaction. There were 72 transcripts with a significant treatment effect, yet without a significant strain or interaction effect (Fig. 4B, Supplementary table 4). The expression of these transcripts was significantly affected by TCE treatment, but not the subject's genotype, and we posit that these transcripts could serve as population-based biomarkers of response to TCE. The cell death, necrosis, and inflammatory-mediated response molecular network (Ingenuity score 20) centered on tumor necrosis factor α and myelocytomatosis oncogene Myc was enriched among these population-based transcripts (Fig. 4C). Transcription factor analysis of these genes was performed using the Pscan (Zambelli et al., 2009) tool and Transfac database (Biobase GmbH, Wolfenbuettel, Germany) to determine common regulators. Interestingly, inflammation-related regulatory proteins, Myc-associated zinc finger protein-related factor (MAZR), activating enhancer-binding protein 2 alpha (AP2α), and specificity protein 1 (SP1), may be common regulators of the strain-independent TCE-induced inflammatory response in mouse liver (Table 1). Indeed, changes in c-Myc protein levels between strains were in accord with mRNA levels (Fig. 4D).
FIG. 4.
Strain-independent effects of TCE on mouse liver transcriptome. (A) Venn diagram showing genes that were significant in the ANOVA comparing the effects of strain, treatment, and strain × treatment interaction. (B) A heatmap of the 72 genes identified as significant for the effect of TCE treatment, but not significant for strain or interaction effects. These genes can be regarded as TCE-responsive independent of the individual's genetic background. See Supplementary table 4 for a complete list of genes, gene expression values, and significance values. (C) A network representation of the 72 genes identified as significantly associated with TCE treatment. Molecular interactions between the genes in cell death, liver necrosis, and inflammatory-mediated response are visualized. Upregulated genes are identified in red and downregulated in green. (D) Protein levels of c-Myc were determined using Western immunoblotting in representative strains.
TABLE 1.
Transcription Factors Identified as Enriched in TCE-Induced Gene Expression Profiles
| Transcription factor | p Value | Bonferroni adjusted p value | |
| TCE-induced gene expression signature without a significant strain or interaction effect | |||
| MAZR | 1.48 × 10−05 | 0.004 | |
| SP1 | 5.15 × 10−05 | 0.015 | |
| AP2α | 7.24 × 10−05 | 0.020 | |
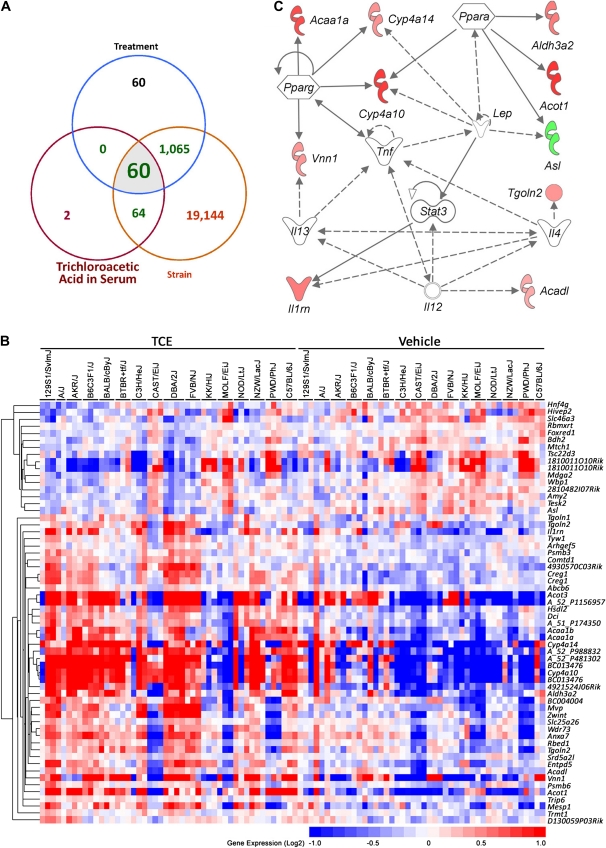
We also examined whether some transcripts were significantly associated with TCE metabolism. Correlation coefficients were calculated for each transcript with serum concentrations of TCA and DCA, two main oxidative metabolites of TCE. Glutathione conjugates were not used in this analysis because they were not detectable at 24 h after treatment. The correlation analysis with serum levels of DCA did not yield any significant transcripts; however, there were 126 transcripts found to have a significant correlation with TCA levels in serum at 24 h (Fig. 5A). Most of these transcripts (124 of 126) also had a significant strain effect, consistent with the observed strain-dependent differences in TCE metabolism (Fig. 1). Of these, 60 transcripts exhibited a significant treatment effect (Fig. 5B, Supplementary table 5). These can be regarded as genetic background–dependent markers of TCE metabolism. The fatty acid metabolism canonical pathway (Ingenuity score 19), which includes PPARα, γ, and other transcripts involved in lipid metabolism, small molecule biochemistry, and molecular transport, was the top network enriched in these 60 genes (Fig. 5C). Similarly, the gene ontology biological process GO:0006631 (fatty acid metabolic process) was the only significant (FDR < 0.05) pathway enriched in this gene list.
FIG. 5.
Strain-dependent gene expression signature, which correlates with TCA levels in serum. (A) Venn diagram showing genes that were significant in the ANOVA with effects for strain and treatment and correlate with serum TCA (24 h after TCE treatment) levels. (B) A heatmap of the 60 genes identified as significant for both TCE treatment and levels of TCA in serum following exposure. These genes are also significant for the strain effect, which suggests that TCE metabolism is dependent on the individual's genetic background. See Supplementary table 5 for a complete list of genes, gene expression values, and significance values. (C) A network representation of the 60 genes identified as significantly associated with TCE treatment and serum TCA level.
DISCUSSION
Studies of species differences in liver effects of TCE are important for assessing the risk of human exposure (Lash et al., 2002; Lumpkin et al., 2003; Sano et al., 2009). It is widely accepted that oxidative metabolism of TCE is a key event for liver toxicity, and there are important quantitative differences among species with regard to formation of two major metabolites, TCA and DCA (NRC, 2006). These molecules are hepatocarcinogenic themselves in the mouse, yet are thought to operate through different molecular pathways (Corton, 2008). TCA-induced mouse liver tumors have molecular features similar to those induced by typical peroxisome proliferators (Bull et al., 2002; Latendresse and Pereira, 1997). Even though TCE induces peroxisome proliferation, TCE-induced tumors exhibit a number of important dissimilarities as compared with TCA-induced tumors, which led to a suggestion that the mode of action for TCE hepatocarcinogenesis is different from that of TCA (Corton, 2008). Thus, the issue of quantitative assessment of TCE metabolism is an important challenge in relating adverse health effects in rodents to humans (NRC, 2006). Importantly, we show that in addition to differences in TCE metabolism between species, there are important differences between individuals within the same species.
The relative amounts of TCA and DCA being formed as a result of TCE administration has been determined in several studies. Although some still debate whether DCA is formed in vivo in quantifiable levels (Merdink et al., 2008), several recent studies showed that DCA can be detected; however, serum concentrations are more than 1000-fold lower than those for TCA (Delinsky et al., 2005; Kim et al., 2009a,b; Sano et al., 2009). Our study confirms this and shows that DCA and TCA serum concentrations do not correlate with each other, supporting the pharmacokinetic model suggesting that the metabolism of TCA to DCA is not a major pathway for the formation of DCA in the mouse (Kim et al., 2009b). At the same time, we show that the interstrain differences in serum TCA correlate well with liver Cyp2e1, an enzyme which is known to be a major oxidase for metabolizing TCE to TCA (Lash et al., 2000a).
Furthermore, this study provides additional evidence that, in the mouse, the glutathione pathway of TCE metabolism plays a minor role. We show that very low levels (∼10,000-fold less than TCA) of DCVG and DCVC are formed, that this phenomenon is reproducible across multiple inbred mouse strains, and that these metabolites are rapidly eliminated after a single large dose of TCE when administered in corn oil vehicle. It has been suggested that the extent of formation of glutathione conjugates from TCE in humans is much higher than in rodents based on high blood concentrations of DCVG reported in humans after inhalation of TCE (Lash et al., 1999). The same group studied formation of glutathione conjugates in rats after single large oral dose of TCE and reported that DCVG concentrations in blood and urine were high, yet not dose or time dependent (Lash et al., 2006). Other groups have reported a very low rate of glutathione conjugates’ formation in vivo in rats (Dekant et al., 1990) and mice (Kim et al., 2009a,b). Thus, the findings reported herein provide additional important information for rodent-to-human extrapolations with regard to TCE metabolism, which is critical for dose-response analysis and derivation of the reference values in risk assessment.
To better understand the differences in the molecular events elicited by TCE in the mouse liver, we evaluated both strain-dependent and independent gene expression changes. Genetic polymorphisms play a major role in influencing gene expression in liver (Gatti et al., 2007) and other tissues (Bystrykh et al., 2005; Chesler et al., 2005), an effect that is often stronger than that of a toxicant (Harrill et al., 2009). When genetic background–dependent gene expression differences between strains are not taken into the account, the population-wide TCE-induced gene expression changes were found to be related to the peroxisomal proliferation mode of action. Indeed, TCE is well known to cause many liver effects in mice in a PPARα-dependent manner (Laughter et al., 2004; Nakajima et al., 2000). Even though we did not find PPAR genes themselves to be induced 24 h after treatment with TCE, similar to the observation of Sano et al. (2009), the targets of these transcription factors were strongly upregulated in the overwhelming majority of the strains. This is indicative of the fact that the peroxisomal proliferation effect of TCE is occurring consistently in all strains.
Interestingly, when correlations between TCE metabolism to TCA or to gene expression changes were considered in the context of strain, we found that almost all the genes were also significantly impacted by the genetic background effect. This observation is in agreement with the fact that major strain-dependent differences in TCE metabolism exist. Induction of a lipid and drug metabolism network of genes centered on PPARs was the major molecular signature of this correlation analysis, consistent with the fact that TCA was the major metabolite of TCE in all strains and that it is a ligand for PPARs (Maloney and Waxman, 1999).
The interstrain variability in TCE metabolism observed in this study resulted in little observable liver toxicity (marginal increases in ALT observed in 2 out of 15 strains) when liver histopathology was considered. Acute single-dose exposures have not been associated with liver injury. The studies of Ramdhan et al. are the shortest that showed liver toxicity in mice following 7 days of inhalation exposure to TCE, an effect that was dependent on Cyp2e1 (Ramdhan et al., 2008), but not PPARα (Ramdhan et al., 2010).
The mechanism for TCE-induced liver damage has not been well studied, but it has been suggested that induction of nuclear factor-kappa-light-chain enhancer of activated B cells-activated cytokine pathways may be responsible for hepatotoxicity (Ramdhan et al., 2008, 2010). Thus, it is interesting that we find that cell death, liver necrosis, and inflammatory-mediated response networks are altered by TCE treatment, and it is genetic background independent. This further suggests that liver damage may not depend solely on TCE metabolism to potentially hepatotoxic intermediates, such as chloral hydrate or DCA, and that sufficient amounts of hepatotoxic intermediates are formed in all strains in spite of a four- to sixfold difference in TCE metabolites.
The transcription factor analysis of the genes that were altered by TCE independent of genetic background revealed several inflammation-related regulatory proteins, MAZR, AP2α, and SP1, which may be common regulators involved in TCE-induced inflammatory response in mouse liver. MAZR transactivates the Myc gene in B cells and is important for the development of B cells in association with Bach2 (Kobayashi et al., 2000). AP2 is involved in a number of inflammatory pathways including NFκB, cyclooxygenase-2, and inducible nitric oxide synthase, as well as PPARγ (Makowski et al., 2005). SP1 is one of key regulators of acute-phase response inflammatory genes in hepatocytes (Cantwell et al., 1998). Because these transcription factors have been shown to be associated with activation of macrophages and lymphocytes, the gene expression signature reported in this study suggests that TCE may have an effect on Kupffer cells, a hypothesis which needs to be tested.
It has been suggested that PPARα may be protective against liver injury, at least in the short-term exposure scenarios (Ramdhan et al., 2008). Interestingly, a recent study investigating the effects of TCE in Pparα-null and humanized mice showed that PPARα and PPARγ may be important factors in TCE-induced lipid accumulation in the liver but that TCE-induced liver toxicity was largely independent of PPARα status and also occurred in humanized mice (Ramdhan et al., 2010). The authors also reported that human PPARα may afford only weak protection against TCE-mediated effects as compared with mouse PPARα (Ramdhan et al., 2010). This observation is of importance for consideration of species-specific differences in TCE toxicity as data exist from human occupational exposure studies suggesting that exposures to TCE may lead to acute and chronic liver injury (Pantucharoensri et al., 2004; Thiele et al., 1982). Our observation that induction of PPARα-mediated pathways by TCE is dependent on strain and thus an individual's genetic background and further supports a need for additional studies elucidating the role of PPARα beyond peroxisome proliferation effects.
We also observed that Myc was induced by TCE. This observation is in agreement with the studies of Tao et al. (2000) who found decreased methylation in the promoter regions of the Jun and Myc genes and increased levels of their mRNA and proteins in mice exposed to TCE, DCA, and TCA. It has been suggested that a rapid increase in proliferation caused by most peroxisome proliferators would prevent the methylation of the newly synthesized strands of DNA (Ge et al., 2001). Indeed, the temporal relationship between increased cell proliferation and DNA hypomethylation of the Myc gene was observed in a study after single administration of another peroxisome proliferative agent WY-14643 to mice (Ge et al., 2001). Importantly, this short-term effect is not sustained because subchronic and chronic administration of WY-14643 has no effect on Myc methylation; however, major changes in DNA methylation occurred (Pogribny et al., 2007, 2008). Thus, it must be established whether upregulation of Myc by TCE is playing a role in long-term effects and what is a temporal relationship between cell proliferation and its promoter methylation and expression levels.
In conclusion, this study shows that important interindividual differences exist in TCE metabolism and molecular signaling in the liver. The biologically relevant exposure metrics collected in this study, both assessment of TCE metabolites and toxicogenomic-based evaluation, allow for better understanding of the association between key events and an individual's genotype. Although it is widely accepted that toxicogenomics has great potential to yield a new generation of exposure metrics and mode of action information (Cui and Paules, 2010), it is also important to understand which of the gene expression changes may be indicative of the population-wide versus an individual's response. Although further research is needed to find genetic and genomic markers that could identify individuals susceptible to TCE toxicity, this study provides important clues which point to liver inflammatory responses as being a population-wide response, although the extent of TCE metabolism and peroxisomal proliferation, at least in the mouse, may be dependent on an individual's genotype. A careful evaluation of gene expression–based biomarkers of response through multistrain experiments can assist in understanding the molecular basis of interindividual differences in metabolism and toxicity.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institutes of Health grant (P42 ES005948).
Acknowledgments
The views expressed in this paper do not necessarily represent those of the U.S. Food and Drug Administration.
References
- Bammler T, Beyer RP, Bhattacharya S, Boorman GA, Boyles A, Bradford BU, Bumgarner RE, Bushel PR, Chaturvedi K, Choi D, et al. Standardizing global gene expression analysis between laboratories and across platforms. Nat. Methods. 2005;2:351–356. doi: 10.1038/nmeth754. [DOI] [PubMed] [Google Scholar]
- Barclay TB, Peters JM, Sewer MB, Ferrari L, Gonzalez FJ, Morgan ET. Modulation of cytochrome P-450 gene expression in endotoxemic mice is tissue specific and peroxisome proliferator-activated receptor-alpha dependent. J. Pharmacol. Exp. Ther. 1999;290:1250–1257. [PubMed] [Google Scholar]
- Barton HA, Clewell HJ., III Evaluating noncancer effects of trichloroethylene: dosimetry, mode of action, and risk assessment. Environ. Health Perspect. 2000;108(Suppl. 2):323–334. doi: 10.1289/ehp.00108s2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- Bull RJ. Mode of action of liver tumor induction by trichloroethylene and its metabolites, trichloroacetate and dichloroacetate. Environ. Health Perspect. 2000;108(Suppl. 2):241–259. doi: 10.1289/ehp.00108s2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RJ, Orner GA, Cheng RS, Stillwell L, Stauber AJ, Sasser LB, Lingohr MK, Thrall BD. Contribution of dichloroacetate and trichloroacetate to liver tumor induction in mice by trichloroethylene. Toxicol. Appl. Pharmacol. 2002;182:55–65. doi: 10.1006/taap.2002.9427. [DOI] [PubMed] [Google Scholar]
- Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, Wiltshire T, Su AI, Vellenga E, Wang J, Manly KF, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat. Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol. Cell. Biol. 1998;18:2108–2117. doi: 10.1128/mcb.18.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Wang J, Lu L, Qu Y, Manly KF, Williams RW. Genetic correlates of gene expression in recombinant inbred strains: a relational model system to explore neurobehavioral phenotypes. Neuroinformatics. 2003;1:343–357. doi: 10.1385/NI:1:4:343. [DOI] [PubMed] [Google Scholar]
- Cooper GS, Makris SL, Nietert PJ, Jinot J. Evidence of autoimmune-related effects of trichloroethylene exposure from studies in mice and humans. Environ. Health Perspect. 2009;117:696–702. doi: 10.1289/ehp.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corton JC. Evaluation of the role of peroxisome proliferator-activated receptor alpha (PPARalpha) in mouse liver tumor induction by trichloroethylene and metabolites. Crit. Rev. Toxicol. 2008;38:857–875. doi: 10.1080/10408440802209796. [DOI] [PubMed] [Google Scholar]
- Cui X, Kerr MK, Churchill GA. Transformations for cDNA microarray data. Stat. Appl. Genet. Mol. Biol. 2003;2 doi: 10.2202/1544-6115.1009. 1–20. [DOI] [PubMed] [Google Scholar]
- Cui Y, Paules RS. Use of transcriptomics in understanding mechanisms of drug-induced toxicity. Pharmacogenomics. 2010;11:573–585. doi: 10.2217/pgs.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekant W, Koob M, Henschler D. Metabolism of trichloroethene—in vivo and in vitro evidence for activation by glutathione conjugation. Chem. Biol. Interact. 1990;73:89–101. doi: 10.1016/0009-2797(90)90110-9. [DOI] [PubMed] [Google Scholar]
- Delinsky AD, Delinsky DC, Muralidhara S, Fisher JW, Bruckner JV, Bartlett MG. Analysis of dichloroacetic acid in rat blood and tissues by hydrophilic interaction liquid chromatography with tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19:1075–1083. doi: 10.1002/rcm.1890. [DOI] [PubMed] [Google Scholar]
- Dongol B, Shah Y, Kim I, Gonzalez FJ, Hunt MC. The acyl-CoA thioesterase I is regulated by PPARalpha and HNF4alpha via a distal response element in the promoter. J. Lipid Res. 2007;48:1781–1791. doi: 10.1194/jlr.M700119-JLR200. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- Gatti D, Maki A, Chesler EJ, Kirova R, Kosyk O, Lu L, Manly KF, Williams RW, Perkins A, Langston MA, et al. Genome-level analysis of genetic regulation of liver gene expression networks. Hepatology. 2007;46:548–557. doi: 10.1002/hep.21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti DM, Harrill AH, Wright FA, Threadgill DW, Rusyn I. Replication and narrowing of gene expression quantitative trait loci using inbred mice. Mamm. Genome. 2009;20:437–446. doi: 10.1007/s00335-009-9199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge R, Wang W, Kramer PM, Yang S, Tao L, Pereira MA. Wy-14,643-induced hypomethylation of the c-myc gene in mouse liver. Toxicol. Sci. 2001;62:28–35. doi: 10.1093/toxsci/62.1.28. [DOI] [PubMed] [Google Scholar]
- Harrill AH, Ross PK, Gatti DM, Threadgill DW, Rusyn I. Population-based discovery of toxicogenomics biomarkers for hepatotoxicity using a laboratory strain diversity panel. Toxicol. Sci. 2009;110:235–243. doi: 10.1093/toxsci/kfp096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (IARC) Dry Cleaning, Some Chlorinated Solvents and Other Industrial Chemicals. Lyon, France: IARC Press; 1995. [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Joliffe IT. Principal Component Analysis. 2nd ed. New York, NY: Springer; 2002. [Google Scholar]
- Kim S, Collins LB, Boysen G, Swenberg JA, Gold A, Ball LM, Bradford BU, Rusyn I. Liquid chromatography electrospray ionization tandem mass spectrometry analysis method for simultaneous detection of trichloroacetic acid, dichloroacetic acid, S−(1,2-dichlorovinyl)glutathione and S-(1,2-dichlorovinyl)-L-cysteine. Toxicology. 2009a;262:230–238. doi: 10.1016/j.tox.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Pollack GM, Collins LB, Rusyn I. Pharmacokinetic analysis of trichloroethylene metabolism in male B6C3F1 mice: formation and disposition of trichloroacetic acid, dichloroacetic acid, S-(1,2-dichlorovinyl)glutathione and S-(1,2-dichlorovinyl)-L-cysteine. Toxicol. Appl. Pharmacol. 2009b;238:90–99. doi: 10.1016/j.taap.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Yamagiwa H, Hoshino H, Muto A, Sato K, Morita M, Hayashi N, Yamamoto M, Igarashi K. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 2000;20:1733–1746. doi: 10.1128/mcb.20.5.1733-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop DR. Hydroxylation of p-nitrophenol by rabbit ethanol-inducible cytochrome P-450 isozyme 3a. Mol. Pharmacol. 1986;29:399–404. [PubMed] [Google Scholar]
- Lash LH, Fisher JW, Lipscomb JC, Parker JC. Metabolism of trichloroethylene. Environ. Health Perspect. 2000a;108(Suppl. 2):177–200. doi: 10.1289/ehp.00108s2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash LH, Parker JC, Scott CS. Modes of action of trichloroethylene for kidney tumorigenesis. Environ. Health Perspect. 2000b;108(Suppl. 2):225–240. doi: 10.1289/ehp.00108s2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash LH, Putt DA, Brashear WT, Abbas R, Parker JC, Fisher JW. Identification of S-(1,2-dichlorovinyl)glutathione in the blood of human volunteers exposed to trichloroethylene. J. Toxicol. Environ. Health A. 1999;56:1–21. doi: 10.1080/009841099158204. [DOI] [PubMed] [Google Scholar]
- Lash LH, Putt DA, Parker JC. Metabolism and tissue distribution of orally administered trichloroethylene in male and female rats: identification of glutathione- and cytochrome P-450-derived metabolites in liver, kidney, blood, and urine. J. Toxicol. Environ. Health A. 2006;69:1285–1309. doi: 10.1080/15287390500360133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash LH, Qian W, Putt DA, Hueni SE, Elfarra AA, Sicuri AR, Parker JC. Renal toxicity of perchloroethylene and S-(1,2,2-trichlorovinyl)glutathione in rats and mice: sex- and species-dependent differences. Toxicol. Appl. Pharmacol. 2002;179:163–171. doi: 10.1006/taap.2001.9358. [DOI] [PubMed] [Google Scholar]
- Latendresse JR, Pereira MA. Dissimilar characteristics of N-methyl-N-nitrosourea-initiated foci and tumors promoted by dichloroacetic acid or trichloroacetic acid in the liver of female B6C3F1 mice. Toxicol. Pathol. 1997;25:433–440. doi: 10.1177/019262339702500501. [DOI] [PubMed] [Google Scholar]
- Laughter AR, Dunn CS, Swanson CL, Howroyd P, Cattley RC, Corton JC. Role of the peroxisome proliferator-activated receptor alpha (PPARalpha) in responses to trichloroethylene and metabolites, trichloroacetate and dichloroacetate in mouse liver. Toxicology. 2004;203:83–98. doi: 10.1016/j.tox.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Lumpkin MH, Bruckner JV, Campbell JL, Dallas CE, White CA, Fisher JW. Plasma binding of trichloroacetic acid in mice, rats, and humans under cancer bioassay and environmental exposure conditions. Drug Metab. Dispos. 2003;31:1203–1207. doi: 10.1124/dmd.31.10.1203. [DOI] [PubMed] [Google Scholar]
- Makowski L, Brittingham KC, Reynolds JM, Suttles J, Hotamisligil GS. The fatty acid-binding protein, aP2, coordinates macrophage cholesterol trafficking and inflammatory activity. Macrophage expression of aP2 impacts peroxisome proliferator-activated receptor gamma and IkappaB kinase activities. J. Biol. Chem. 2005;280:12888–12895. doi: 10.1074/jbc.M413788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney EK, Waxman DJ. Trans-activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol. Appl. Pharmacol. 1999;161:209–218. doi: 10.1006/taap.1999.8809. [DOI] [PubMed] [Google Scholar]
- Marron JS, Todd MJ, Ahn J. Distance weighted discrimination. J. Am. Stat. Assoc. 2007;102:1267–1271. doi: 10.1198/jasa.2010.tm08487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdink JL, Robison LM, Stevens DK, Hu M, Parker JC, Bull RJ. Kinetics of chloral hydrate and its metabolites in male human volunteers. Toxicology. 2008;245:130–140. doi: 10.1016/j.tox.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Kamijo Y, Usuda N, Liang Y, Fukushima Y, Kametani K, Gonzalez FJ, Aoyama T. Sex-dependent regulation of hepatic peroxisome proliferation in mice by trichloroethylene via peroxisome proliferator-activated receptor alpha (PPARalpha) Carcinogenesis. 2000;21:677–682. doi: 10.1093/carcin/21.4.677. [DOI] [PubMed] [Google Scholar]
- National Research Council (NRC) Assessing the Human Health Risks of Trichloroethylene: Key Scientific Issues. Washington, DC: The National Academies Press; 2006. [Google Scholar]
- National Research Council (NRC) Review of the Environmental Protection Agency's Draft IRIS Assessment of Tetrachloroethylene. Washington, DC: The National Academies Press; 2010. [PubMed] [Google Scholar]
- Pantucharoensri S, Boontee P, Likhitsan P, Padungtod C, Prasartsansoui S. Generalized eruption accompanied by hepatitis in two Thai metal cleaners exposed to trichloroethylene. Ind. Health. 2004;42:385–388. doi: 10.2486/indhealth.42.385. [DOI] [PubMed] [Google Scholar]
- Pastino GM, Yap WY, Carroquino M. Human variability and susceptibility to trichloroethylene. Environ. Health Perspect. 2000;108(Suppl. 2):201–214. doi: 10.1289/ehp.00108s2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Tryndyak VP, Boureiko A, Melnyk S, Bagnyukova TV, Montgomery B, Rusyn I. Mechanisms of peroxisome proliferator-induced DNA hypomethylation in rat liver. Mutat. Res. 2008;644:17–23. doi: 10.1016/j.mrfmmm.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Tryndyak VP, Woods CG, Witt SE, Rusyn I. Epigenetic effects of the continuous exposure to peroxisome proliferator WY-14,643 in mouse liver are dependent upon peroxisome proliferator activated receptor alpha. Mutat. Res. 2007;625:62–71. doi: 10.1016/j.mrfmmm.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdhan DH, Kamijima M, Yamada N, Ito Y, Yanagiba Y, Nakamura D, Okamura A, Ichihara G, Aoyama T, Gonzalez FJ, et al. Molecular mechanism of trichloroethylene-induced hepatotoxicity mediated by CYP2E1. Toxicol. Appl. Pharmacol. 2008;231:300–307. doi: 10.1016/j.taap.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdhan DH, Komijima M, Wang D, Ito Y, Naito H, Yanagiba Y, Hayashi Y, Tanaka N, Aoyama T, Gonzalez FJ, et al. Differential response to trichloroethylene-induced hepatosteatosis in wild-type and PPARa-humanized Mice. Environ. Health Perspect. 2010;118:1557–1563. doi: 10.1289/ehp.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusyn I, Gatti DM, Wiltshire T, Kleeberger SR, Threadgill DW. Toxicogenetics: population-based testing of drug and chemical safety in mouse models. Pharmacogenomics. 2010;11:1127–1136. doi: 10.2217/pgs.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y, Nakashima H, Yoshioka N, Etho N, Nomiyama T, Nishiwaki Y, Takebayashi T, Oame K. Trichloroethylene liver toxicity in mouse and rat: microarray analysis reveals species differences in gene expression. Arch. Toxicol. 2009;83:835–849. doi: 10.1007/s00204-009-0431-1. [DOI] [PubMed] [Google Scholar]
- Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Effect of trichloroethylene and its metabolites, dichloroacetic acid and trichloroacetic acid, on the methylation and expression of c-Jun and c-Myc protooncogenes in mouse liver: prevention by methionine. Toxicol. Sci. 2000;54:399–407. doi: 10.1093/toxsci/54.2.399. [DOI] [PubMed] [Google Scholar]
- Thiele DL, Eigenbrodt EH, Ware AJ. Cirrhosis after repeated trichloroethylene and 1,1,1-trichloroethane exposure. Gastroenterology. 1982;83:926–929. [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services/Public Health Service/National Toxicology Program (DHHS/PHS/NTP) In: Report on Carcinogens. 11th ed. 2005. Trichloroethylene; pp. 261–263. U.S. DHHS/PHS/NTP. Washington, DC. [Google Scholar]
- Yuan R, Tsaih SW, Petkova SB, Marin de E, Xing S, Marion MA, Bogue MA, Mills KD, Peters LL, Bult CJ, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8:277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.