Abstract
We report here a new small molecule fluorogen and RNA aptamer pair for RNA labeling. The small-molecule fluorogen is designed based on fluorescently quenched sulforhodamine dye. The SELEX (Systematic Evolution of Ligands by EXponential enrichment) procedure and fluorescence screening in E. coli have been applied to discover the aptamer that can specifically activate the fluorogen with μM binding affinity. The systematic mutation and truncation study on the aptamer structure determined the minimum binding domain of the aptamer. A series of rationally modified fluorogen analogs have been made to probe the interacting groups of fluorogen with the aptamer. These results led to the design of a much improved fluorogen ASR 7 that displayed a 33-fold increase in the binding affinity for the selected aptamer in comparison to original ASR 1 and an 88-fold increase in the fluorescence emission after the aptamer binding. This study demonstrates the value of combining in vitro SELEX and E. coli fluorescence screening with rational modifications in discovering and optimizing new fluorogen/RNA aptamer labeling pairs.
INTRODUCTION
Site-specific labeling of proteins with molecular tags has been widely used to study structure and function of proteins, and for the direct visualization of protein dynamics, localization and interactions in single living cells (1–5). The proteins of interest can be labeled by genetic fusions to fluorescent proteins, or chemical reactions with fluorescent dyes. With the important role of RNA molecules in biology, there is strong interest in applying the specific labeling strategy to RNA molecules (6–8). This extension is however challenging since there is no RNA counterpart of green fluorescent proteins (GFPs) and there are limited chemical functional groups in RNA molecules available for specific chemical labeling.
The straightforward RNA tagging approach is to attach fluorophores to oligonucleotides complementary to the target RNA molecules. This technique is commonly used in fluorescent in-situ hybridization (FISH) for imaging target mRNA in fixed cells and tissues (9–13). To improve the sensitivity in detecting a low abundant RNA target, oligonucleotides may be labeled with an additional quencher or dye molecule to form “molecular beacons” so that the probe is quenched in an “off” state; it turns on after forming a complex with the target RNA (6, 8, 14–17). Another strategy similar to molecular beacons uses the target RNA as the template to direct the ligation of two complementary oligonucleotides, resulting in the activation of the probe from fluorescently quenched to emitting states (18, 19). Because of the negatively charged backbone of the nucleotides, this type of fluorescent probes is generally not cell-permeable and demonstrates poor delivery for live-cell imaging. To address these challenges, we have developed a trans-splicing ribozyme based approach that offers high sensitivity for imaging target RNA molecules through signal amplification (20). Guided by the complementary oligonucleotides, the RNA reporter will generate an mRNA of a reporter enzyme (e.g. β-lactamase or firefly luciferase) via trans-splicing, which will be translated into reporter enzymes for imaging with their substrates. This approach has been successfully demonstrated with imaging target RNA in live cells and live animals, but the enzyme-based amplification compromises the ability to reveal the spatial information of target RNA in subcellular locations.
GFPs have also been used for RNA tagging in a genetic fusion with RNA binding proteins (RBPs). The binding of RBPs to the RBP-specific aptamer genetically attached to the target RNA leads to the formation a protein-RNA complex containing GFP as the fluorescent tag. Several pairs of RBPs and RNA aptamers have been reported with GFP or split GFP (21, 22) as the fluorescent reporter to study RNA localization (23–26), RNA dynamics (27–29), and transcriptional profiling (28, 30). The size of this genetically encoded tag is generally large, sometimes larger than the RNA itself, which may result in unnatural restriction of the movement and function of the target RNA.
A small-molecule based RNA tagging approach has been of enormous interest because of its small molecular weight and ability to tune its cell permeability and fluorescent property via chemical modifications. These small-molecule probes can be designed, such that the fluorescence is initially quenched, but is activated after binding to a specific RNA aptamer, thus promising high sensitivity (6). A number of small-molecule fluorogenic probe and RNA aptamer binding pairs have been reported in the literature (31–36), but these probes haven’t been successfully demonstrated for live-cell imaging. There is still a strong need for developing new small-molecule probes and discovering RNA aptamers for RNA labeling.
In this paper, we describe our effort towards developing a new set of small-molecule fluorogenic probe and RNA aptamer pairs for RNA labeling. We designed a quenched sulforhodamine analogue (ASR) and performed an in vitro SELEX (Systematic Evolution of Ligands by EXponential enrichment) followed by fluorescence screening in E. coli to discover an RNA aptamer that can bind the probe ASR and activate its fluorescence with a maximal increase of more than 135-fold in the quantum yield. To elucidate the interaction between the ASR probe and the RNA aptamer, we systematically modified both the ASR structure and the aptamer sequence and identified the structural elements important for binding affinity and fluorescence activation. A rationally designed ASR analogue displayed a 33-fold improvement in the binding affinity compared to the original ASR probe.
RESULTS AND DISCUSSION
Design and synthesis of ASR probe
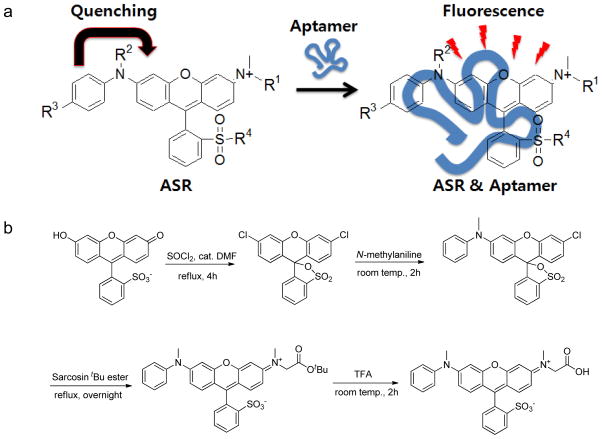
We derivatized one of the sulforhodamine amino groups with an aromatic moiety aniline, resulting a quenched, non-fluorescent analogue aniline-substituted sulforhodamine (ASR). We hypothesized that the binding of an RNA aptamer would activate the fluorescence emission of ASR (Figure 1a). The second amino group of sulforhodamine was alkylated to introduce a carboxylate group for enabling further biotinylation for immobilization on the beads for in vitro SELEX selection.
Figure 1.
Design and synthesis of ASR fluorogenic probes for RNA tagging. a) The general structure of ASR and the scheme for the fluorescence enhancement of ASR by RNA aptamer binding (R1 = CH2CO2−, R2 = Me, R3 = H, R4 = O− for ASR 1 as the SELEX target). b) Synthetic scheme of the ASR 1 preparation.
The synthesis of ASR 1 is straightforward as outlined in the Figure 1b (37, 38). The first step involves the conversion of sulfofluorescein to dichlorosulfofluorscein. Subsequently, one chloro substituent was displaced by aniline, followed by a second substitution by sarcosine to yield the disubstituted sulforhodamine. ASR 1 shows an absorbance maximum at 555 nm and a very low quantum yield in an aqueous buffer (ΦWater = 0.0017). The mechanism for quenching the sulforhodamine fluorescence emission is believed to operate through photoinduced electron transfer (PET) (39–42). The electron in the highest occupied molecular orbital (HOMO) of the aniline group in ASR 1 can transfer to the sulforhodamine HOMO, and prevent the electron at the excited state of sulforhodamine from returning to its ground state via the fluorescence mechanism, resulting in the decrease in the quantum yield of the fluorophore. The PET quenching would become inefficient if the relative geometries of the HOMOs of the fluorophore and aniline are not optimum due to steric restrictions such as the aptamer binding (41). Consistence with this mechanism, the quantum yield of ASR 1 increases by more than 400-fold to 0.71 in 90% glycerol solution (Supplementary Figure 1).
In vitro SELEX and fluorescence screening in E. coli
The selection strategy combines an in vitro SELEX affinity screening (Figure 2a) and E. coli fluorescence imaging (Figure 2b) to discover aptamers with high binding affinity for ASR 1 and large fluorescence activation.
Figure 2.
Discovery of ASR binding aptamers via in vitro SELEX and E. coli fluorescence screening. a) Schematic of the selection strategy. b) Representative fluorescence image of a bacterial plate with 1 μM of ASR 1 in the LB Agar gel. Clones containing aptamers that enhance fluorescence of ASR 1 show increased fluorescence signal in the light-tight chamber of an IVIS 200 fluorescence imager: a colony with high fluorescence signal is marked with a red arrow.
A library of 100mer RNA aptamers (estimated to be 1013) containing 20 constant nucleotides at each end and 60 randomized nucleotide sequences in the middle was transcribed and mixed with the biotinylated analog of ASR 1 (Supporting Information) immobilized on the avidin-agarose beads. It was then washed three times with PBS buffer to remove non-bound RNA aptamers, and eluted by excess free ASR 1, followed by reverse transcription and amplification by PCR. The PCR product was then transcribed for the second round of SELEX. These steps were repeated three times to enrich the RNA sequence with high affinity to ASR 1 (43–48).
Next, the selected pool of RNA aptamers was then screened for their light-up property in E. coli by fluorescence imaging. Out of approximately 200 bacterial plates that were screened, 35 colonies that displayed strong fluorescence signal (Figure 2b) were sequenced and contained the following nucleotides at the randomized region: GCAGGACCCT CACCTCGGTG ATGATGGAGG GGCGCAAGGT TAACCGCCTC AGGGTCCTCG
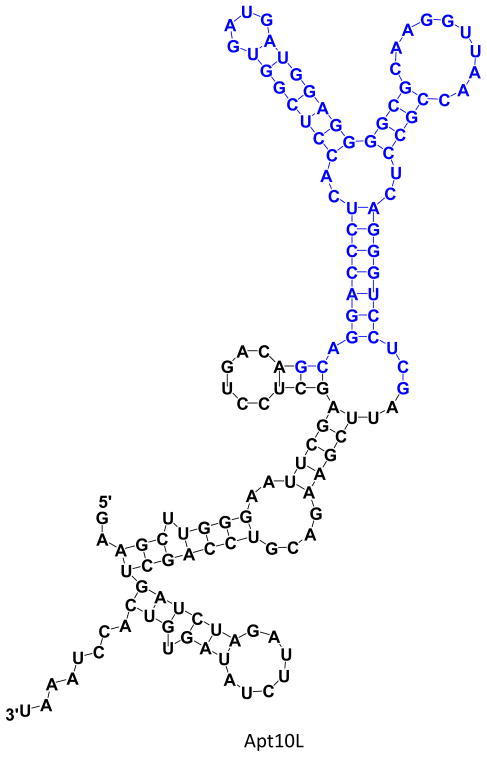
The aptamer containing both this randomized sequence and 20mer primer sequences at each end plus additional flanking sequences for cloning into pBK-CMV-eGFP plasmid vector (HindIII site et al., see Supplementary Table 1 and 2 for the definition and primers) is denoted as Apt10L. The M-fold program predicts 4 secondary structures (Figure 3), and the 60 nucleotides randomized sequence in Apt10L is predicted to have two possible secondary structures (Supplementary Figure 2a).
Figure 3.
Sequence and representative secondary structure of selected aptamer Apt10L as predicted by the M-fold program; total 4 structures are predicted for Apt10L. Sequences in blue represent the region of randomized 60 nucleotides. Apt10L contains long flanking sequences (HindIII site et al.) on 5′- and 3′-region for cloning into pBK-CMV-eGFP plasmid vector for fluorescence screening in E. coli.
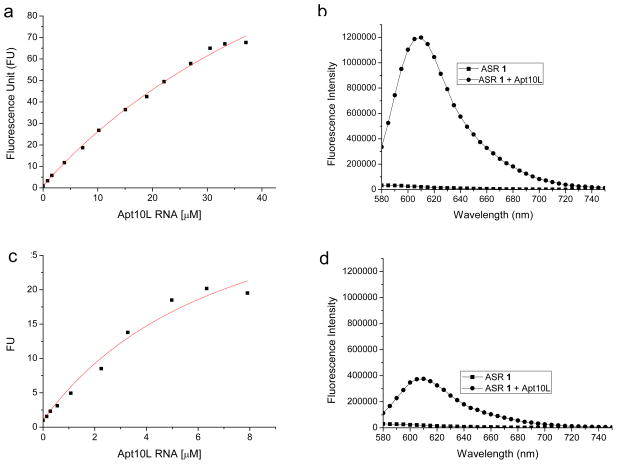
The Kd value and the fluorescence enhancement activity of Apt10L for ASR 1 were determined with fluorescence titration (Figure 4). Fluorescence intensity of ASR 1 at 610 nm increased upon the RNA aptamer binding. The kinetics of binding between the Apt10L and ASR 1 was rather fast: the fluorescence intensities increased instantaneously to the stable values after mixing during the titration. We found that the method used for preparing Apt10L significantly affected the value of Kd. The Kd value of Apt10L for ASR 1 was 39.1 ± 7.6 μM and Fmax (expected fluorescence fold enhancement at saturation) was 135 ± 15 (Figure 4a) if Apt10L was purified with G-25 column without denaturing and renaturing subsequent to in vitro transcription. On the other hand, if Apt10L was purified using 6% polyacrylamide-7M urea gel, which is a common method for RNA purification, the value of Kd was 10-fold lower but the value of Fmax was much reduced (Kd = 3.5 ± 1.5 μM, Fmax = 29.0 ± 5.1; Figure 4c, d). This result may be explained by the fact that after the PAGE purification Apt10L may not refold into the same conformation formed during in vitro transcription. Considering that the conformation of in vitro transcribed RNA may be closer to its likely structure in vivo, we used G-25-purified RNA apatmers for the binding affinity determination in our study.
Figure 4.
Fluorescence titration of Apt10L RNA aptamer against ASR 1 (1 μM) in PBS buffer (pH 7.4) containing 1 mM MgCl2. Excitation and emission wavelength were 555 nm and 610 nm, respectively. a) The Kd value of Apt10L against ASR 1 is 39.1 ± 7.6 μM and Fmax (expected fluorescence enhancement at saturation) is 135 ± 15. Fluorescence intensity was normalized as fluorescence unit (FU) against the fluorescence intensity of ASR 1 in the absence of RNA. b) Fluorescence spectrum of ASR 1 in the absence or presence of several tens of μM Apt10L RNA. 610 nm is the maximum peak after RNA binding. c) and d) Fluorescence titrations were carried out using PAGE (6% polyarylamide-7M urea gel) purified Apt10L RNA at the same experimental conditions as in a) and b). The Kd value of Apt10L against ASR 1 is 3.5 ± 1.5 μM and Fmax is 29.0 ± 5.1.
Mutation and truncation analysis to probe the interaction between ASR and Apt10L
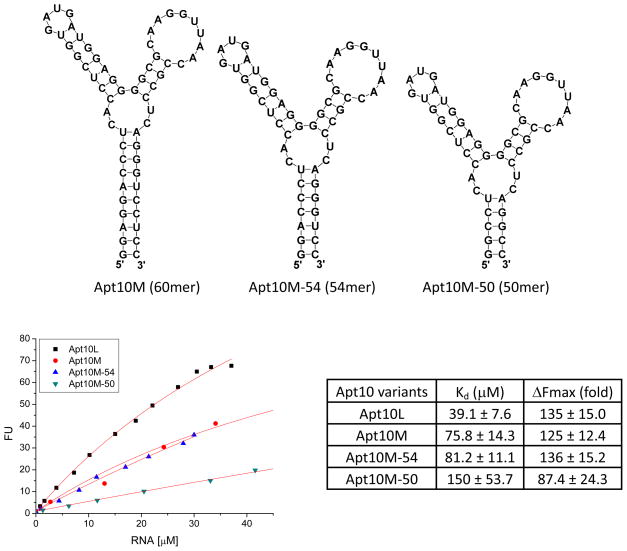
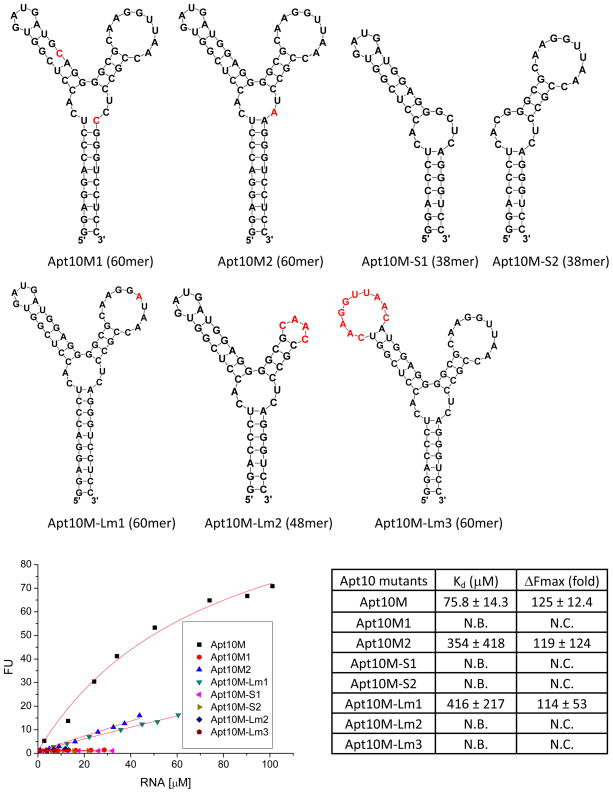
To understand the interaction between ASR 1 and Apt10L for further optimization of the binding, a systematic truncation and mutation study was performed. We first designed Apt10M (60mer) with all the flanking sequences but the 60 random sequences removed. In addition, two mutations were introduced at the bottom of its stem region to form two GC base pairs for in vitro transcription and for enhanced stabilization of the stem (Figure 5). The Kd value of Apt10M for ASR 1 was 75.8 ± 14.3 μM and the estimated Fmax was 125 ± 12.4 (Figure 5). Compared to Apt10L, a mere two-fold decrease in the binding affinity of Apt10M indicates that the structure of the randomized sequence area is primarily responsible for the binding to ASR 1. On the other hand, the flanking sequences in Apt10L also contribute to both the binding and light-up properties.
Figure 5.
Fluorescence titration of Apt10M variants against ASR 1 (1 μM) in PBS buffer (pH 7.4) containing 1 mM MgCl2. Excitation and emission wavelength were 555 nm and 610 nm, respectively.
To examine whether the aptamer binding sequence can be further shortened, Apt10M-54 (54mer) and Apt10M-50 (50mer) variants with three and five less base pairs on the stem regions, respectively, were designed (Figure 5). The binding affinity of these variants for ASR 1 appears to be dependent on the length of the stem: with a shorter stem, the affinity of Apt10M-50 for ASR 1 decreased by two-fold in comparison to Apt10M, but the affinity of Apt10M-54 for ASR 1 only slightly reduced and the maximal fluorescence enhancement slightly increased (Kd = 81.2 ± 11.2 μM, Fmax = 136 ± 15.2). On the other hand, further increase in the length of the stem did not necessarily increase the binding affinity and fluorescence enhancement. For example, variants Apt10M-66, -70, -76, -80 (Supplementary Figure 2b) all showed a lower affinity for ASR 1 than Apt10M except Apt10M-80 that displayed a slightly improved binding affinity but a reduced Fmax value (Supplementary Figure 3a).
Both of the two stem-loop structures in Apt10M are required for ASR binding and fluorescence activation. Deleting either stem-loop abolished the binding activity of the mutants (Apt10M-S1 and Apt10M-S2) (Figure 6). Mutants Apt10M-Lm2 (the size of the right loop was decreased by deleting 5 nucleotides in the middle) and Apt10M-Lm3 (the left loop was replaced by the right loop) displayed undetectable fluorescence enhancement upon the addition of ASR 1, further suggesting the cooperative interaction between the two stem-loop structures (Figure 6).
Figure 6.
Fluorescence titration of Apt10M mutants against ASR 1 (1 μM) in PBS buffer (pH 7.4) containing 1 mM MgCl2. Excitation and emission wavelength were 555 nm and 610 nm, respectively. N.C.: Not Calculable; N.B.: No Binding. Nucleotides in red stand for mutations.
The binding of Apt10M to ASR 1 is highly sensitive to mutations in the stem-loop region. Single or double mutations were introduced to stabilize the representative secondary structures as predicted by the M-fold program, for example, Apt10M1 (containing mutations G27C and A51C), Apt10M2 (containing a mutation C50A), and Apt10M-Lm1 (containing a U39A mutation on the loop region) (Figure 6). All these mutants either do not bind to ASR 1 or have a largely reduced affinity, suggesting the specific interaction with ASR 1 through the loop.
Influence of the flanking sequence on the binding
The M-fold program predicts several secondary structures for Apt10L (Supplementary Figure 2a), suggesting that the Apt10L structure is unstable and flexible and that this structural instability could affect its binding affinity to ASR. We examined the role of the flanking sequence on the binding affinity by designing truncated Apt10L variants (T variants). The deletion of some of the flanking sequences decreases the possibility of forming multiple secondary structures. Each of the three Apt10T variants (with the total length of 108, 107 and 88 nucleotides) has similar binding affinity to ASR 1 compared to Apt10L (Supplementary Figure 2c & 3b), suggesting that the flanking sequences have only an insignificant effect on the binding affinity.
Mutations were also made in the flanking sequence to strengthen the base-pairing on the stems of each conformation of Apt10L predicted by the M-fold program to stabilize these secondary structures. Both Apt10L1 and Apt10L2 (each with 2, or 3 mutations in the flanking sequences, respectively; Supplementary Figure 2a) showed an identical affinity compared to the Apt10L whereas the Apt10L3 with four mutations showed a 2-fold decrease in the binding affinity (Supplementary Figure 3c). These results further suggest that the interaction of the flanking sequence to the ASR 1 is not as specific as that in the randomized region, although overall these flanking sequences contribute to a 2-fold increase in the binding affinity.
Improvement of Kd and light-up activity by ASR analogues
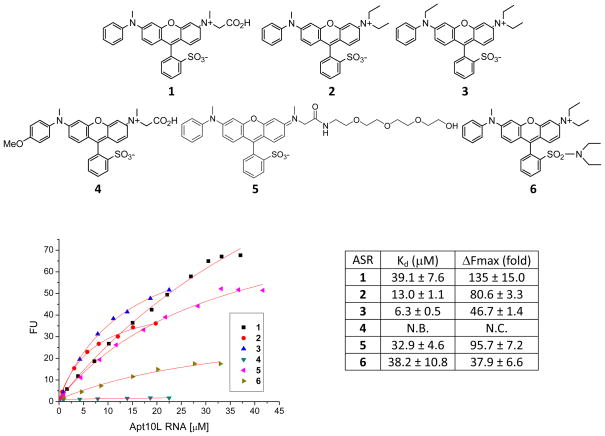
To identify the functional groups on ASR 1 that interact with the aptamer, we systematically varied the substitutions on the two amino groups and sulfonate group on ASR 1 to generate a series of analogues. A similar fluorescence titration assay was carried out in the presence of 1 μM of each ASR analogue to estimate its binding affinity with Apt10L (Figure 7).
Figure 7.
Fluorescence titration of Apt10L against ASR analogues (1 μM) in PBS buffer (pH 7.4) containing 1 mM MgCl2. Excitation and emission wavelength were 555 nm and 610 nm, respectively. N.C.: Not calculable; N.B.: No binding.
The elimination of the carboxylate group on ASR 1 and the replacement of the methyl group with a diethyl substitution on the amine led to a 3-fold increase in the binding affinity (Kd = 13.0 ± 1.1 μM, Fmax = 80.6 ± 3.3 for ASR 2). A substitution of the methyl group on the other amine with an ethyl group produces another 2-fold increase in the binding affinity but a largely reduced Fmax (Kd = 6.3 ± 0.5 μM, Fmax = 46.7 ± 1.4 for ASR 3). Interestingly, the introduction of a methoxy group to the quencher moiety, aniline, of ASR 4 abolished the light-up activity, although the quenching effect still remains; it confirms that the quencher moiety provides one of the interaction sites for the Apt10L binding and fluorescence activation.
A long tetra-ethylene glycol group introduced on ASR 5 showed minimal effects on the Kd, which is consistent with the SELEX experiment design that used an ASR analogue biotinylated at the same position as ASR 5. ASR 3 and 6 share similar structures except that the sulfonate group is converted to sulfonamide in ASR 6. This conversion results in a 6-fold decrease in the binding affinity, suggesting that the sulfonate group is also in some contact with Apt10L.
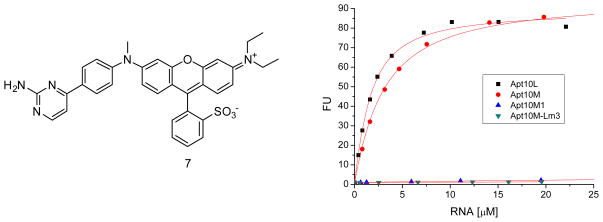
These results suggest that consistent with our design, both methyl aniline group and the sulfonate substituted benzene make important contacts with Apt10L, resulting in the fluorescence activation after binding. Based on these observations, we designed ASR 7 as an improved analogue (Figure 8), which has a substitution on the amine moiety in the same manner as ASR 2 and a biaryl quencher to increase the contact interface with the aptamer. Indeed, ASR 7 showed a 33-fold increase in the binding affinity for Apt 10L compared to ASR 1 (Kd = 1.2 ± 0.1 μM, Fmax = 88.6 ± 1.7 for ASR 7). In addition, ASR 7 binds tightly to Apt10M with a 2-fold less affinity than Apt10L (Kd = 2.6 ± 0.2 μM, Fmax = 95.1 ± 1.5), as is the case with ASR 1. Despite the highly improved binding affinity by the introduction of a biaryl quencher, ASR 7 does not bind to mutants like Apt10M1 and Apt10M-Lm3, suggesting that the binding specificity was not affected by a biaryl quencher. This result indicates that rational modifications of these functional groups on ASR effectively improve the binding affinity to aptamer and fluorogenic properties.
Figure 8.
ASR 7 containing a biaryl quencher has a highly improved binding affinity for Apt10L. Fluorescence titration was carried out in PBS buffer (pH 7.4) containing 1 mM MgCl2. Increasing concentrations of Apt10L RNA or selected Apt10M variants (Apt10M, Apt10M1 or Apt10M-Lm3) were added in the presence of 1 μM of ASR 7. Excitation and emission wavelength were 555 nm and 610 nm, respectively. Kd of ASR 7 for Apt10L RNA is 1.2 ± 0.1 μM and Fmax value is 88.6 ± 1.7. Kd of ASR 7 for Apt10M RNA is 2.6 ± 0.2 μM and Fmax value is 95.1 ± 1.5. Like ASR 1, ASR 7 does not bind Apt10M1 and Apt10M-Lm3 mutants, suggesting that the binding specificity not affected by the introduction of a biaryl quencher.
Both the binding affinity and fluorescence activation with ASR 7 and Apt10L compare well to previously reported fluorogen-RNA aptamer pairs (31–36) whose binding constants range from tens of μM to sub-μM, and whose fluorescence increase is generally less than 100-fold except that malachite green displays 2360-fold. Through mutation and truncation analysis and chemical modifications, we have gained a better understanding of the structural interactions between the ASR fluorogen and the aptamer, which can guide us in continuous optimization to further improve both the binding affinity and the fluorescence activation in this new RNA labeling pair.
In summary, we report here a new small-molecule fluorogen and RNA aptamer pair for RNA labeling. The SELEX procedure and fluorescence screening in E. coli have been applied to discover the aptamer that can specifically activate the fluorogen. The systematic mutation and truncation study on the aptamer structure determined the minimum binding domain of aptamer specificity to the fluorogen. A series of rationally modified fluorogen analogs have been made to probe the interacting groups of the fluorogen with the aptamer. These results allow us to design ASR 7 that displayed a 33-fold increase in the binding affinity for the selected aptamer compared to original ASR 1 and an 88-fold increase in the fluorescence emission after the aptamer binding. The further refining of the probe structure and aptamer selection could eventually lead to an ideal labeling pair that can be used for in vivo RNA live-cell imaging.
METHODS
Synthesis
Detailed synthetic procedures and characterizations of all the fluorophores ASR 1–7 are described in Supporting Information.
Quantum Yield Determination
The quantum yield of the ASR (Φx) dye was determined relative to a known standard dye rhodamine B (Φst= 0.31) (37). The integrated fluorescence emission of the ASR dye in 90% glycerol (w/v) with refractive index (nx = 1.45839) (49) and rhodamine B in water with refractive index (nst = 1.00) was plotted against the absorbance to measure slope of the plot for rhodamine B (mst) and ASR (mx). The quantum yield of the ASR was obtained by using the slopes of the plot in the following formula: Φx = Φst * (mx/mst) * (nx/nst)2.
RNA Preparation
DNA templates for the random N60 library with 20 flanking primer sequences were ordered from Integrated DNA Technologies (See Supplementary Figure 2 for the list of primers). The template was amplified by standard PCR using forward primer containing T7 RNA polymerase promoter and reverse primer in standard GoTaq green solution by promega. The PCR product was purified by QIAprep Spin Miniprep Kit from QIAGEN. The PCR product was transcribed into RNA using in vitro RNA transcription kit (T7 Ribo Max; Promega). The transcribed RNA was purified by G-25 spin column (GE healthcare). The purified RNA was quantified by measuring its UV absorption (expected ε260 = 972,100 L M−1 cm−1).
RNA Selection (SELEX)
A biotin-conjugated analogue of ASR 1 was synthesized as described in supplementary methods. It was then immobilized on avidin-agarose beads and incubated with in vitro transcribed library (random 60mer), washed three times with PBS and was subsequently reverse transcribed PCR. The PCR product was transcribed into RNA that is used in the second round of SELEX. After four rounds of enrichment, aptamer library with high affinity to ASR 1 was generated from the original RNA pool containing about 1013 random sequences. The RT-PCR products from this secondary aptamer library were then cloned into pBK-CMV-eGFP vector using restriction enzyme HindIII and XbaI, and transformed into competent E. coli cells. Under the lac promoter on the vector, the RNA aptamers will be transcribed in the E. coli and bind to the cell membrane permeable probe subsequently. If the aptamers can functionally enhance fluorescence intensity of the probe, the clones containing these aptamers will be captured in the light-tight chamber of an IVIS 200 fluorescence imager. After validation of the affinity and light-up activity of the aptamers isolated from the positive colonies by in vitro titration, the plasmids containing these aptamers were sequenced.
In vitro RNA Fluorescence Measurement
Fluorescence titration was carried out in PBS buffer (pH 7.4) containing 1 mM MgCl2 using Horiba FluoroMax-3 fluorometer. Increasing concentrations of Apt10L variants were added in the presence of 1 μM of each ASR analogue. The concentration of ASR was calculated by measuring absorption at 555 nm (for example, extinction coefficient of ASR 1 was determined to be 75,000 M−1 cm−1). Maximal excitation and emission wavelength were 555 nm and 610 nm, respectively.
Determination of Dissociation Constants and Fmax
Dissociation constant Kd (in μM) was determined by measuring fluorescence intensity (F) versus concentration of the aptamer ([RNA] in μM) at 1 μM dye concentration. The result was analyzed by the nonlinear fit function of OriginPro 8 using the following formula,
where Fmax = Fluorescence intensity at infinite RNA concentration in μM, and F0 = Fluorescence intensity at 0 μM RNA.
Supplementary Material
Acknowledgments
This work has been supported by a Young Investigator Award from Human Science Frontier Program and a research grant from NIGMS (1R01GM086196-01).
Footnotes
Supporting Information Available: This material is available free of charge via the Internet.
References
- 1.Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Marks KM, Nolan GP. Chemical labeling strategies for cell biology. Nat Methods. 2006;3:591–596. doi: 10.1038/nmeth906. [DOI] [PubMed] [Google Scholar]
- 3.Johnsson N, Johnsson K. Chemical tools for biomolecular imaging. ACS Chem Biol. 2007;2:31–38. doi: 10.1021/cb6003977. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 5.Dragulescu-Andrasi A, Rao J. Chemical labeling of protein in living cells. ChemBioChem. 2007;8:1099–1101. doi: 10.1002/cbic.200700158. [DOI] [PubMed] [Google Scholar]
- 6.Tyagi S. Imaging intracellular RNA distribution and dynamics in living cells. Nat Methods. 2009;6:331–338. doi: 10.1038/nmeth.1321. [DOI] [PubMed] [Google Scholar]
- 7.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gang Bao WJR, Andrew Tsourkas. Fluorescent probes for live-cell RNA detection. Annu Rev Biomed Eng. 2009;11:25–47. doi: 10.1146/annurev-bioeng-061008-124920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paillasson S, vandeCorput M, Dirks RW, Tanke HJ, RobertNicoud M, Ronot X. In situ hybridization in living cells: Detection of RNA molecules. Exp Cell Res. 1997;231:226–233. doi: 10.1006/excr.1996.3464. [DOI] [PubMed] [Google Scholar]
- 10.Molenaar C, Marras SA, Slats JCM, Truffert JC, Lemaitre M, Raap AK, Dirks RW, Tanke HJ. Linear 2’ O-Methyl RNA probes for the visualization of RNA in living cells. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.17.e89. art. no. e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molenaar C, Abdulle A, Gena A, Tanke HJ, Dirks RW. Poly(A)(+) RNAs roam the cell nucleus and pass through speckle domains in transcriptionally active and inactive cells. J Cell Biol. 2004;165:191–202. doi: 10.1083/jcb.200310139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dirks RW, Tanke HJ. Advances in fluorescent tracking of nucleic acids in living cells. Biotechniques. 2006;40:489–496. doi: 10.2144/000112121. [DOI] [PubMed] [Google Scholar]
- 13.Dirks RW, Molenaar C, Tanke HJ. Methods for visualizing RNA processing and transport pathways in living cells. Histochem Cell Biol. 2001;115:3–11. doi: 10.1007/s004180000214. [DOI] [PubMed] [Google Scholar]
- 14.Tyagi S, Marras SAE, Kramer FR. Wavelength-shifting molecular beacons. Nat Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 15.Tyagi S, Kramer FR. Molecular beacons: Probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 16.Tyagi S, Bratu DP, Kramer FR. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 17.Tyagi S, Alsmadi O. Imaging native beta-actin mRNA in motile fibroblasts. Biophys J. 2004;87:4153–4162. doi: 10.1529/biophysj.104.045153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Franzini RM, Kool ET. Efficient nucleic acid detection by templated reductive quencher release. J Am Chem Soc. 2009;131:16021–16023. doi: 10.1021/ja904138v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman AP, Kool ET. Detecting RNA and DNA with templated chemical reactions. Chem Rev. 2006;106:3775–3789. doi: 10.1021/cr050057+. [DOI] [PubMed] [Google Scholar]
- 20.So MK, Gowrishankar G, Hasegawa S, Chung JK, Rao J. Imaging target mRNA and siRNA-mediated gene silencing in vivo with ribozyme-based reporters. ChemBioChem. 2008;9:2682–2691. doi: 10.1002/cbic.200800370. [DOI] [PubMed] [Google Scholar]
- 21.Valencia-Burton M, McCullough RM, Cantor CR, Broude NE. RNA visualization in live bacterial cells using fluorescent protein complementation. Nat Methods. 2007;4:421–427. doi: 10.1038/nmeth1023. [DOI] [PubMed] [Google Scholar]
- 22.Ozawa T, Natori Y, Sato M, Umezawa Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat Methods. 2007;4:413–419. doi: 10.1038/nmeth1030. [DOI] [PubMed] [Google Scholar]
- 23.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 24.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 25.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 26.Singer RH. RNA localization: Visualization in real-time. Curr Biol. 2003;13:R673–R675. doi: 10.1016/s0960-9822(03)00605-5. [DOI] [PubMed] [Google Scholar]
- 27.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 28.Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shav-Tal Y, Singer RH, Darzacq X. Imaging gene expression in single living cells. Nat Rev Mol Cell Biol. 2004;5:855–862. doi: 10.1038/nrm1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: Real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 32.Constantin TP, Silva GL, Robertson KL, Hamilton TP, Fague K, Waggoner AS, Armitage BA. Synthesis of new fluorogenic cyanine dyes and incorporation into RNA fluoromodules. Org Lett. 2008;10:1561–1564. doi: 10.1021/ol702920e. [DOI] [PubMed] [Google Scholar]
- 33.Pei R, Rothman J, Xie YL, Stojanovic MN. Light-up properties of complexes between thiazole orange-small molecule conjugates and aptamers. Nucleic Acids Res. 2009;37:e59. doi: 10.1093/nar/gkp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sparano BA, Koide K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J Am Chem Soc. 2005;127:14954–14955. doi: 10.1021/ja0530319. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C, Szostak JW. Isolation of a fluorophore-specific DNA aptamer with weak redox activity. Chem Biol. 1998;5:609–617. doi: 10.1016/s1074-5521(98)90289-7. [DOI] [PubMed] [Google Scholar]
- 36.Sando S, Narita A, Hayami M, Aoyama Y. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem Comm. 2008:3858–3860. doi: 10.1039/b808449a. [DOI] [PubMed] [Google Scholar]
- 37.Magde D, Rojas GE, Seybold PG. Solvent dependence of the fluorescence lifetimes of xanthene dyes. Photochem Photobiol. 1999;70:737–744. [Google Scholar]
- 38.Woodroofe CC, Lim MH, Bu WM, Lippard SJ. Synthesis of isomerically pure carboxylate- and sulfonate-substituted xanthene fluorophores. Tetrahedron. 2005;61:3097–3105. [Google Scholar]
- 39.Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, Nagano T. Evolution of fluorescein as a platform for finely tunable fluorescence probes. J Am Chem Soc. 2005;127:4888–4894. doi: 10.1021/ja043919h. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka K, Miura T, Umezawa N, Urano Y, Kikuchi K, Higuchi T, Nagano T. Rational design of fluorescein-based fluorescence probes, mechanism-based design of a maximum fluorescence probe for singlet oxygen. J Am Chem Soc. 2001;123:2530–2536. doi: 10.1021/ja0035708. [DOI] [PubMed] [Google Scholar]
- 41.Miura T, Urano Y, Tanaka K, Nagano T, Ohkubo K, Fukuzumi S. Rational design principle for modulating fluorescence properties of fluorescein-based probes by photoinduced electron transfer. J Am Chem Soc. 2003;125:8666–8671. doi: 10.1021/ja035282s. [DOI] [PubMed] [Google Scholar]
- 42.deSilva AP, Gunaratne HQN, Gunnlaugsson T, Huxley AJM, McCoy CP, Rademacher JT, Rice TE. Signaling recognition events with fluorescent sensors and switches. Chem Rev. 1997;97:1515–1566. doi: 10.1021/cr960386p. [DOI] [PubMed] [Google Scholar]
- 43.Bartel DP, Szostak JW. Isolation of new ribozymes from a large pool of random sequences. Science. 1993;261:1411–1418. doi: 10.1126/science.7690155. [DOI] [PubMed] [Google Scholar]
- 44.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 45.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded-DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 46.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 47.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment - RNA ligands to bacteriophage-T4 DNA-polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 48.Wilson C, Szostak JW. In vitro evolution of a self-alkylating ribozyme. Nature. 1995;374:777–782. doi: 10.1038/374777a0. [DOI] [PubMed] [Google Scholar]
- 49.http://www.dow.com/glycerine/resources/table12_020.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.