Abstract
RELATED TO UBIQUITIN (RUB) modification of CULLIN (CUL) subunits of the CUL-RING ubiquitin E3 ligase (CRL) superfamily regulates CRL ubiquitylation activity. RUB modification requires E1 and E2 enzymes that are analogous to, but distinct from, those activities required for UBIQUITIN (UBQ) attachment. Gene duplications are widespread in angiosperms, and in line with this observation, components of the RUB conjugation pathway are found in multiples in Arabidopsis. To further examine the extent of redundancy within the RUB pathway, we undertook biochemical and genetic characterizations of one such duplication event- the duplication of the genes encoding a subunit of the RUB E1 into AUXIN RESISTANT1 (AXR1) and AXR1-LIKE1 (AXL1). In vitro, the two proteins have similar abilities to function with E1 C-TERMINAL-RELATED1 (ECR1) in catalyzing RUB1 activation and RUB1-ECR1 thioester formation. Using mass spectrometry, endogenous AXR1 and AXL1 proteins were found in complex with 3HA-RUB1, suggesting that AXR1 and AXL1 exist in parallel RUB E1 complexes in Arabidopsis. In contrast, AXR1 and AXL1 differ in ability to correct phenotypic defects in axr1-30, a severe loss-of-function AXR1 mutant, when the respective coding sequences are expressed from the same promoter, suggesting differential in vivo functions. These results suggest that while both proteins function in the RUB pathway and are biochemically similar in RUB-ECR1 thioester formation, they are not functionally equivalent.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-011-9750-8) contains supplementary material, which is available to authorized users.
Keywords: RELATED TO UBIQUITIN, AUXIN RESISTANT1, AXR1-LIKE1, RUB-activating enzyme, Functional divergence, Arabidopsis thaliana
Introduction
The UBIQUITIN (UBQ) conjugation pathway consists of an activating enzyme (E1), a conjugating enzyme (E2), and a ligase (E3) that function in concert to catalyze covalent attachment of UBQ to cellular proteins, often followed by UBQ chain assembly. The attachment of UBQ can affect substrate protein longevity, activity, subcellular localization, and/or protein–protein interactions [reviewed in (Petroski and Deshaies 2005; Welchman et al. 2005; Dreher and Callis 2007; Vierstra 2009; Miura and Hasegawa 2010)]. UBQ is the founding member of the UBQ-like protein (UBL) superfamily, members of which share a conserved structure and are conjugated to their respective substrates by similar conjugation systems [reviewed in (Dye and Schulman 2007; Schulman and Harper 2009)].
RELATED TO UBIQUITIN (RUB), called neuronal precursor cell-expressed developmentally downregulated 8 (NEDD8) in animals and fission yeast, is a UBL whose covalent ligation to its substrate is mechanistically identical to UBQ ligation, but unique enzymes are required. Initiating RUB attachment is a heterodimeric RUB E1 that is composed of two 50-60 kDa subunits sharing identity with either the N-terminal or C-terminal half of the UBQ E1 [reviewed in (Downes and Vierstra 2005; Hotton and Callis 2008; Schulman and Harper 2009)]. In Arabidopsis, a single gene encodes the C-terminal RUB E1 subunit, E1 C-TERMINAL-RELATED1 (ECR1) (del Pozo et al. 1998; del Pozo et al. 2002; Woodward et al. 2007). Two genes, AUXIN RESISTANT1 (AXR1) and AXR1-LIKE1 (AXL1), encode N-terminal RUB E1 subunits of 540 and 523 amino acids, respectively, with ~ 80% amino acid identity (del Pozo et al. 1998; Dharmasiri et al. 2003; Dharmasiri et al. 2007). The heterodimer AXR1/ECR1 catalyzes RUB-adenylate formation and subsequent thioester formation between the RUB C-terminal carboxyl group and an ECR1 cysteinyl sulfhydryl group, and genetic data suggests that AXL1 can function in place of AXR1 in the RUB E1 complex (del Pozo et al. 1998; Dharmasiri et al. 2007).
Required next is a RUB E2, or RUB-conjugating enzyme (RCE), encoded by one of two closely related genes, RCE1 and RCE2, in Arabidopsis (del Pozo and Estelle 1999; Dharmasiri et al. 2003). Finally, a RUB-specific E3 ligase, or RUB E3, is responsible for catalyzing transfer of the activated RUB to its substrate, though the RUB E3 has remained ill-defined in Arabidopsis. In defining the RUB E3, one must first consider the substrate being modified, as the E3 is the specificity factor that both recognizes a specific substrate and brings the E2 in close proximity to the substrate, prior to UBL modification [reviewed in (Petroski and Deshaies 2005; Dye and Schulman 2007)]. In contrast to UBQ, which covalently attaches to a large number of diverse proteins, only a limited number of proteins have been identified as substrates of the RUB pathway. The best characterized and most evolutionarily conserved RUB substrates are the CULLIN (CUL) subunits of the CUL-RING, or CRL, class of UBQ E3 ligases [reviewed in (Petroski and Deshaies 2005)], and attachment occurs near the C-terminus within the short consensus sequence VRIMK via the ε-amino group of the lysyl residue (del Pozo and Estelle 1999; Wada et al. 1999). In Arabidopsis, CUL1, CUL3a/b and CUL4 are known to be RUB-modified and function in distinct CRLs [reviewed in (Hotton and Callis 2008)].
RING BOX1 (RBX1) [also called regulator of cullins 1 (ROC1) and high-level expression reduces Ty3 transposition 1 (HRT1) in mammals and yeast] interacts with CUL proteins to serve as a docking site for UBQ E2 and RUB E2, and its importance to RUB conjugation has been genetically characterized in Arabidopsis (Seol et al. 1999; Gray et al. 2002; Lechner et al. 2002; Morimoto et al. 2003). Advances in Saccharomyces cerevisiae, Caenorhabditis elegans and mammals suggest that a component of the cullin-specific RUB E3 is the protein defective in cullin neddylation (DCN) [also called squamous cell carcinoma related oncogene (SCCRO) in humans], which serves a scaffold function in the transfer of RUB to its substrate CUL protein (Kurz et al. 2005; Kim et al. 2008; Kurz et al. 2008; Meyer-Schaller et al. 2009). Three DCN1 homologs have been identified in Arabidopsis, but their role in the RUB pathway remains to be elucidated (Kurz et al. 2005; Biswas et al. 2007).
Regarding UBL conjugation pathways, it was widely held until recently that one activating enzyme (E1) charges a handful of conjugating enzymes (E2) that then interact with a multiplicity of ligases (E3s); the E3s specifically recognize then modify a vast number of substrates with a UBL [reviewed in (Pickart and Eddins 2004)]. This view has shifted with the characterization of a second UBQ E1, ubiquitin-like modifier activating enzyme 6 (UBA6), found in vertebrates and sea urchins. UBA6 functions with a single E2, presumably to interact with a subset of E3s, indicating a split in the UBQ conjugation pathway from the first activation step (Chiu et al. 2007; Jin et al. 2007; Pelzer et al. 2007). Also of interest is the expansion of NEDD8 E2s in metazoans, where a newly characterized E2, ubiquitin conjugating enzyme E2F (UBE2F), was found to differ in enzymatic activity and to preferentially interact with RBX2 and CUL5, compared to UBE2M, which interacts with RBX1 and the remaining CUL proteins (Huang et al. 2009).
In angiosperms, gene duplications and thus genetic redundancy are common [reviewed in (Paterson et al. 2010)]; the RUB pathway is no exception (Rao-Naik et al. 1998; del Pozo and Estelle 1999; Gray et al. 2002; Lechner et al. 2002; Dharmasiri et al. 2003). In light of recent advances in metazoan systems, we examined whether expansion in Arabidopsis RUB E1 subunits, AXR1 and AXL1, remains simple genetic redundancy, or if these proteins have functionally diverged, as seen for other organisms and UBL conjugation systems. Here we present biochemical data that show AXR1 and AXL1 have similar activity in vitro. Additionally, a mass spectrometric approach was taken that identified both endogenous AXR1 and AXL1 in complex with 3HA-RUB1 in Arabidopsis, suggesting concurrent functions of AXR1 and AXL1 in the plant. Finally, complementation experiments, using the AXR1 5′ intergenic region and either AXR1 or AXL1 coding sequences, suggest that the two proteins are not functionally equivalent in vivo.
Results
AXL1 catalyzes thioester formation between ECR1 and RUB1 in vitro
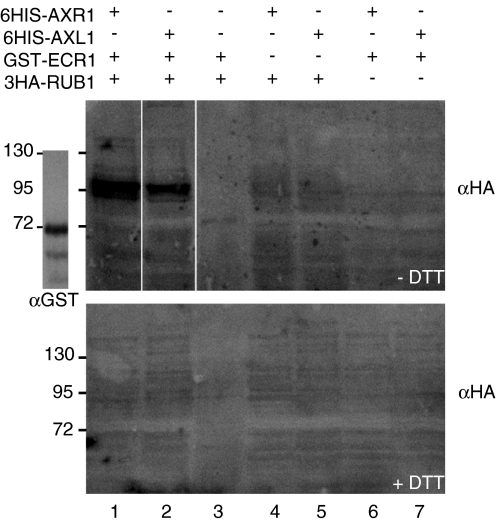
Sequence similarity between AXR1 and AXL1 suggests that AXL1 has the same, or similar, biochemical activity to AXR1 (Dharmasiri et al. 2007). Previous in vitro assays show AXR1 to catalyze formation of a thioester linkage between the C-terminus of RUB1 and the catalytic cysteine, C215, of ECR1 (del Pozo et al. 1998). However, the biochemical activity of AXL1 has not been demonstrated directly. Utilizing an in vitro thioester assay, we show RUB1-ECR1 thioester formation is also catalyzed by AXL1 (Fig. 1). Recombinant GST-ECR1 and 6HIS-3HA-RUB1 were incubated with 6HIS-AXL1 or 6HIS-AXR1, in the presence of ATP. Each reaction was then split: one-half was added to stop buffer lacking dithiothreitol (Fig. 1, −DTT, upper panel), and the other half was added to stop buffer containing DTT (Fig. 1, +DTT, lower panel). Thioester linkages are reduced by DTT, while oxyester and amide linkages are resistant to DTT-mediated cleavage. After separation by SDS–PAGE, conjugated 6HIS-3HA-RUB1 was visualized by anti-HA immunoblot analysis. In these experiments unconjugated 6HIS-3HA-RUB1 is 17 kDa in size and is not present on the blots. GST-ECR1 alone migrates at 72 kDa (Fig. 1, left panel, αGST). An anti-HA band migrating approximately 20 kDa larger than GST-ECR1 was visible in complete reactions containing either 6HIS-AXR1 or 6HIS-AXL1 and stopped in the absence of DTT (Fig. 1, lanes 1 and 2, compare –DTT and +DTT), indicative of a RUB1-ECR1 thioester conjugate. GST-ECR1 and 6HIS-3HA-RUB1, in reactions lacking 6HIS-AXR1 and 6HIS-AXL1, were insufficient to support RUB1-ECR1 thioester formation (Fig. 1, lane 3). No slow-migrating HA-immunoreactive protein was visible when either GST-ECR1 or 6HIS-3HA-RUB1 was omitted from reactions containing 6HIS-AXR1 or 6HIS-AXL1 (Fig. 1, lanes 4–7). Thus, AXL1 is required for conjugation of RUB1 to ECR1 in an in vitro thioester reaction, as seen for AXR1 under the same conditions.
Fig. 1.
AXL1 catalyzes RUB1-ECR1 thioester formation like AXR1. In vitro thioester assays with recombinant 6HIS-AXR1, 6HIS-AXL1, GST-ECR1, and 6HIS-3HA-RUB1 are shown. An anti-GST blot of purified GST-ECR1 at 72 kDa is included for reference. Anti-HA immunoblot analysis detected 3HA-RUB1-GST-ECR1 thioester formation under non-reducing conditions (−DTT) only, catalyzed by 6HIS-AXR1 (lane 1), or by 6HIS-AXL1 (lane 2). Lanes 3–7 for both upper and lower panels show reactions lacking 6HIS-AXL1 and 6HIS-AXR1 (lane 3), GST-ECR1 (lanes 4 and 5), or 6HIS-3HA-RUB1 (lanes 6 and 7), as negative controls. All reactions were stopped under non-reducing (−DTT, upper panel) or reducing (+DTT, lower panel) conditions and separated by non-reducing SDS–PAGE. White spaces represent positions where unnecessary lanes were removed, or where lanes were moved for alignment purposes. Size markers are in kDa
Additionally, enzyme specificity for RUB1 over UBQ was examined for AXL1 in comparison to AXR1. 6HIS-AXR1 and 6HIS-AXL1 again supported 6HIS-3HA-RUB1 thioester formation to GST-ECR1, but neither was able to support HA-UBQ attachment to GST-ECR1, while the same HA-UBQ formed an UBQ-E1 thioester conjugate (Online Resource 1). In conclusion, AXL1 and AXR1 support RUB1, but not UBQ, thioester formation, demonstrating that AXL1 has biochemical activity and specificity similar to AXR1.
AXL1 and AXR1 have similar biochemical activities in vitro
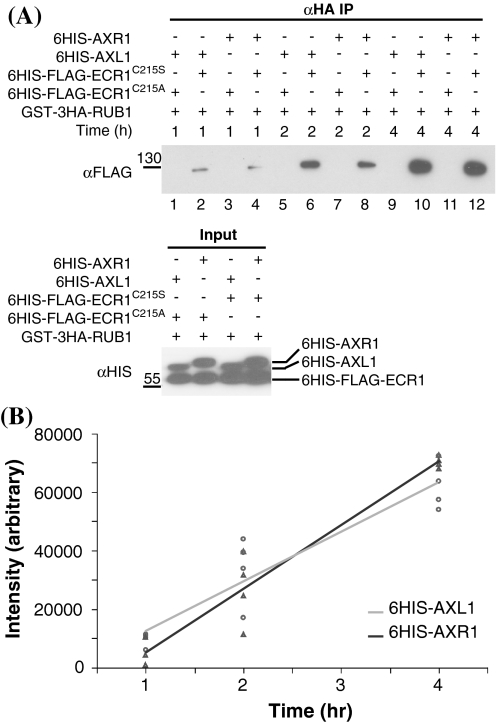
While having the same selectivity for RUB1, it is possible that AXR1 and AXL1 differ subtly in ability to catalyze RUB1-ECR1 thioester formation. To further examine the biochemical activities of AXR1 and AXL1 we set up an in vitro time course assay to compare the ability of each enzyme to catalyze covalent attachment of RUB1 to ECR1. Here we utilized ECR1C215S that carries a cysteine to serine mutation at the active site, in order to stabilize the bond for visualization purposes. Recombinant 6HIS-FLAG-ECR1C215S and GST-3HA-RUB1 were incubated with either 6HIS-AXL1 or 6HIS-AXR1 for 1, 2, or 4 h. At each time point, reaction aliquots were split and both halves were subjected to anti-HA-agarose immunoprecipitation to capture GST-3HA-RUB1 and interacting proteins.
One-half of each reaction was kept under neutral conditions to maintain the RUB1-ECR1C215S oxyester bond (Fig. 2a, αHA IP, even-numbered lanes), while the other half reaction was treated with base to destroy the oxyester bond (data not shown) to demonstrate the nature of the RUB1-ECR1 linkage. In addition, parallel reactions were done with ECR1C215A [described in (del Pozo et al. 1998)] that carries a cysteine to alanine mutation at the active site (Fig. 2a, αHA IP, odd-numbered lanes). This protein should not support covalent RUB attachment, and it serves to verify the specificity of the reaction. Presence of 6HIS-AXL1, 6HIS-AXR1, 6HIS-FLAG-ECR1C215S, and 6HIS-FLAG-ECR1C215A in the reactions was verified by anti-HIS immunoblot analysis (Fig. 2a, Input). To visualize the RUB1-ECR1 oxyester bond, anti-FLAG immunoblots were done (Fig. 2a, αHA IP). Only reactions that included 6HIS-FLAG-ECR1C215S, not 6HIS-FLAG-ECR1C215A, supported RUB1-ECR1 oxyester bond formation.
Fig. 2.
AXL1 and AXR1 have similar biochemical activities in vitro. a Time course for various oxyester assays with recombinant 6HIS-AXR1, 6HIS-AXL1, 6HIS-FLAG-ECR1C215S, 6HIS-FLAG-ECR1C215A, and GST-3HA-RUB1. After anti-HA immunoprecipitation, anti-FLAG immunoblot analysis was used to detect GST-3HA-RUB1-6HIS-FLAG-ECR1C215S oxyester formation, catalyzed by 6HIS-AXL1 (lanes 2, 6, 10), or by 6HIS-AXR1 (lanes 4, 8, 12), for various lengths of time. As a negative control, parallel reactions were done with the catalytic mutant 6HIS-FLAG-ECR1C215A and 6HIS-AXL1 (lanes 1, 5, 9) or 6HIS-AXR1 (lanes 3, 7, 11). Input (fraction of total for each sample, removed prior to start of time course) was checked with anti-HIS immunoblot to verify addition of 6HIS-AXL1, 6HIS-AXR1, 6HIS-FLAG-ECR1C215S, and 6HIS-FLAG-ECR1C215A. Size markers are in kDa. b GST-3HA-RUB1-6HIS-FLAG-ECR1C215S band intensity plotted against time, comparing catalytic activity of 6HIS-AXL1 (circles) and 6HIS-AXR1 (triangles) for n = 4 replicates. Regression lines for 6HIS-AXL1 (light gray) and 6HIS-AXR1 (dark gray) are included. At α = 0.05, testing for an E1*time interaction is not significant (factorial ANOVA, P = 0.0755)
To compare relative activities of 6HIS-AXL1 and 6HIS-AXR1, immunoblots were quantified and RUB1- ECR1C215S bond formation was plotted against time (Fig. 2b). Regression line slopes for ECR1C215S reactions containing 6HIS-AXR1 or 6HIS-AXL were not statistically different (factorial ANOVA, P = 0.0755). The marginal P-value does not exclude the possibility that a subtle difference in catalytic abilities might exist between AXR1 and AXL1, but a more precise assay would be required to discern such a difference. In this assay, we could not detect a difference between AXR1 and AXL1 in ability to support RUB1-ECR1C215S bond formation over time.
AXR1 and AXL1 interact with RUB1 in vivo
To assess whether AXL1 functions in native RUB E1 complexes, we used a mass spectrometry (MS) approach. Transgenic plant lines that express 3HA-RUB1 under control of an inducible promoter were utilized [described in (Bostick et al. 2004)] and protein extracts were made from seedlings induced for 3HA-RUB1 expression. Proteins that interact with 3HA-RUB1 were immunoprecipitated with anti-HA-agarose beads, digested with trypsin, and analyzed by high-accuracy MS. Results were compiled from five biological replicates; recovered peptides were compared between Columbia expressing 3HA-RUB1 and wild-type Columbia, prepared in parallel. Table 1 combines data from replicates where peptide and protein standards were met, specifically a minimum 95% peptide probability, using the Peptide Prophet algorithm, and protein probability of minimally 95% with 2 unique peptides, using the Protein Prophet algorithm (Keller et al. 2002; Nesvizhskii et al. 2003). Peptides were identified from AXR1, AXL1, and other known RUB-conjugation components, including RUB1, ECR1, RCE1, CUL1, and CUL4. These results suggest that AXL1, as found naturally in cells, interacts with 3HA-RUB1, despite the presence of AXR1, suggesting that AXL1 forms functional E1 complexes in vivo and is active concurrent with AXR1.
Table 1.
RUB conjugation components recovered by LC–MS/MS analysis of 3HA-RUB1 immunoprecipitations
| Gene | Uniprot ID | AGI | Mass (Da) | Unique peptidesa | Total unique hits | Protein ID probabilityb (%) | % Coveragec |
|---|---|---|---|---|---|---|---|
| AXL1 | Q9ZV69 | At2g32410 | 57924.1 | 12 | 63 | 100 | 33 |
| AXR1 | P42744 | At1g05180 | 60018.1 | 20 | 212 | 100 | 56 |
| CUL1 | Q94AH6 B9DGE3 | At4g02570 | 86287.4 | 7 | 11 | 100 | 12 |
| CUL4 | Q8LGH4 | At5g46210 | 91457.7 | 3 | 5 | 100 | 5 |
| ECR1 | O65041 | At5g19180 | 50524.4 | 18 | 264 | 100 | 48 |
| RCE1 | Q9SDY5 | At4g36800 | 20768.9 | 4 | 45 | 100 | 28 |
| RUB1 | Q9SHE7 | At1g31340 | 8872.8 | 3 | 460 | 100 | 50 |
aPeptides are designated as unique if they differ in primary amino acid sequence and satisfy rules of parsimony
bProtein ID probability is for the highest probability observed in 5 biological replicates
c% coverage is cumulative for 5 biological replicates
Additionally, single peptide-based protein identifications were made for CUL1, CUL3a, CUL4, and RCE2, as individual peptides met MS criteria, but within a particular biological replicate, protein criteria were not met (lack of 2 unique peptides and/or below 95% protein probability threshold). Spectra from single peptide-based protein identifications are presented in Online Resource 2, all recovered ions are presented in Online Resource 3, and data are summarized in Table 2. Of interest, peptides are recovered for both RCE2 and RCE1, suggesting both are functional RUB E2s.
Table 2.
Single peptide-based protein identifications from LC–MS/MS analysis of 3HA-RUB1 immunoprecipitations
| Index | Gene | AGI | Sequence identified | Peptide ID probability (%) | −Log(e) peptide score | Precursor Ion m/z | Charge state | Sample |
|---|---|---|---|---|---|---|---|---|
| 1 | CUL1 | At4g02570 | (R)EAFEEYINSTVLPALR(E) | 95 | 4.23 | 926.48 | 2 | C |
| 2 | CUL1 | At4g02570 | (K)IPLPPVDER(K) | 95 | 3.96 | 518.30 | 2 | B |
| 3 | CUL1 | At4g02570 | (K)IPLPPVDER(K) | 95 | 3.52 | 518.30 | 2 | B |
| 4 | CUL3a | At1g26830 | (K)EIEQATEIPAADLK(R) | 95 | 6.52 | 764.40 | 2 | A |
| 5 | CUL4 | At5g46210 | (K)VLSHTLLITELFQQLK(F) | 95 | 6.42 | 628.37 | 3 | D |
| 6 | RCE2 | At2g18600 | (K)DISELNLPK(S) | 95 | 2.82 | 514.78 | 2 | A |
| 7 | RCE2 | At2g18600 | (K)DISELNLPK(S) | 95 | 3.13 | 514.79 | 2 | A |
AXL1 and AXR1 proteins accumulate to similar levels when expressed exogenously in axr1-30 under the same promoter
Loss-of-function mutations in AXR1 result in a well-described phenotype; adult plants are dwarfed and show reduced apical dominance, leaf curling and reduced fertility; axr1 seedlings are also less sensitive to exogenous auxin (Lincoln et al. 1990; Leyser et al. 1993). Loss-of-function mutation in AXL1 has little or no phenotypic effect in plants that carry two AXR1 wild-type alleles, but in plants that carry a mutation in one or both AXR1 alleles, the effect of axl1 is synergistic (Dharmasiri et al. 2007). A simple explanation for the differential contribution of each gene could be differences in transcript expression, as mRNA levels of AXL1 are lower than AXR1 (Dharmasiri et al. 2007).
To examine the biochemical and functional equivalence of AXR1 and AXL1 in vivo, expression cassettes for the respective coding sequences, under the control of the same 5′ flanking region, were introduced into axr1-30, a severe AXR1 mutant [phenotypically characterized in (Gilkerson et al. 2009)] that is similar in phenotype to axr1-12, a well-characterized severe allele (Lincoln et al. 1990; Leyser et al. 1993). axr1-30 does not produce detectable authentic mRNA and thus appears to be a loss-of-function allele. Additionally, loss of AXR1 does not affect the expression of AXL1 (Online Resource 4). The expression cassettes contain the 5′ intergenic region for AXR1, hereafter referred to as the AXR1 promoter (AXR1p), sequence for an N-terminal 10xMYC epitope tag, and the respective coding sequence. These constructs were introduced into AXR1/axr1-30 plants and then plants were brought to homozygosity for both the axr1-30 allele and the respective transgene.
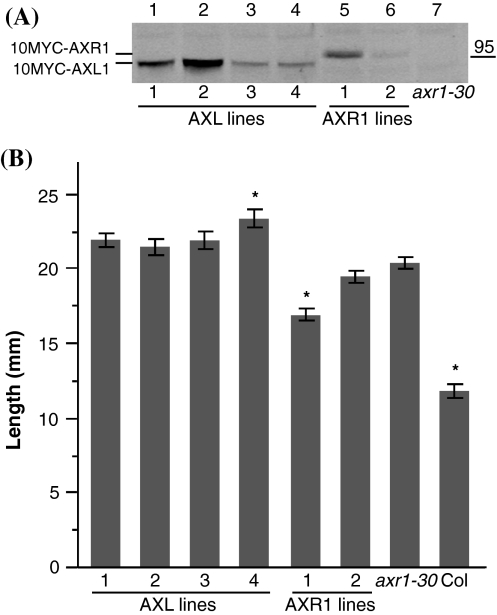
To make comparisons, 10MYC-AXR1 and 10MYC-AXL1 protein levels were examined in all AXR1p:10MYC-AXR1 (called AXR1 lines) and AXR1p:10MYC-AXL1 (called AXL lines) transgenic lines. 10MYC-AXL1 protein levels were found to be similar to or exceeding 10MYC-AXR1 protein levels (Fig. 3a and data not shown). As elucidated by crystal structures for the human equivalent of RUB E1, the N-terminal RUB E1 subunit provides structural support and binding surfaces for RUB and ATP, in complex with ECR1 that houses the active catalytic cysteine and adenylation domains (Walden et al. 2003a; Walden et al. 2003b). Both 10MYC-AXR1 and 10MYC-AXL1 maintain the ability to interact with 6HIS-3HA-RUB1, when co-expressed transiently in tobacco, confirming functionality of these epitope-tagged proteins (Online Resource 5).
Fig. 3.
Protein levels for AXR1 and AXL1 are similar in axr1-30 transgenic lines, but only AXR1 line 1 shows moderate correction of axr1-30 phenotype in seedlings. a Protein levels in 8-day old seedlings. Based on anti-MYC immunoblot analyses, 10MYC-AXL1 (lanes 1–4) and 10MYC-AXR1 (lanes 5 and 6) accumulate protein to similar levels above background (lane 7), in characterized axr1-30 transgenic lines. 150 μg total protein was loaded for all samples. b Root length in 9-day old seedlings. Transgenic lines were plated on GM agar and grown for 9 days with germination marked on day 2. On day 9, seedlings were removed from plates, photographed, and root length was measured. Student’s t-tests with Bonferonni adjustment (α = 0.00385) were performed on combined data from three replicates. Plant lines marked with an asterisk are significantly different from axr1-30. Error bars represent SE with minimally n = 41 measurements per line
AXR1 corrects axr1-30 phenotypic defects more than AXL1 throughout development
After recovering homozygous axr1-30 lines that were also homozygous for either the AXR1p:10MYC-AXR1 (AXR1 lines) or AXR1p:10MYC-AXL1 (AXL lines) expression cassette, we sought to measure the effect of transgene expression by comparing growth of transgenic lines to axr1-30 and wild-type Columbia. Surprisingly, AXR1p:10MYC-AXR1 and AXR1p:10MYC-AXL1 expression cassettes differentially corrected axr1-30 phenotypic defects. Of five AXR1 lines analyzed, all showed moderate to strong complementation of axr1-30 phenotypic defects in adult plants. In contrast, of nine AXL lines analyzed, none showed strong complementation of axr1-30 phenotypic defects. Four AXL lines were then characterized in detail and compared to two AXR1 lines. For each set of phenotypic observations (i.e. root length, response to auxin, rosette diameter, and inflorescence height), Student’s t-tests with Bonferroni correction for multiple comparisons were performed to compare all lines to axr1-30 and wild type (Columbia). Additional statistical tests were then done to make comparisons among those lines that had phenotypes intermediate between axr1-30 and Columbia.
First, root length and response to the synthetic auxin, 2,4-D, were measured in 9-day old seedlings. axr1-30 roots, averaging 20.3 mm in length, are significantly longer than Columbia roots, which average 11.8 mm (Fig. 3b and Online Resource 6). Most lines are not significantly different from axr1-30, with only AXR1 line 1 that averages 16.8 mm showing a phenotype intermediate between axr1-30 and Columbia (Fig. 3b and Online Resource 6). Of the two AXR1 lines, line 1 expresses more 10MYC-AXR1 (Fig. 3a). Unexpectedly, AXL line 4 with average length of 23.3 mm is significantly longer than axr1-30, suggesting a potential ectopic phenotype, resulting from transgene over-expression or site of transgene insertion (Fig. 3b and Online Resource 6). When auxin responsiveness of seedling roots is measured, axr1-30 is auxin resistant whereas Columbia is auxin sensitive. The roots of AXL and AXR1 lines are auxin resistant, and thus they remain like axr1-30 regarding this phenotype (Online Resources 7, 8).
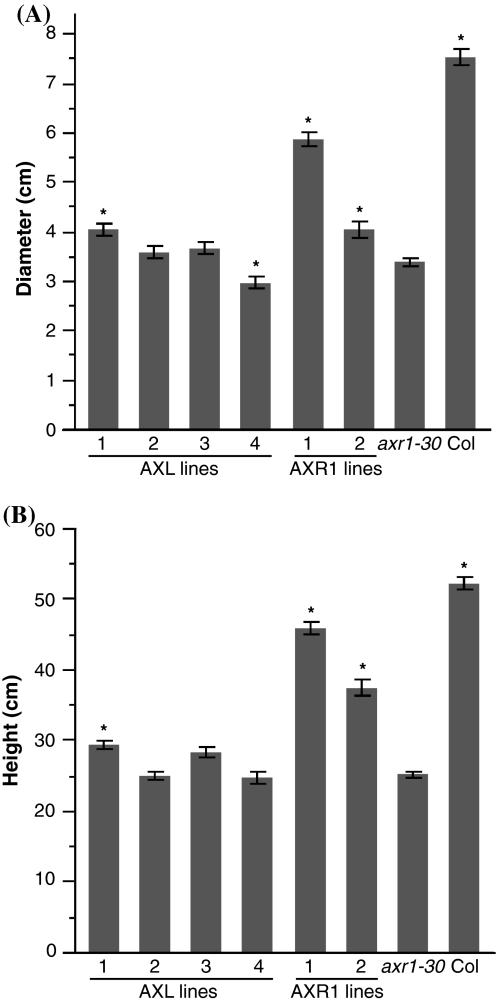
Next, rosette diameter at four weeks was measured and statistical analyses were performed. axr1-30 plants are dwarfed with small leaves; their rosette diameter is 45% that of Columbia at the same age (Fig. 4a). At this point in development, correction of the axr1-30 phenotype is readily apparent for AXR1 lines, but is not for AXL lines. The average rosette diameters for AXL line 1 and AXR1 lines 1 and 2 are 4.0 cm, 5.9 cm, and 4.0 cm, respectively, statistically larger than for axr1-30, which averages 3.4 cm in diameter. However, none are equivalent to Columbia, which averages 7.5 cm in diameter (Fig. 4a and Online Resources 9, 10). Of these three lines that are significantly larger than axr1-30, AXR1 line 1 is significantly larger than the other two (P < 0.0001), and AXL line 1 and AXR1 line 2 are indistinguishable (P = 0.7165), utilizing a Student’s t-test with Bonferroni correction and α = 0.00313. At four weeks, AXL line 1 is the only AXL line that is significantly larger than axr1-30, whereas both AXR1 lines are distinguishable from axr1-30.
Fig. 4.
AXR1 corrects axr1-30 adult phenotypic defects more than AXL1. a Rosette diameter measurements on 28-day old plants. Transgenic lines were grown for 28 days, then each was photographed and diameter was measured. Student’s t-tests with Bonferonni adjustment (α = 0.00313) were performed on log-transformed data combined from two replicates. Plant lines marked with an asterisk are significantly different from axr1-30. Error bars represent SE with minimally n = 29 measurements per line. b Inflorescence height measurements on 70-day old plants. Transgenic lines were grown for 70 days and height was measured. Student’s t-tests with Bonferonni adjustment (α = 0.00313) were performed on log-transformed data combined from two replicates. Plant lines marked with an asterisk are significantly different from axr1-30. Error bars represent SE with minimally n = 24 measurements per line
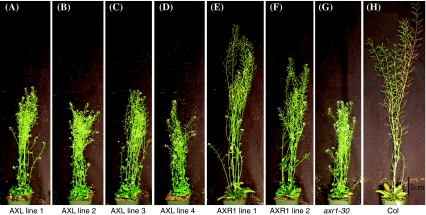
Finally, inflorescence height at ten weeks was measured for all lines. Here the difference in ability to restore the phenotype to Columbia between AXR1 and AXL1 expression is most apparent. Both AXR1 lines show good restoration of height, averaging 45.8 cm and 37.3 cm, compared to axr1-30 and Columbia, which average 25.2 cm and 52.1 cm, respectively (Figs. 4b, 5). Of the AXL lines, only line 1 that averages 29.3 cm in height shows a moderate correction of axr1-30 height. Statistical analyses confirm that AXL line 1, AXR1 line 1, and AXR1 line 2 are significantly taller than axr1-30, though none are the same height as Columbia (Figs. 4b, 5 and Online Resource 11). Both AXR1 lines (P < 0.0001) are significantly taller than AXL line 1, and AXR line 1 is taller than AXR1 line 2 (P < 0.0001), using a Student’s t-test with Bonferroni correction and α = 0.00313.
Fig. 5.
At day 70 post-plating, AXR1 corrects the inflorescence height defect of axr1-30 more than AXL1. AXR1 and AXL lines were grown for 70 days and height was measured. Representative pictures of AXL lines (a–d), AXR1 lines (e–f), axr1-30 (g), and Columbia (h) are shown. Scale bar represents 5 cm
In summary, we show that the in vivo functions of AXR1 and AXL1 are not equal. When AXL1 is expressed at a similar or higher level than AXR1, AXL1 does not have equivalent ability to correct axr1-30 phenotypic defects, suggesting that the two proteins differ in function at either the biochemical level, or with preference for downstream interacting partners.
Discussion
Here it is shown that AXL1 supports RUB1-ECR1 thioester formation and is unable to support UBQ conjugation to ECR1 (Fig. 1 and Online Resource 1), providing biochemical evidence for AXL1 as a RUB E1 subunit. The presence of AXL1 in RUB-containing complexes in vivo also supports its role in the RUB pathway (Table 1). Additionally, in vitro AXL1 and AXR1 have comparable abilities to support RUB1-ECR1C215S bond formation over time (Fig. 2). These data are consistent with the genetic data of Dharmasiri and colleagues (2007) wherein they suggest biochemical equivalence of AXR1 and AXL1 by showing that 35S:AXL1 can rescue a weak loss-of-function allele of AXR1, axr1-3.
Based on our in vitro biochemical data and the extensive complementation of axr1-3 by 35S:AXL1 (Dharmasiri et al. 2007), there was the initial expectation that the AXR1p:10MYC-AXL1 expression cassette should correct axr1-30 phenotypes to a similar degree as AXR1p:10MYC-AXR1. Contrary to this, the data presented in this paper suggest that AXL1 and AXR1 are not functionally equal, as AXR1p:10MYC-AXL1 largely fails to correct the strong axr1-30 mutant phenotype, whereas AXR1p:10MYC-AXR1 is readily able to correct axr1-30 phenotypic defects (Figs. 3, 4, 5). Whereas Dharmasiri and colleagues (2007) addressed the question of biochemical functionality by overexpression of 35S:AXL1 in the weak axr1-3 mutant, we examined the ability of AXL1 to function in place of AXR1, when expressed at levels comparable to AXR1, in an axr1 null background.
The differences in our results from those of Dharmasiri et al. (2007) are not implausible, considering the severity in phenotype of axr1-30 [null mutation due to T-DNA insertion (Online Resource 4)], as compared to axr1-3 [non-synonymous point mutation resulting in partial loss-of-function phenotype, as described in (Leyser et al. 1993)], and relative transgene expression under 35S and AXR1 promoters. One can hypothesize that the protein levels for the 35S:AXL1 construct exceeds a threshold level that allows for correction of the relatively mild axr1-3 phenotype, considering the comparable in vitro biochemical activities for the two proteins (Figs. 1, 2). In contrast, experiments presented here address protein functionality at lower levels of expression, in a null AXR1 background, where correction of axr1-30 phenotypic defects is only partial for both AXR1p:10MYC-AXL1 and AXR1p:10MYC-AXR1 constructs. Relative in vivo function is more comparable under such circumstances, and a difference in AXR1 and AXL1 is then apparent. Thus, AXL1 protein, expressed in similar location and levels to AXR1 protein, is not equivalent to AXR1 (Figs. 3, 4, 5).
Neither AXR1p:10MYC-AXL1 nor AXR1p:10MYC-AXR1 completely compensated for lack of AXR1 in axr1-30 plants. Two possible explanations for this observation are— (1) in selecting the intergenic region 5′ to the AXR1 locus as the promoter sequence, we left out essential enhancer elements, or (2) the N-terminal 10xMYC epitope interfered with in vivo biochemical functions of AXR1 and AXL1 in RUB E1 complexes. Alternative expression constructs could be used to examine these questions. To rule out the later possibility, one could express AXR1 and AXL1 without an epitope tag, raising antibodies specific to each endogenous protein. Alternative epitope tags, or their placement relative to the expressed protein sequence, could also address this question. However, in this study we were able to distinguish 10MYC-AXL1 function from 10MYC-AXR1 because all comparisons were made relative to the control, correction of axr1-30 phenotypic defects by 10MYC-AXR1.
To further the hypothesis that AXR1 and AXL1 function in normal RUB E1 complexes, an MS approach was taken to look at interacting partners with 3HA-RUB1. Both AXR1 and AXL1 are present in complexes with 3HA-RUB1 (Table 1), suggesting that they function concurrently in RUB E1 complexes. Additionally, peptides were recovered from RCE1 and RCE2, suggesting that both function as RUB E2s (Tables 1, 2).
Multiple angiosperms, including soybean (Glycine max), maize (Zea mays), and poplar (Populus trichocarpa) appear to have two AXR1 homologs, but the respective pairs are nearly identical in protein sequence, ranging from 92 to 96% identity among pairs, and distinguishable AXR1 or AXL1 homologs are not obvious. Others, like Medicago (Medicago trunculata), rice (Oryza sativa), and sorghum (Sorghum bicolor), only have one AXR1 homolog. Arabidopsis lyrata appears to have a similar genome structure to Arabidopsis thaliana with discernable AXL1 and AXR1 gene homologs (www.phytozome.net). Thus, the occurrence of AXL1 may be rather limited in plant genomes.
Why two functional RUB E1s exist remains a question, but as seen with the UBQ E1s, UBE1 and UBA6, or the NEDD8 E2s, UBE2M and UBE2F, the purpose of two Arabidopsis RUB E1s could be to preferentially interact with different downstream proteins (Jin et al. 2007; Huang et al. 2009). Alternatively, subtle differences in enzyme activity, not detected by our methods of analysis, could exist and AXL1 could function in RUB E1 complexes during certain developmental stages or cellular conditions, as also observed for UBE2M and UBE2F (Huang et al. 2009). Identification of additional downstream RUB substrates might help elucidate functional differences between the two RUB E1 subunits.
Materials and methods
Thioester reactions
Arabidopsis ECR1 (At5g19180.1) coding sequence [described in (del Pozo and Estelle 1999)] was used to create pDEST15-ECR1 (Gateway, Invitrogen, www.invitrogen.com) for production and purification of recombinant GST-ECR1 using glutathione-Sepharose (GE Healthcare, www.gehealthcare.com). Arabidopsis AXL1 (At2g32410.1) coding sequence was isolated from mRNA, recombined into pDONR201 (Invitrogen), sequence verified (to TAIR v7), expressed as 6HIS-AXL1 from the pDEST17 (Invitrogen) vector and used from a bacterial extract. A bacterial extract expressing recombinant 6HIS-AXR1 (derived from At1g05180.1) from pQE30-AXR1 was used (del Pozo et al. 1998). 3HA-RUB1 (derived from At1g31340.1) coding sequence [described in (Bostick et al. 2004)] was used to create pDEST17-3HA-RUB1 for production and purification of 6HIS-3HA-RUB1 using Ni–NTA-Sepharose (GE Healthcare). Recombinant 6HIS-HA-ubiquitin was expressed from pDEST17-HA-UBQ, derived from HA-UBQ [described in (Nishikawa et al. 2004)]. Recombinant ubiquitin E1 (from yeast) was cloned into pTYB2 (NEB, www.neb.com), then expressed and purified using the IMPACT-CN (NEB) system.
For thioester reactions, proteins were incubated in 2 mM ATP, 0.1 mM DTT, 10 mM MgCl2, and 100 mM TRIS (pH 7.5) buffer at 37°C for 5 min. Reactions were then split in half and stopped in the presence or absence of DTT (5.3% SDS, 13.3% glycerol, 5.3 M urea, ±133 mM DTT) for 15 min at RT. The proteins were separated on a non-reducing 8% tris–glycine gel at 4°C, transferred to Immobilon-P membrane (Millipore, www.Millipore.com), and analyzed by anti-HA (monoclonal rat anti-HA-HRP; Roche, www.roche.com) and anti-GST (polyclonal rabbit anti GST(Z-5); Santa Cruz Biotech., www.scbt.com) immunoblot analysis.
Oxyester reactions
Vectors used in this assay include: pDEST17-AXR1, pDEST17-AXL1, pDEST17-FLAG-ECR1C215S, pDEST17-FLAG-ECR1C215A, and pDEST15-HARUB1. All vectors were cloned using Gateway cloning technology; sources for original sequences are as described above. In brief, all coding sequences were first cloned into pDONR201 and sequenced, then moved into appropriate expression vectors. pDEST17-FLAG has been modified to have a FLAG sequence 3′ to 6HIS and 5′ to att1 sequences. All proteins were induced in BL21(DE3)pLysS cells and the soluble protein fraction was recovered by centrifugation after cells were lysed by sonication. Anti-HIS (monoclonal mouse anti-HIS; GE Healthcare) immunoblot analysis and/or Coomassie blue staining were used to establish relative protein concentrations.
For oxyester assays, proteins were incubated in 2 mM ATP, 0.1 mM DTT, 10 mM MgCl2, and 100 mM TRIS (pH 7.5) buffer at 30°C for the duration of the time course (1 h, 2 h, 4 h). At each time point, 10 μl of sample were removed from a 25 μl reaction for base and neutral conditions and mixed with 10 μl EZ-view anti-HA-agarose beads (Sigma–Aldrich, www.sigmaaldrich.com) and 90 μl of 50 mM TRIS (pH 7.5). Immunoprecipitation proceeded for 45 min, then beads were washed 3 × 5 min in 50 mM TRIS (pH 7.5). Finally, beads were mixed with 10 μl of 50 mM TRIS (pH 7.5) and were subjected to base or neutral conditions. For base conditions, 20 μl TRIS/beads were incubated with 10 μl 0.4 M NaOH for 20 min at 30°C, then 25 μl of 5× loading buffer [125 mM TRIS (pH 6.8), 20% glycerol, 5% SDS, 5% BME] were added, followed by 1 μl concentrated HCl (to acidify the reaction). Reactions were then titrated back to neutral pH with 1 M NaOH. For neutral conditions, 20 μl TRIS/beads were mixed with 25 μl 5× loading buffer and water was added to make volume equivalent with base treatment. Samples were then boiled for 5 min and run on reducing 8% tris–glycine gels at RT, transferred to Immobilon-P membrane, and analyzed by anti-FLAG-HRP (monoclonal mouse anti-FLAG M2-HRP; Sigma–Aldrich). Loading was checked by analysis of inputs (fraction of total for each sample), which were not subjected to immunoprecipitation, using anti-HA-HRP (monoclonal rat anti-HA-HRP; Roche) and anti-HIS (monoclonal mouse anti-HIS; GE Healthcare) immunoblot analysis. Time course experiments were done in quadruplicate.
Immunoblots were scanned using a flatbed scanner at 600 dpi, with no auto-correction. Blots were then analyzed in ImageJ (version 1.43u, National Institutes of Health, http://rsb.info.nih.gov/ij), using the Gel Analyzer program. In brief, all bands on a blot were boxed in uniform rectangles, then intensity peaks were plotted, and the absolute intensity of each band was calculated as the area under each peak. These values were then plotted against time and analyzed using the JMP (version 8.0.1, SAS Institute Inc., http://www.jmp.com) statistical package.
Mass spectrometry of 3HA-RUB1-interacting proteins
In preparation for MS analysis, a minimum of 0.25 g seeds dexamethasone (DEX)-inducible 6xUAS gal4 :3HA-RUB1 in Columbia [experimental line; described in (Bostick et al. 2004)] and Columbia (control) were bleach-sterilized and then cold-treated minimally overnight before plating. For each line, seeds were evenly distributed over 4-section culture plates and 1 ml of germination media (GM; 1× MS salts, 1% sucrose, 1× B-vitamins, 0.05% MES) was added to each quadrant. Plates were then grown under continuous light for 7 days. Plates were then removed from lights, excess GM was removed and 1 ml of fresh GM +30 μM DEX was added. Plates were returned to lights and seedlings were DEX-treated overnight, before seedlings were collected and flash-frozen in liquid N2.
Protein was extracted in aqueous buffer [50 mM TRIS (pH 8), 150 mM NaCl, 20 mM EDTA, 10% glycerol, 0.15% NP-40, 1 mM PMSF, 0.5 mM 1,10-phenanthroline, 1× protease inhibitors (Roche)] with 1.5% PVPP (w/w) by grinding with mortar and pestle in liquid N2. Samples were then clarified at 4°C by centrifugation at 16,060g and 17,369g. Between each centrifugation step, the soluble fractions were moved to new collection tubes. We then proceeded with analysis of the soluble fraction and determined protein concentration by Bradford assay. Samples were brought to equal concentration, and anti-HA immunoprecipitation proceeded overnight at 4°C from minimally 65 mg soluble protein with minimally 0.25 ml equilibrated EZ-view anti-HA-agarose beads (Sigma–Aldrich). Anti-HA-agarose beads were then washed with 450× bead volume of various buffers [60× bead volume buffer 1 (50 mM TRIS (pH 8), 150 mM NaCl, 0.15% NP-40, 0.5× protease inhibitors); 60× bead volume buffer 2 (50 mM MES (pH 6), 150 mM NaCl, 0.5× protease inhibitors); 150× bead volume buffer 3 (50 mM TRIS (pH 8), 150 mM NaCl); 180 × bead volume buffer 4 (100 mM ammonium bicarbonate (pH 8)]. Then (next day) samples were trypsin digested, using minimally 1.6 μg Trypsin (Promega, www.promega.com), for 6–8 h at 37°C. We recovered minimally 5 μg digested peptides per sample by A280 measurement.
The peptides from approximately 50% of each sample were separated by reverse phase chromatography using a Waters nanoACQUITY UPLC system (www.waters.com) and a Waters BEH C18 1.7 μm, 100 μm × 10 cm column, using a gradient of 1–80% acetonitrile with 0.1% formic acid maintained throughout the gradient. Peptides were directly eluted into a ThermoElectron LTQ-FT mass spectrometer (www.thermo.com) with a nano-electrospray ionization source at a flow rate of 1 μl min−1 for detection of the intact peptides’ mass to charge (m/z) ratios. Data-dependent software parameters allowed the top four most abundant ions to be selected with a 30 s exclusion list time and repeat count of 2. Singly-charged ions were disallowed for collision-induced dissociation.
Tandem mass spectra were extracted with ReADW (version 4.0.2, Seattle Proteome Center, www.proteomecenter.org), with centroiding (-c) enabled. All MS/MS samples were analyzed using X! Tandem [version TORNADO (2010.01.01.4)] from the Global Proteome Machine (www.thegpm.org). X! Tandem was set up to search the Uniprot Arabidopsis thaliana complete proteome database (downloaded from www.uniprot.org March 29, 2010, 31672 entries), along with common contaminants (cRAP database downloaded from www.thegpm.org April 30, 2010, 101 entries), assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.40 Da and a precursor ion tolerance of 10.0 p.p.m. Variable oxidation of methionine, oxidation of tryptophan to formylkynurenin, acetylation of the n-terminus, and diglycine modification of lysine (+114.042927 Da at K, rubbylation/ubiquitylation footprint) were specified as variable modifications in X! Tandem. One missed cleavage was allowed.
Scaffold (version Scaffold_2_06_02, Proteome Software Inc., www.proteomesoftware.com) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm (Keller et al. 2002). Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least 2 identified unique peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al. 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Production of axr1-30 complementation lines
The intergenic region (called “AXR1 promoter”) 5′ to the ATG start codon of AXR1 (At1g05180.1) was amplified from genomic DNA by PCR that added HindIII and XbaI restriction sites to 5′ and 3′ ends, respectively. This sequence was ligated into pGWB21 (Nakagawa et al. 2007), replacing its 35S CaMV promoter; the construct was designated pGWB21-AXR1p. The vector was sequenced to confirm sequence and orientation of the inserted promoter. The pGWB21 vector confers an N-terminal 10xMYC epitope and is compatible with Gateway cloning technology (Nakagawa et al. 2007). AXR1 (At1g05180.1) and AXL1 (At2g32410.1) coding sequences were first amplified by PCR and moved into pDONR201, as described above, before being moved into pGWB21-AXR1p, using Gateway cloning technology. The cloned constructs were designated AXR1p:10MYC-AXR1 and AXR1p:10MYC-AXL1.
AXR1p:10MYC-AXR1 and AXR1p:10MYC-AXL1 constructs, which carry genes for resistance to kanamycin and hygromycin, were introduced into plants heterozygous for AXR1/axr1-30 (carrying a gene for glufosinate resistance), using the floral dip method with Agrobacterium strain AGL1 (Clough and Bent 1998). In this experiment, we utilized axr1-30 mutants in preference to other severe axr1 alleles because axr1-30 carries a selectable marker and is thus readily followed when screening for complementation. T1 transformants were selected on kanamycin plates and then sprayed with Finale (1% glufosinate-ammonium; Bayer CropScience, www.bayercropscience.com) to eliminate plants with the AXR1/AXR1 genotype. T2 seedlings were then analyzed to recover lines that carried a single AXR1p:10MYC-AXR1 or AXR1p:10MYC-AXL1 transgene. Individuals homozygous for axr1-30/axr1-30 and the respective transgene were recovered in T3 and later generations.
Starting at the T2 generation, plant lines were screened for complementation of axr1-30 phenotypic defects in adult plants; 10MYC-AXR1 and 10MYC-AXL1 protein levels were also evaluated. Nine AXL lines were recovered where 10MYC-AXL1 protein accumulated to levels equivalent to or higher than 10MYC-AXR1 protein; however complementation of axr1-30 phenotypic defects was not obvious in these lines. Though 10MYC-AXL1 largely failed to correct axr1-30 phenotypic defects in adult plants, screening for additional AXL lines was not pursued as 10MYC-AXL1 protein was readily expressed. Five AXR1 lines were recovered where 10MYC-AXR1 protein was visible and complementation of axr1-30 phenotypic defects was readily apparent.
Phenotypic analyses of axr1-30 complementation lines
For seedling analyses, seeds were surface-sterilized with 30% bleach and cold-treated for a minimum of 24 h, then plated on GM (0.8% agar added). 2,4-D (50 μM stock) was added to plates, prior to solidification, to a final concentration of 0.025 μM or 0.05 μM with solvent (i.e. ethanol) made equivalent across all plates. Based on preliminary experiments, we found these concentrations of 2,4-D result in a linear decrease in length for Columbia and little change in axr1-30 root length. Seedlings were grown for 9 days, with germination (radicle emergence noted on dissecting scope) marked on day 2, in continuous light (average = 43 μmol s−1 m−2) at 20°C. Seedlings were then removed from GM plates and roots were photographed. Root length was then determined using the segmented line tool in ImageJ, for minimally n = 41 individuals per line per treatment. Three experimental replicates were combined and statistical analyses were done using the JMP statistical package with a log-transformation applied to correct for heterogeneity of variance, as needed.
For analyses of adult plants, seeds were surface-sterilized with 30% bleach and cold-treated for a minimum of 48 h. Seeds were then plated on GM and grown for 7 days in continuous light (average = 46 μmol s−1 m−2) at 22°C, before transplanting seedlings to soil and continued growth in 16 h light/8 h dark (average = 116 μmol s−1 m−2) at 18°C and minimally 50% humidity. Plants were photographed for rosette diameter measurements at 28 days post-plating. Diameter measurements were then made in ImageJ, using the ellipse function to draw the smallest circle that would encompass the whole rosette, for minimally n = 29 individuals per line. The circle’s diameter was then taken as a measure of the rosette’s broadest diameter. Measurements of inflorescence height were made 70 days post-plating. The longest measurement from rosette base to inflorescence tip was used as a measure of height. Measurements were made with a ruler, for minimally n = 24 individuals per line. Plants representative of average height were also photographed. Two experimental replicates were combined and statistical analyses were done using the JMP statistical package with a log-transformation applied to correct for heterogeneity of variance, as needed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors gratefully acknowledge the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (contracts DE-FG02-03ER15416 and DE-FG02-09ER16077). In addition, the work was supported in part by a graduate research fellowship from the National Science Foundation to S. Hotton (NSF-GRFP) and the Paul K. and Ruth R. Stumpf Endowed Professorship in Plant Biochemistry to J. Callis. We wish to acknowledge the UC-Davis Controlled Environment Facility (CEF) for assistance in the propagation of transgenic plants, Syngenta for the axr1-30 T-DNA insertion line (SAIL_904_E06), M. Estelle for pQ30-AXR1 and ECR1 cDNA sequences, and T. Ohta for pHA-UBQ sequences. In addition, we thank S. Lochhead for technical assistance, the Callis lab undergraduate students for general assistance and care of Arabidopsis plants, and members of the J. Callis lab for critical comments and helpful discussions in preparation of this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Biswas KK, Ooura C, Higuchi K, Miyazaki Y, Van Nguyen V, Rahman A, Uchimiya H, Kiyosue T, Koshiba T, Tanaka A, Narumi I, Oono Y. Genetic characterization of mutants resistant to the antiauxin p-chlorophenoxyisobutyric acid reveals that AAR3, a gene encoding a DCN1-like protein, regulates responses to the synthetic auxin 2, 4-dichlorophenoxyacetic acid in Arabidopsis roots. Plant Physiol. 2007;145:773–785. doi: 10.1104/pp.107.104844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostick M, Lochhead SR, Honda A, Palmer S, Callis J. Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell. 2004;16:2418–2432. doi: 10.1105/tpc.104.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Sun Q, Chen ZJ. E1–L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27:1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsisthaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Estelle M. The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc Natl Acad Sci USA. 1999;96:15342–15347. doi: 10.1073/pnas.96.26.15342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M. The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science. 1998;280:1760–1763. doi: 10.1126/science.280.5370.1760. [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Dharmasiri S, Hellmann H, Walker L, Gray WM, Estelle M. AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell. 2002;14:421–433. doi: 10.1105/tpc.010282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri S, Dharmasiri N, Hellmann H, Estelle M. The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J. 2003;22:1762–1770. doi: 10.1093/emboj/cdg190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Weijers D, Karunarathna N, Jurgens G, Estelle M. AXL and AXR1 have redundant functions in RUB conjugation and growth and development in Arabidopsis. Plant J. 2007;52:114–123. doi: 10.1111/j.1365-313X.2007.03211.x. [DOI] [PubMed] [Google Scholar]
- Downes B, Vierstra RD. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans. 2005;33:393–399. doi: 10.1042/BST0330393. [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Gilkerson J, Hu J, Brown J, Jones A, Sun TP, Callis J. Isolation and characterization of cul1–7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics. 2009;181:945–963. doi: 10.1534/genetics.108.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotton SK, Callis J. Regulation of cullin RING ligases. Annu Rev Plant Biol. 2008;59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, Schulman BA. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kim AY, Bommelje CC, Lee BE, Yonekawa Y, Choi L, Morris LG, Huang G, Kaufman A, Ryan RJ, Hao B, Ramanathan Y, Singh B. SCCRO (DCUN1D1) is an essential component of the E3 complex for neddylation. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Lechner E, Xie D, Grava S, Pigaglio E, Planchais S, Murray JA, Parmentier Y, Mutterer J, Dubreucq B, Shen WH, Genschik P. The AtRbx1 protein is part of plant SCF complexes, and its down-regulation causes severe growth and developmental defects. J Biol Chem. 2002;277:50069–50080. doi: 10.1074/jbc.M204254200. [DOI] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Britton JH, Estelle M. Growth and development of the axr1 mutants of Arabidopsis. Plant Cell. 1990;2:1071–1080. doi: 10.1105/tpc.2.11.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Schaller N, Chou YC, Sumara I, Martin DD, Kurz T, Katheder N, Hofmann K, Berthiaume LG, Sicheri F, Peter M. The human Dcn1-like protein DCNL3 promotes Cul3 neddylation at membranes. Proc Natl Acad Sci USA. 2009;106:12365–12370. doi: 10.1073/pnas.0812528106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8–E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301:392–398. doi: 10.1016/S0006-291X(02)03051-6. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Freeling M, Tang H, Wang X. Insights from the comparison of plant genome sequences. Annu Rev Plant Biol. 2010;61:349–372. doi: 10.1146/annurev-arplant-042809-112235. [DOI] [PubMed] [Google Scholar]
- Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, Scheffner M, Schmidtke G, Groettrup M. UBE1L2, a novel E1 enzyme specific for ubiquitin. J Biol Chem. 2007;282:23010–23014. doi: 10.1074/jbc.C700111200. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Rao-Naik C, delaCruz W, Laplaza JM, Tan S, Callis J, Fisher AJ. The rub family of ubiquitin-like proteins. Crystal structure of Arabidopsis RUB1 and expression of multiple RUBs in Arabidopsis. J Biol Chem. 1998;273:34976–34982. doi: 10.1074/jbc.273.52.34976. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Wada H, Yeh ET, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257:100–105. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12:1427–1437. doi: 10.1016/S1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Schulman BA. Insights into the ubiquitin transfer cascade from the structure of the activating enzyme for NEDD8. Nature. 2003;422:330–334. doi: 10.1038/nature01456. [DOI] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- Woodward AW, Ratzel SE, Woodward EE, Shamoo Y, Bartel B. Mutation of E1-CONJUGATING ENZYME-RELATED1 decreases RELATED TO UBIQUITIN conjugation and alters auxin response and development. Plant Physiol. 2007;144:976–987. doi: 10.1104/pp.107.100404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.