Abstract
The phytohormone abscisic acid (ABA) and reactive oxygen species (ROS) play critical roles in mediating abiotic stress responses in plants. It is well known that ABA is involved in the modulation of ROS levels by regulating ROS-producing and ROS-scavenging genes, but the molecular mechanisms underlying this regulation are poorly understood. Here we show that the expression of maize ABP9 gene, which encodes a bZIP transcription factor capable of binding to the ABRE2 motif in the maize Cat1 promoter, is induced by ABA, H2O2, drought and salt. Constitutive expression of ABP9 in transgenic Arabidopsis leads to remarkably enhanced tolerance to multiple stresses including drought, high salt, freezing temperature and oxidative stresses. ABP9 expressing Arabidopsis plants also exhibit increased sensitivity to exogenously applied ABA during seed germination, root growth and stomatal closure and improved water-conserving capacity. Moreover, constitutive expression of ABP9 causes reduced cellular levels of ROS, alleviated oxidative damage and reduced cell death, accompanied by elevated expression of many stress/ABA responsive genes including those for scavenging and regulating ROS. Taken together, these results suggest that ABP9 may play a pivotal role in plant tolerance to abiotic stresses by fine tuning ABA signaling and control of ROS accumulation.
Electronic supplementary material
The online version of this article (doi:10.1007/s11103-011-9732-x) contains supplementary material, which is available to authorized users.
Keywords: Transcription factor ABP9, ABA, Reactive oxygen species, Stress tolerance, Gene expression
Introduction
Plants are frequently exposed to a variety of abiotic stresses such as drought, salt, and extremes of temperature, which constrain plant growth and crop productivity worldwide. Understanding how plants tolerate these stresses is a vital prerequisite for developing strategies to improving plant stress tolerance. Plants sense and adapt to dynamic stresses by altering their genome expression profiling, growth pattern and cellular metabolism and mobilizing various defense mechanisms (Bartels and Sunkar 2005). A common metabolic change provoked by stresses is the accelerated generation/accumulation of reactive oxygen species (ROS) in various cellular compartments (Mittler 2002). While excess amount of ROS can cause oxidative damage to cells during abiotic stresses, recent studies have shown that ROS at low concentrations function as signaling molecules to regulate plant protective stress responses including ABA-induced activation of Ca2+ channel and stomatal closure and induction of defense gene expression (Desikan et al. 2001; Pei et al. 2000; Rizhsky et al. 2002; Zhang et al. 2001). The dual roles of ROS imply the necessity to tightly control the cellular levels of ROS. Plants possess a ROS gene network encoding components of both ROS-producing and ROS-scavenging pathways to secure an appropriate ROS level and signaling while minimizing their toxic effects during environmental stresses (Apel and Hirt 2004; Mittler et al. 2004). Elucidating the molecular mechanisms that regulate ROS homeostasis and signaling in response to abiotic stress could therefore provide useful insights for generating crop plants with improved tolerance to various environmental stress conditions.
The phytohormone abscisic acid (ABA) is critical for plant growth and development and plays a pivotal role in controlling stress responses through regulating a set of ABA/stress-responsive gene expression including ROS network genes such as superoxide dismutase (SOD), ascorbate peroxidase (APX) and catalase (CAT) (Finkelstein et al. 2002; Jiang and Zhang 2002; Yamaguchi-Shinozaki and Shinozaki 2006; Zhu and Scandalios 1994). Functional dissection of ABA-responsive gene promoters has identified several types of ABA-responsive cis-elements (ABREs) among which the G-box like sequence (C/T)ACGTGGC functions as a strong one (Busk and Pagès 1998). Several ABRE-binding transcription factors such as ABF/AREB/ABI5 have been identified and shown to play important roles in modulating ABA and stress responses (Finkelstein and Lynch 2000; Kim 2006; Uno et al. 2000). However, it is still largely unclear about how ABA signaling regulates the cellular levels of ROS in response to various abiotic stresses.
In maize, CAT genes encode three biochemically distinct isozymes including CAT1, CAT2 and CAT3, which catalyze the decomposition of H2O2 and thus act as a group of major enzymatic scavengers of ROS. It has been shown that a 9-bp G-box-containing sequence (5′-CCACGTGGA-3′) in the maize Cat1 promoter, designated ABRE2, is responsible for ABA-induced Cat1 expression (Guan and Scandalios 1998; Guan et al. 2000). In our previous work, we isolated an ABRE2-binding factor, named ABP9 (for ABRE Binding Protein 9) by using ABRE2 of maize Cat1 promoter as a bait in yeast one-hybrid screening of a maize cDNA library. We showed that ABP9 encodes a transcription factor of bZIP family capable of trans-activating the expression of a downstream reporter gene by specific binding to the ABRE2 motif in yeast cells (Wang et al. 2002). Further, we showed that ABP9 expression improves the photosynthetic capacity of transgenic Arabidopsis plants under stress conditions (Zhang et al. 2008). In this study, we show that ABP9 overexpressing transgenic Arabidopsis plants exhibit enhanced tolerance to various abiotic stresses, including drought, salt, freeze temperature and oxidative stress. Our results suggest that ABP9 plays a critical role in regulating ABA responses and modulation of the cellular levels of ROS.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants were grown aseptically in a growth chamber (22°C, ~120 μmol photos m−2 s−1 under a 16-h light/8-h darkness photocycle). Seeds were surface sterilized in 75% ethanol containing 0.05% (v/v) Tween-20 for 10 min followed by three times rinse with 95% ethanol and then plated on half strength Murashige-Skoog (1/2 MS) medium (Sigma–Adrich) (Murashige and Skoog 1962) solidified with 0.8% agar. 1 or 2-week-old seedlings were transferred from plates into pots filled with compost soil. Unless stated otherwise, the 1/2 MS medium was supplemented with 1% sucrose.
Maize (Zea mays L.) inbred line Qi319 was used in this work. Before sowing, the seeds were imbibed and incubated on moisture filter paper at 28°C in the dark for 48 h, then were sown in compost soil and grown in a growth chamber (28°C, 12-h light/12-h darkness photocycle).
Electrophoretic mobility shift assays
To prepare ABP9 recombinant protein, the full-length cDNA of ABP9 (Genebank assession No. GU237073) was PCR amplified using anchor primers from pPC86-ABP9 (Wang et al. 2002). The PCR fragment with XbaI and XhoI restriction sites at 5′ and 3′ ends, respectively, was cloned into pGEM®-T Easy (Promega) vector resulting in pGEM®-T Easy-ABP9. Then the full-length cDNA of ABP9 was released by XbaI digestion of pGEM®-T Easy-ABP9 and blunt-ended with Klenow enzyme followed by NotI digestion, and then cloned in frame into SmaI/NotI-digested prokaryote expression vector pGEX-4T-1 (GE Healthcare) to generate the construct pGEX-4T-1-ABP9. pGEX-4T-1-ABP9 was transformed into the Escherichia coli (E.coli) stain BL21 cells to produce a GST fusion protein. The GST-ABP9 fusion protein was purified using MicroSpin® GST purification module (GE Healthcare). The 19 bp oligos containing ABRE2 (5′-GAAGTCCACGTGGAGGTGG-3′) or its mutant form mABRE2 (5′-GAAGTaacatgttcGGTGG-3′) was annealed and labeled with [γ-32P]-ATP using DNA 5′ End-Labeling system (Promega). Binding reaction containing 4 μg purified ABP9 protein, labeled probe DNA (20,000–50,000 cpm), 25 mM Hepes–KOH (pH 8.0), 50 mM KCl, 10% glycerol, and 1 mM dithiothreitol in 20 μl volume was incubated on ice for 15 min. When performing a competition assay, unlabeled probes were added 30 min prior to the addition of the 32P-labeled probes. Binding reactions were separated on 5% non-denaturing PAGE gels in 0.5× Tris–borate–EDTA buffer. After electrophoresis, gels were dried and visualized by autoradiography.
Trans-activation assay in maize cells
To prepare the effector plasmid pAHC25-ABP9, the full-length cDNA of ABP9 was isolated from pGEM®-T Easy-ABP9 by XbaI/XhoI double digestion, followed by blunt-ending with Klenow enzyme, then cloned into the SmaI/Ecl136II sites of the vector pAHC25 to replace the GUS-coding region. To construct the reporter plasmid pIGA, pIG46 (Ono et al. 1996), which consists of the CaMV 35S minimal promoter and GUS gene, digested with XhoI and filled-in with Klenow, was ligated with 4 copies of ABRE2. Effector plasmid pAHC25-ABP9 and reporter plasmid pIGA were co-transformed into maize Black Mexican Sweet (BMS) suspension cells by biolistic method. Briefly, the cells maintained in 50 ml liquid MS medium (Sigma–Adrich) supplemented with 2.0 mg l−1 2,4-D and 3% sucrose were subcultured into fresh culture and treated with 3% PEG 8000 at 26–28°C in the dark on the shaker of 150 rpm for overnight before bombardment. The plasmids were introduced into osmotic-treated cells on the glass microfibre filters (Whatman, GF/A) using 1100 psi rupture pressure. After bombardment, the glass microfibre filters with the bombarded cells were transferred to MS agar medium and cultured at 26–28°C in the dark for 2 days. In situ assay of GUS activity was determined as described (Jefferson et al. 1987).
Generation of Arabidopsis transgenic plants
The full-length cDNA of ABP9 was isolated by XhoI-digestion of pGEM®-T Easy-ABP9 and blunt-ended by the Klenow enzyme, followed by XbaI digestion. The cDNA fragment was then cloned into XbaI and Ecl136II double digested pBI121 vector to generate the pBI121-ABP9 plasmid, in which ABP9 is under the control of the cauliflower mosaic virus 35S promoter (CaMV 35S). pBI121-ABP9 was introduced into the Agrobacterium tumefaciens strain GV3101 through electroporation and used to transform Arabidopsis thaliana (ecotype Columbia) using a floral dip method (Bechtold et al. 1993). Transgenic plants were selected by kanamycine resistance and confirmed by PCR analysis. Two representative T3 homozygous lines (5P2 and 5P3) with different expression level of ABP9 were chosen for detailed analyses.
Stress tolerance assays
One-week-old young seedlings aseptically grown on 1/2 MS agar medium were transferred into pots filled with the same amount of compost soil and grown for another 1 or 2 weeks before stress treatments were applied. For drought tolerance assay, soil-grown plants were fully watered, and then withheld irrigation for 4 weeks, followed by rewatering plants. Survival rates were scored 1 week after rewatering. For salt tolerance assay, soil-grown plants were treated with progressively applied high salt stress by irrigating plants with NaCl solutions of stepwisely increasing concentrations (50, 100, and 200 mM) every 4 days and lasting at the concentration of 200 mM for 12 days when chlorophyll contents were measured according to the method (Lichtenthaler 1987). Cold treatment was performed by transferring the plants into a low temperature incubator for desired durations (−4°C, Sanyo, MIR-253). After 7 days-recovery under normal growth conditions, the survival rates were recorded. For oxidative stress treatment, leaves of similar developmental stages (7th and 8th rosette leaves) were detached from 3 or 4 week-old plants aseptically grown on 1/2 MS agar medium and floated abaxial side up in 2 μM methyl viologen (MV, Sigma–Adrich) solution under controlled growth conditions. Chlorophyll contents of the detached leaves were measured as described (Lichtenthaler 1987).
ABA response and water loss assays
For germination assay, seeds were surface-sterilized and sown on MS medium (3% sucrose and 0.8% agar, pH 5.7) supplemented with or without different concentrations of ABA (Sigma–Aldrich). Seeds were stratified at 4°C for 3 days before growing at 22°C under light conditions. To score seed germination, the percentages of seeds with fully emerged radicles were determined in three independent experiments.
For root growth assay, seeds were germinated after stratification and grown for 48 h on ABA-free MS medium, seedlings were then transferred to MS medium supplemented with different concentrations of ABA and grown vertically. The primary root lengths were measured 10 days after the transfer. The root growth ratio refers to the root length on ABA medium normalized to that on ABA-free medium.
Stomatal aperture assays were performed essentially as previously described (Pei et al. 1997). Briefly, detached rosette leaves were first floated in a buffer containing 20 mM KCl, 5 mM Mes-KOH, pH 6.15, and 1 mM CaCl2 at 150 μmol photos m−2 s−1 for 2 h followed by addition of 10 μM ABA. 2 h later, apertures were recorded on epidermal strips using a digital video camera (AxioCam HR) attached to an inverted microscope (Leica DMR) equipped with image analysis software (Axio Vision 4.0).
For water loss assay, detached rosette leaves of the same developmental stage were placed abaxial side up in open Petri dishes and left on the lab bench. The fresh weight was monitored at the indicated time points.
Detection of ROS and cell death
A histochemical staining procedure was used to detect superoxide and hydrogen peroxide in situ as described (Fryer et al. 2002). Rosette leaves sampled from seedlings with or without ABA or NaCl treatment were infiltrated with nitroblue tetrazolium (NBT) (Amresco, 1 mg ml−1 NBT in 10 mM sodium azide and 10 mM phosphate buffer, pH 7.8) or 3, 3′-diaminobenzidine (DAB) solutions (Sigma–Aldrich, 1 mg ml−1 DAB-HCl, pH 3.8), respectively. The stained leaves were imaged after removal of chlorophylls by boiling in 96% ethanol for 10 min. Cell death was examined by Evans Blue staining as described (Kim et al. 2003) with minor modifications. Briefly, detached leaves from plants untreated or treated with 50 mM NaCl for 1 h were vacuum-infiltrated in 0.1% Evans Blue (w/v; Sigma–Aldrich) for 15 min and then kept under vacuum for 6 h, then washed three times (15 min each) with a phosphate-buffered saline containing 0.05% (v/v) Tween 20 followed by boiling in 96% ethanol for 10 min to remove chlorophylls.
ROS production in intact guard cells was detected using 2, 7-dichlorofluorescin diacetate (H2DCF-DA, Fluka) as described (Pei et al. 2000). Epidermal peels were floated in an incubation buffer (30 mM KCl, 10 mM Mes-KOH, pH 6.15) for 2 h under the light of 150 μmol photos m−2 s−1 at 22°C to induce stomatal opening and then loaded with 50 μM H2DCF-DA. After 10 min of dye loading, the epidermal strips were washed three times with the incubation buffer. For ABA treatment, 10 μM ABA was added to the incubation buffer and the epidermal strips were further incubated for 2 h before dye loading. Guard cells were visualized by confocal laser microscope (Leica TCS SP2) with excitation at 488 nm and emission at 525 nm and images were acquired using the Leica Confocal Software version 2.6 Build 1537. The DCF fluorescence emission of guard cells was quantified using the Leica Qwin software.
Determination of cellular levels of ROS by Electron Spin Resonance (ESR) spectroscopy was performed essentially according to the protocol as described (Capani et al. 2001) with minor modifications. Briefly, plants of the same developmental stage grown aseptically on 1/2 MS agar medium were collected, weighed and homogenized on ice in 600 μl ice-cold spin trap solution containing 100 mM α-phenyl-N-tert-butylnitron (PBN, Sigma–Adrich) and 2 mM diethylenetriamine-pentacetic acid (DPTA) in phosphate-buffered saline (PBS, pH 7.4). After homogenization, 250 μl ethyl acetate were added, vortexed for 30 s and centrifuged at 12,000 rpm for 15 min at 4°C for complete phase separation. Then the ethyl acetate phase was transferred to a clean tube for ESR measurement. The Brucker 200D SRC ESR spectrometer was set as followings: center field, 3385 Gauss; scan width, 400 Gauss; modulation amplitude, 3.2 G; microwave power, 20 mW; receiver gain, 4.0 × 105.
RNA analysis
Reverse transcriptase-coupled PCR (RT-PCR) analysis was performed to analyze ABP9 expression in Maize. For dosage analysis of NaCl, H2O2 and ABA effects, roots of soil-grown maize seedlings with three fully expanded leaves were submerged after the removal of soil into the NaCl, H2O2 and ABA solutions of different concentrations for 3 days and 24 h, respectively. While for time course analysis, plants were put into NaCl, H2O2 and ABA solutions of given concentrations for 12 h and sampled at the desired time points. Drought treatment was performed by transferring plants to pots with different soil water contents (SWC) for 3 days. Pots were covered with saran wrap and weighed daily to maintain the SWC of 13% (0.13 g H2O g−1 dry soil), 10% (0.10 g H2O g−1 dry soil) and 8% (0.08 g H2O g−1 dry soil). For dehydration treatment, plants after the removal of soil were kept on tissue paper and left on bench for 6 h. Plants grown on soil without treatment were used as control 1 (CK1), and plants treated with distilled water were used as control 2 (CK2). Total RNAs were isolated from samples treated with different stresses using RNAgents® Total RNA Isolation System (Promega). RNA samples were reverse transcribed with SuperScriptTM II Reverse Transcriptase (Invitrogen). PCR was conducted at linearity phase of the exponential reactions with ABP9-specific primer pairs as follows: forward 5′-catgacgctggaggacttcct -3′ and reverse 5′-ttgacgaaaacacagagc-3′. Transcripts of maize actin1 (GenBank accession No. J01238) amplified with specific primers (forward 5′-caccttctacaacgagctccg-3′ and reverse 5′-taatcaagggcaacgtaggca-3′) were used as the internal control.
RNA gel blotting was performed to analyze the expression of ABP9 in transgenic Arabidopsis plants. Total RNA was isolated from 3-week-old seedlings untreated or treated with 200 mM NaCl for 1 h and 2 h, 100 μM ABA for 3 h or 30% PEG for 3 h using Trizol reagent (Sigma–Aldrich), then was subjected to electrophoresis on formaldehyde/agarose gel and transferred to Zeta-probe nylon membrane (Bio-Rad) according to standard protocols. The amount of ethidium bromide stainied rRNA was used as the control for RNA loading. RNA blots were probed with the 32P-labled ABP9 full length cDNA (Prime-a-Gene® Labeling System, Progema). Hybridization was performed according to the method of Church and Gilbert (Church and Gilbert 1984).
Real-time quantitative reverse transcriptase-PCR (qRT-PCR) was performed to validate the expression changes of ABP9-regulated genes in ABP9 transgenic Arabidopsis plants. The first-strand cDNA was synthesized using 1 μg of total RNA and ReverTra Ace-α® (Toyobo) kit. PCR amplification was done in a 20 μl reaction volume containing primers, SYBR Premix Ex Tag™ (TaKaRa) and diluted cDNA templates on a DNA Engine Optico 2 thermal cycler (MJ Research). Gene-specific primer pairs used here were listed in Supplementary material 6. The cycle threshold (C T) values, corresponding to the PCR cycle number at which fluorescence emission reaches a threshold above baseline emission, were determined and the relative fold differences were calculated by 2−ΔΔC T method using the actin2 gene as an endogenous reference and the wild type as a calibrator (Livak and Schmittgen 2001). Samples were run in triplicate on each 96-well plate and were repeated three times for each experiment. Two experiments on independently grown plant materials were performed to confirm the reproducibility of the results.
Results
ABP9 encodes a transcription activator that directly binds in a sequence-specific manner to ABRE2 of maize Cat1 promoter in vitro and trans-activates gene expression via specific interaction with ABRE2 in plant cells
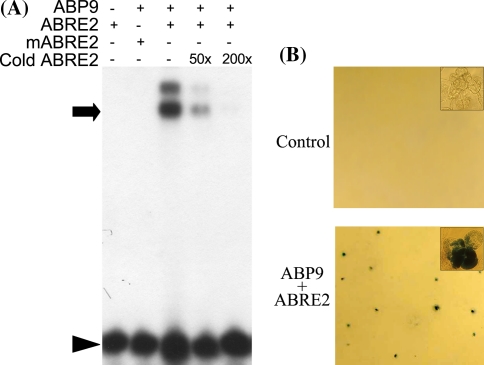
We previously showed that in yeast cells ABP9 was able to trans-activate a reporter gene expression through specific binding to the ABRE2 motif of maize Cat1 promoter (Wang et al. 2002). To test whether ABP9 can specifically in vitro bind to ABRE2, we conducted an electrophoretic mobility shift assay (EMSA) using purified GST-ABP9 fusion protein and radio-labeled ABRE2 or mABRE2 as the probes. As shown in Fig. 1a, the mobility of radio-labeled wild-type ABRE2 was retarded by GST-ABP9 (not by GST itself, data not shown) and this interaction was disrupted by competition of unlabeled ABRE2. In contrast, GST-ABP9 failed to change the mobility of radio-labeled mABRE2. This result indicates that ABP9 indeed physically binds to ABRE2 and the binding is sequence-specific. Interestingly, two shift bands appeared in the ABP9-ABRE2 interaction, suggesting that ABP9 may function as a homo-dimmer.
Fig. 1.
ABRE2-binding specificity and trans-activation activity of ABP9. a EMSA assay showing that ABP9 binds specifically to ABRE2. Purified ABP9 protein was added to reactions containing 32P-labeled ABRE2 or mABRE2 probe with or without unlabeled probe (cold ABRE2). The unlabeled ABRE2 as competitors were added as 50 or 200 times of the labeled ABRE2 probe. Arrow marks the ABP9-ABRE2 binding complex, arrowhead indicates free probe. b Histochemical assay of trans-activation activity of ABP9 in maize suspension cells. The GUS reporter construct containing ABRE2 in the promoter was co-transformed by bombardment into maize cells with ABP9 effector plasmid (lower panel) or with empty vector as the background control (upper panel) and stained with X-Gluc buffer. The insets within the panels show a magnified picture of stained cells
We next tested whether ABP9 can activate gene expression through binding to ABRE2 in plant cells. We co-transformed an effector construct (pAHC25-ABP9) in which ABP9 is driven by an ubiquitin promoter and a reporter plasmid (pIGA) where GUS is under the control of ABRE2-35S mini chimeric promoter, into maize BMS suspension cells. Histochemical staining assay showed that the GUS reporter gene was activated in maize cells by ABP9 via interaction with ABRE2 motif in the chimeric promoter (Fig. 1b). These data clearly indicate that ABP9 also functions as an ABRE-dependent transcription activator in plant cells.
Expression of ABP9 in maize is inducible by abiotic stresses and ABA
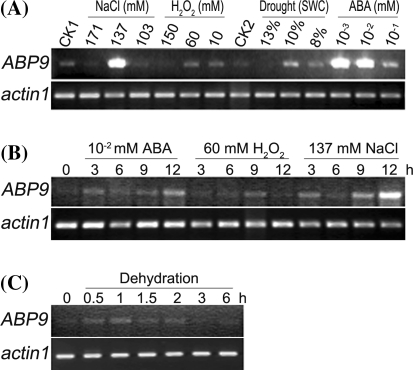
To determine whether ABP9 is regulated by abiotic stresses and ABA, we performed semi-quantitative RT-PCR with maize seedlings treated with different stresses or ABA. Compared to well-watered soil-grown (CK1) or water-treated (CK2) maize seedlings, the expression of ABP9 is significantly induced by treatments with medium levels of NaCl (137 mM) or ABA (10−3 or 10−2 mM) and slightly induced by H2O2 (10 and 60 mM) and drought (8 and 10% soil water content) treatments (Fig. 2a). Interestingly, higher levels of NaCl (171 mM), H2O2 (150 mM) and ABA (10−1 mM) were not effective in inducing ABP9 expression in maize seedlings (Fig. 2a). Next we conducted a time course experiment to examine the kinetics of ABP9 expression by various abiotic stresses. Induction of ABP9 by ABA (10−2 mM) and NaCl (137 mM) can be clearly detected 3 h after the treatments, whereas induction by H2O2 (60 mM) became clear 9 h after the treatment (Fig. 2b). Notably, induction of ABP9 by dehydration was most obvious during the first 2 h of treatment and became undetectable thereafter (Fig. 2c). These results suggest that ABP9 may be involved in ABA and abiotic stress responses in plants.
Fig. 2.
ABP9 expression in maize is inducible by ABA and abiotic stresses. a RT-PCR analysis of ABP9 expression under NaCl, H2O2, ABA and drought treatments at different dosages. CK1 and CK2 represent the untreated soil-grown and water-grown control plants, respectively. b, c Time course examination of ABP9 expression under treatments of 10 μM ABA, 60 mM H2O2, 137 mM NaCl (b) and dehydration (c). Pre-treatments of maize seedlings with ABA and stresses were described in the “Materials and methods” section. Maize actin1 was used as an internal control. Experiments were repeated at least 5 times with independent biological samples showing consistent results
Constitutive expression of ABP9 in Arabidopsis leads to enhanced tolerance to multiple stresses
To investigate the function of ABP9 in plant response to abiotic stress, transgenic Arabidopsis plants expressing the ABP9 cDNA under the control of the CaMV 35S promoter (35S-ABP9) were generated and eight independent T3 homozygous lines were obtained. Northern blot analysis revealed that ABP9 expressing transgenic plants accumulated higher levels of ABP9 mRNA under either normal growth or stress conditions with the treatment of 200 mM of NaCl (1 or 2 h), 100 μM ABA (3 h) or 30% PEG (3 h), indicating that ABP9 is constitutively expressed in these transgenic plants (Fig. 3a, b). Two representative lines (5P2 and 5P3), with medium and high expression levels of ABP9 were selected for this study.
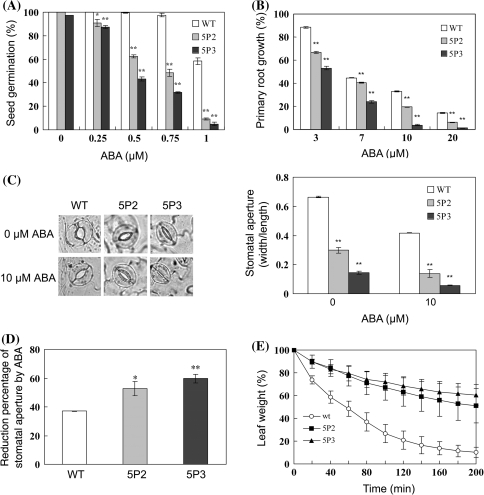
Fig. 3.
Constitutive expression of ABP9 enhances tolerance to drought, salt, cold and oxidative stresses in transgenic Arabidopsis. a, b RNA gel blot analysis of ABP9 expression in wild type (WT) and 35S-ABP9 transgenic plants (5P2 and 5P3) under normal growth and salt stress (a), ABA and PEG treatment conditions (b). Plants grown on 1/2 MS agar plates were untreated or treated with 200 mM NaCl for 1 h and 2 h, 100 μM ABA or 30% PEG for 3 h. Equal amounts (8 μg) of total RNAs separated by electrophoresis on formaldehyde-agarose gels and rRNA was used as loading control. c Drought tolerance in 35S-ABP9 transgenic plants. Intact plants were drought stressed by withholding water for 4 weeks, then were rewatered for 1 week before photograph was taken. Survival rates were calculated from the results of three independent experiments (n ≥ 30 for each experiment). Untreated plants grown under normal conditions as control were shown in the top panels. d Capacity of 35S-ABP9 transgenic plants to tolerate salt stress. The plants were treated with progressively increased concentrations of NaCl solutions ranging from 50 to 200 mM and kept irrigated with 200 mM NaCl for 12 days, then chlorophyll contents were determined. Percentage shown are chlorophyll content of salt-stresses plants relative to that of untreated ones from the results of three independent experiments with 5 intact seedlings determined for each experiment. Untreated plants grown under normal conditions as control were shown in the top panels. e Freeze tolerance of 35S-ABP9 transgenic plants. Plants were exposed to freezing temperature −4°C for 6 h and recovered for 1 week under normal growth conditions. Survival rates were counted from the results of three independent experiments (n ≥ 30 for each experiment). Untreated plants grown under normal conditions as control were shown in the top panels. f Tolerance to oxidative stress in 35S-ABP9 transgenic plants. Rosette leaves from wild-type (WT) plants and transgenic lines (5P2 and 5P3) were floated in 1/2 MS medium containing 2 μM MV for 72 h before photographs were taken. Chlorophyll contents compared to that of mocked-treated samples are shown at the bottom. Shown are representative photographs from 30 seedlings in triplicates
Next, we examined drought stress response of the transgenic plants compared to wild type plants. Drought stress was imposed by continuously withholding water to soil-grown plants. As shown in Fig. 3c, under normal growth conditions, growth and development were somewhat delayed in the ABP9 overexpressing transgenic line 5P2 and 5P3 and this delay is much more significant in 5P3, in which ABP9 expression is 2-3 fold higher than in 5P2 (Fig. 3a, b). During 4-week exposure to drought, almost all the 35S-ABP9 transgenic plants kept growing vigorously, whereas wild-type plants became withered, chlorotic and bleached. After resuming water, 65% of 5P2 and 88% of 5P3 plants survived to maturity. In contrast, more than 60% of wild-type plants were completely dehydrated and failed to recover.
For salt tolerance assay, we examined the performance of transgenic plants under progressively applied and relatively long-term high salt stress conditions. Plants were irrigated with stepwise increasing concentrations of NaCl solution and then lasting at the concentration of 200 mM for 12 days. Most leaves of 35S-ABP9 plants kept green whereas wild-type leaves gradually became yellow and purple. Measurement of chlorophyll contents by the end of treatment revealed that the chlorophyll contents of 5P2 and 5P3 remained ~91 and 93% of untreated plants, respectively, whereas the chlorophyll content of treated wild-type plants dropped to 42% of untreated plants (Fig. 3d).
For cold tolerance assay, plants were exposed to −4°C freezing temperature for 6 h. We noticed that soon after the cold treatment, wild-type plants all collapsed with leaves showing a “boiled” phenotype in color, while most of the transgenic plants kept their original morphological structure and leaf color. After a recovery period of 7 days under normal growth conditions, at least 90% of the transgenic plants resumed their growth and development, whereas only 30% of wild-type plants survived yet grew unhealthily. The other 70% of wild-type plants failed to recover and died (Fig. 3e).
We also analyzed the oxidative stress tolerance of 35S-ABP9 transgenic plants by assessing their response to methyl viologen (MV), a herbicide causing chlorophyll degradation and the leakage of cell membrane by generating ROS (Kurepa et al. 1998). Rosette leaves from transgenic and wild-type plants were incubated in 1/2 MS liquid medium containing 2 μM MV. After 72 h of incubation, the wild-type leaves became partially bleached. By contrast, the transgenic leaves remained greener than wild-type ones (Fig. 3f). Quantitative assay revealed that in comparison with mock-treated control leaves, wild-type leaves decreased by 61% of their chlorophyll contents after MV treatment, whereas the transgenic leaves only decreased by 25% (5P2) and 6% (5P3), respectively.
Taken together, these results indicate that constitutive expression of ABP9 causes enhanced tolerance to drought, salt, low temperature, and oxidative stresses in transgenic plants.
Constitutive expression of ABP9 in Arabidopsis enhances ABA sensitivity
Given the facts that ABP9 is an ABRE2-dependent transcription activator and its expression is induced by ABA, we next examined the effect of ABP9 on other ABA-mediated responses including inhibition of seed germination and root growth, promotion of stomatal closure and water conservation in 35S-ABP9 transgenic plants.
In the absence of exogenously applied ABA, the germination percentage of 35S-ABP9 seeds was similar to that of wild-type seeds when scored at 72 h on MS medium. However, compared to wild-type seeds, germination of 35S-ABP9 seed is more sensitive to exogenously applied ABA (concentrations ranging from 0.25 to 1 μM), and the sensitivity is dependent on the level of ABP9 expression (Fig. 4a). Measurement of root length showed that the ABP9 transgenic plants exhibited a greater reduction in response to exogenously applied ABA, in comparison with wild-type plants (Fig. 4b).
Fig. 4.
Constitutive expression of ABP9 enhances ABA sensitivity and water-conserving capacity of 35S-ABP9 transgenic Arabidopsis plants. a Germination rates of wild-type (WT) and 35S-ABP9 seeds (5P2 and 5P3) exposed to 0, 0.25, 0.5, 0.75, and 1 μM ABA at 72 h after stratification. Data represent means ± SD of three independent experiments with 40 seeds per genotype and experiment. Asterisks indicate significant differences between the WT and 5P2 or 5P3 plants (*P < 0.05, **P < 0.01, Students t test). b Primary root growth of WT, 5P2 and 5P3 seedlings. The germinated seeds plated in ABA-free MS medium for 48 h after stratification were transferred to plates supplemented with 0, 3, 7, 10 and 20 μM ABA and root elongation was scored after 10 days. Each value is the mean ± SD of at least 30 seedlings from three independent experiments. Asterisks indicate significant differences between the WT and 5P2 or 5P3 plants (**P < 0.01, Students t test). c Stomatal aperture measurements of wild-type (WT) and 35S-ABP9 transgenic lines (5P2 and 5P3) in response to 0 and 10 μM ABA. Values are means ± SD of 75 stomata from three independent experiments. Asterisks indicate significant differences between the WT and 5P2 or 5P3 plants (**P < 0.01, Students t test). d Percentage of ABA-induced stomatal closure in (c) of wild-type (WT) and 35S-ABP9 transgenic plants (5P2 and 5P3). Asterisks indicate significant differences between the WT and 5P2 or 5P3 plants (*P < 0.05, **P < 0.01, Students t test). e Transpirational water loss in wild-type (WT) and 35S-ABP9 transgenic plants (5P2 and 5P3). Leaf weight is expressed as the percentage of initial fresh weight (FW). Each data point represents the mean of duplicate measurements (5 detached leaves for each measurement)
In addition, measurement of stomatal aperture showed that 35S-ABP9 transgenic plants had much smaller stomatal aperture than wild type in the absence of exogenously applied ABA (Fig. 4c). When exposure to the treatment of 10 μM ABA, the stomatal aperture of 5P2 and 5P3 plants was reduced by 54 and 60%, respectively, relative to that of untreated ones, whereas the stomatal aperture of wild-type plants only reduced by 37% (Fig. 4d). These results suggest that ABP9 expression confers enhanced sensitivity in ABA-induced stomatal closure to 35S-ABP9 transgenic plants. Consistent with this, the ABP9 expressing transgenic plants also exhibited an enhanced capacity to conserve water, as revealed by delayed loss of fresh-weight measured in detached leaves under dehydration conditions (Fig. 4e).
Taken together, these results demonstrate that ABP9 plays a positive role in regulating ABA-mediated inhibition of seed germination and root growth as well as promotion of stomatal closure.
The ABP9 expressing plants display reduced ROS levels and alleviated oxidative damage and cell death
As ABA is known to be involved in regulating ROS levels and responses in plants, and ABP9 expressing Arabidopsis plants displayed enhanced tolerance to the ROS-generating agent MV (Fig. 3f), we speculated that ABP9 may be involved in the control of ROS levels during normal growth and in response to stress and ABA. To test this, we compared ROS levels in 35S-ABP9 transgenic and wild type plants under normal growth and stress/ABA treatment conditions.
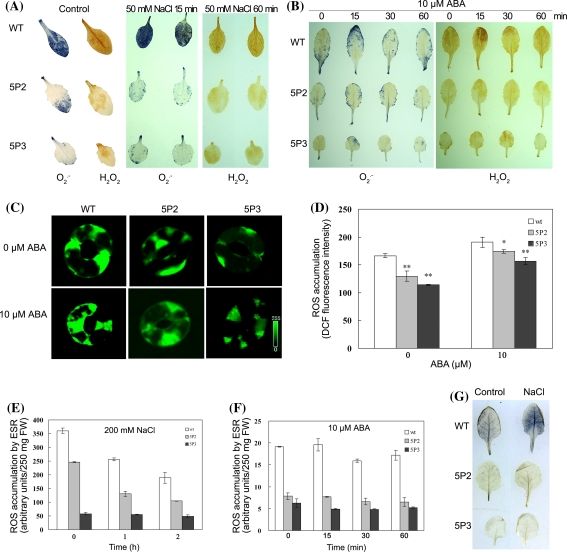
We first examined the cellular levels of O2 ·− and H2O2, two prominent ROS species involved in stress signaling and oxidative injuries, under normal growth and salt stress conditions by NBT and DAB staining, respectively. As shown in Fig. 5a, the steady state levels of both O2 ·− and H2O2, visualized as dark blue and deep brown products, respectively, were largely reduced in leaves of 35S-ABP9 transgenic plants compared to wild-type plants under normal growth and salt stress conditions. Similarly, the accumulation of O2 ·− and H2O2 in ABA-treated 35S-ABP9 transgenic plants were also clearly lower than that in wild-type plants treated with ABA (Fig. 5b).
Fig. 5.
Constitutive expression of ABP9 decreases ROS levels and cell death in 35S-ABP9 transgenic Arabidopsis plants under normal growth, ABA and salt stress conditions. a, b Cellular levels of H2O2 and O2 ·− are reduced in leaves of 35S-ABP9 transgenic plants under normal growth (Control), salt (a) and ABA (b) conditions. Detached leaves from wild-type (WT) and transgenic plants (5P2 and 5P3) pre-treated with 50 mM NaCl or 10 μM ABA were stained with DAB and NBT to visualize H2O2 and O2 ·−, respectively. All experiments were repeated at least 3 times, and about 15 leaves collected from multiple seedlings (4–5-week-old) were inspected in each experiment. c Representative microscopy images of ROS production in intact guard cells indicated by fluorescent dye DCF. Epidermal peels were loaded with H2DCF-DA for 10 min after the incubation with 10 μM ABA for 2 h. The pseudo-color key is shown at the bottom of lower panel and applied to pixel intensity (0–255) for all fluorescence images. d Quantification of ROS production in guard cells in (c) of wild-type and 35S-ABP9 plants following ABA treatment. Data are from measurements of pixel intensity in whole cells determined that are treated with or without ABA in three independent experiments (means ± SD). For the wild type (WT), n = 93 cells without ABA treatment, n = 100 cells with ABA treatment; for 35S-ABP9 transgenic line 5P2, n = 125 cells without ABA treatment, n = 220 cells with ABA treatment; and for 5P3, n = 120 cells without ABA treatment, n = 140 cells with ABA treatment. Asterisks indicate significant differences between the WT and 5P2 or 5P3 plants (*P < 0.05, **P < 0.01, by Students t test). e, f ESR spin trapping detection of carbon-centered free radicals by PBN in wild type and 35S-ABP9 transgenic plants in response to salt (e) and ABA (f). Intensity of the ESR spectra signal in plants untreated and treated with 200 mM NaCl or 10 μM ABA was measured and expressed in arbitrary units. FW, fresh weight. Experiments were performed three times. Shown are means ± SD of three replicates from one experiment. All differences between the WT and 5P2 or 5P3 plants are significant at P < 0.05 using Students t test. g Cell death staining with Evans blue before (Control) and 60 min after the treatment of 50 mM NaCl. The experiment was repeated 3 times, and 10 leaves collected from multiple seedlings (4–5-week-old) were inspected in each experiment
We next measured the cellular level of H2O2 in guard cells with or without ABA treatment using H2DCF-DA, a fluorogenic probe reporting ROS levels within cells by detecting enzymatically formed H2O2. As shown in Fig. 5c and 5d, although ABA treatment significantly increased H2O2 levels in both wild type and 35S-ABP9 transgenic plants, H2O2 levels in guard cells of 35S-ABP9 transgenic plants was much lower than that in wild-type plants either in the presence or absence of 10 μM ABA. Interestingly, quantitative analysis revealed that the level of H2O2 in ABA-treated guard cells of 5P2 and 5P3 transgenic plants increased by 33 and 37%, respectively, relative to the untreated control. In contrast, H2O2 level in ABA-treated guard cells of wild type plants only increased by 14% relative to the untreated control (Fig. 5d). These results suggest that ABP9 expression renders plant cells an enhanced capacity to constrain the absolute amount of H2O2 to a lower level in both the rest and ABA-stimulated cells, but allows a higher relative increase of H2O2 in response to ABA stimulation.
Since highly reactive free radicals such as OH·, resulting from reactions between O2 ·− and H2O2, can cause lipid peroxidation by generating carbon-centered lipid free radicals (Halliwell 2006), our observations that ABP9 down-regulates the total cellular levels of both O2 ·− and H2O2 lead us to speculate that ABP9 may alleviate cellular oxidative damages caused by ROS. To test this, the steady state level of carbon-centered lipid free radicals was detected by ESR as spin adducts of PBN, a lipophilic spin-trap that is commonly used to detect lipid-derived carbon-centered radicals (Garlick et al. 1987). Quantification of ESR signals revealed that the cellular levels of lipid radicals in transgenic lines 5P2 and 5P3 were much reduced in comparison with wild-type plants under both normal growth conditions or treated with salt stress and ABA. Further, the radical levels in 5P2 and 5P3 reversely correlate with the expression levels of ABP9 in these two independent transgenic lines (Fig. 5e, f). These observations suggest that the cellular oxidative damage resulting from secondary reactions of initially-formed reactive oxygen species is alleviated by ABP9. Consistent with this notion, Evans Blue staining assay showed the decreased cell death in 35S-ABP9 transgenic plants under either normal growth or salt stress conditions, compared with wild type plants (Fig. 5g).
Taken together, these results demonstrate that ABP9 plays a critical role in controlling the homeostasis of cellular levels of ROS, thus preventing cells from oxidative damages and decreasing cell death.
Constitutive expression of ABP9 enhances the expression of stress/ABA-responsive genes including ROS-scavenging/signaling genes
To investigate the molecular mechanism by which ABP9 regulates ABA signaling, ROS accumulation and thus stress tolerance, we performed microarray analysis to measure global changes of gene expression in 5P2 and 5P3 transgenic plants in comparison with wild-type plants using the Arabidopsis Affymetrix GeneChip® ATH1 array containing probe sets representing approximately 24000 genes. The chip assay showed that under normal growth conditions, a total of 365 and 607 genes, respectively in 5P2 and 5P3 transgenic plants, were significantly up-regulated (twofold cutoff) relative to wild-type plants (Supplementary material 2, 3). When treated with 200 mM NaCl for 1 h and 2 h, 357 and 373 genes with levels greater than two fold change over wild-type plants were identified in the 5P2 transgenic line, respectively (Supplementary material 4, 5).
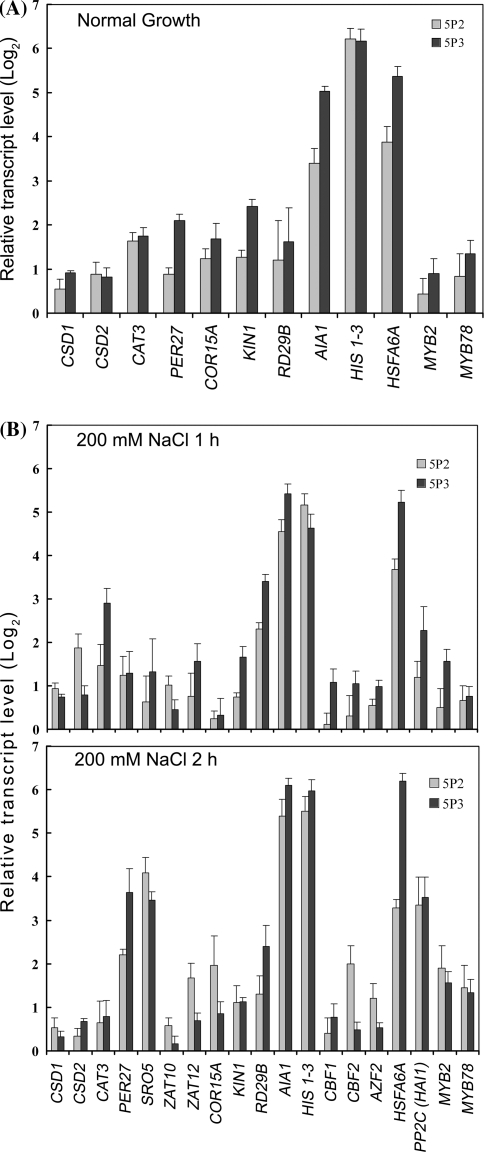
To verify the microarray results, we selected 19 well-documented stress-inducible genes for qRT-PCR analysis. Among which, 14 were shown to be both stress- and ABA-inducible (Supplementary material 7). As shown in Fig. 6a, under normal growth conditions, 12 genes displayed elevated expression in both 5P2 and 5P3 transgenic plants, relative to wild-type plants. These genes are (i) ROS-scavenging enzymes CSD1, CSD2, CAT3 and PER27 (Mittler et al. 2004); (2) stress- and ABA-inducible protective or regulatory genes COR15A, KIN1, RD29B, AIA1, HIS1-3, MYB2, MYB78 (Abe et al. 1997; Fujita et al. 2005; Thomashow 1999; Xin et al. 2005); (3) heat-inducible transcription factor HSFA6A (von Koskull-Döing et al. 2007). It is noteworthy that HIS1-3, AIA1 and HSFA6A show highest induction among these genes. Further, among the 19 selected genes, 7 of them show enhanced expression only under salt treatments (Fig. 6b). These genes include SRO5, Zat10 and Zat12 which are involved in ROS signaling to regulate ROS-scavenging gene expression (Borsani et al. 2005; Mittler et al. 2006; Rizhsky et al. 2004), three stress- and ABA-responsive transcription factors CBF1, CBF2, AZF2 (Sakamoto et al. 2004; Stockinger et al. 1997), and the protein phosphatase PP2C (HAI1)/At5g59220 (Fujita et al. 2009; Xin et al. 2005).
Fig. 6.
Constitutive expression of ABP9 enhances the expression of stress- and ABA- responsive genes. Seedlings grown on agar plates were untreated or treated with 200 mM NaCl for 1 h and 2 h. Transcript level of each gene before (a) and after the treatment (b) detected by qRT-PCR, normalized to the endogenous control (actin2) relative to the calibrator (wild type), is given by 2−ΔΔC T. The Figure was plotted by Log2 values of relative transcript levels vs. genes. For each gene, the expression level in the WT plants was defined as 0. Data represent means ± SD of three replicates
Since ABP9 binds to a G-box type ABRE in a sequence-specific manner, we next searched for candidate ABREs in the promoters (2,000-bp region upstream of the start codon) of the 19 ABP9-regulated genes described above using the ACGT core-containing ABRE motifs available in the plantCARE database (http://sphinx.rug.ac.be:8080/PlantCare/) (Lescot et al. 2002). This analysis identified 13 genes (CSD1, CAT3, SRO5, ZAT10, COR15A, RD29B, KIN1, AIA1, HIS1-3, CBF1, CBF2, AZF2, and PP2C(HAI1)/At5g59220) that contain at least one G-box type ABREs with the sequence (C/T)ACGTG, among which 10 genes (ZAT10, COR15A, RD29B, KIN1, AIA1, HIS1-3, CBF1, CBF2, AZF2 and PP2C(HAI1)/At5g59220) have been reported to be both stress- and ABA-inducible (Supplementary material 7). Our results suggest that ABP9 may mediate enhanced stress tolerance through regulating the expression of these downstream target genes either directly or indirectly.
Discussion
Modulation of ROS to appropriate levels is critical for tolerance to abiotic stress given the facts that ROS act as both signaling molecules at low concentrations (Pei et al. 2000; Prasad et al. 1994) and damaging agents when accumulated in excess in plants (Desikan et al. 1998; Levine et al. 1994). Previous studies have suggested that the cellular ROS levels are modulated by ABA via regulation of ROS-producing and ROS-scavenging genes (Fryer et al. 2003; Kwak et al. 2003; Zhu and Scandalios 1994). However, the transcription factors and mechanisms involved in the regulation of ROS genes and hence ROS levels by ABA remain unclear. In this report, we demonstrated that (1) ABP9, a bZIP protein of maize, in vitro directly binds in a sequence-specific manner to a G-box type ABRE in the promoter of a maize antioxidant gene Cat1, and trans-activates ABRE-dependent expression of downstream reporter gene in plant cells (Fig. 1); (2) the expression of ABP9 in maize is ABA- and stress-inducible (Fig. 2); (3) Constitutive expression of ABP9 in transgenic Arabidopsis leads to enhanced tolerance to multiple stresses, and to alleviate oxidative damage (Fig. 3); (4) Constitutive expression of ABP9 also confers hyper-sensitivity to ABA during germination, root growth, stomatal closure and increased capability to conserve water (Fig. 4); (5) Constitutive expression of ABP9 down-regulates cellular ROS levels induced by ABA and stress, and decreases cell death (Fig. 5); (6) Constitutive expression of ABP9 results in elevated expression of many ABA- and stress-inducible genes and those for scavenging and regulating ROS (Fig. 6). These results clearly indicate that ABP9 is an ABRE-binding transcription activator that acts as a positive regulator of ABA signaling and is involved in modulation of ROS accumulation in plants to confer an enhanced tolerance to various abiotic stresses by reducing the oxidative damages of ROS and cell death.
H2O2 functions as essential signaling molecule in ABA-induced stomatal closure (Pei et al. 2000). Interestingly, we found that although ABA treatment significantly increased H2O2 levels in both 35S-ABP9 trangenic and wild type plants, the net H2O2 increase by ABA as compared to untreated control in guard cells of 35S-ABP9 transgenic plants was much higher than that in wild-type plants (Fig. 5d) accompanied by enhanced ABA-induced stomatal closure in transgenic plants (Fig. 4c, d), and meanwhile the total H2O2 level in guard cells of 35S-ABP9 transgenic plants was much lower than that in wild-type plants either in the absence or presence of exogenously applied ABA (Fig. 5c, d). These results suggest that ABP9, on one hand, induces a higher ROS increase to induce stomatal closure, while on the other hand, constrains the total ROS levels by decreasing total ROS production/accumulation, thus alleviating oxidative damage and cell death under stress conditions. Thus, ABP9 may play a dual role in the harmonious regulation of ROS production and signaling by ABA and abiotic stresses.
Our genome-wide gene expression and qRT-PCR analyses provided further evidence supporting the above notion. We identified 19 genes that were up-regulated in transgenic Arabidopsis lines constitutively expressing ABP9 under normal growth and/or salt stress conditions (Fig. 6). Among them, CSD1, CSD2, CAT3, and PER27 encode major antioxidant enzymes for removing superoxide and hydroperoxide in various cellular compartments. ZAT10, ZAT12 and SRO5 are all regulatory components in ROS signaling, and all can be induced by multiple abiotic stresses including oxidative stress and are found capable of enhancing stress tolerance of plants via elevating the expression of reactive oxygen-defense transcripts (Borsani et al. 2005; Davletova et al. 2005; Mittler et al. 2006; Rizhsky et al. 2004; Vogel et al. 2005). The observation that these genes were up-regulated in ABP9 overexpressing transgenic plants under normal growth and/or salt stress conditions suggests that ABP9 enables the plants to detoxify ROS efficiently and to tightly control ROS accumulation and thus enhances stress tolerance via mobilizing an array of ROS-scavenging enzymes and activating signaling molecules regulating ROS-scavenging genes.
Our transcriptome analysis also revealed that expression of many stress/ABA-responsive protective and regulatory genes, is drastically up-regulated in ABP9 overexpression plants, including COR15A, KIN1, RD29B, AIA1, HIS1-3, ATMYB2, ATMYB78, CBF1, CBF2, AZF2, PP2C (HAI1)/At5g59220, and HSFA6A. All these 12 members were evidenced as both stress- and ABA-inducible (Supplementary material 7), and some of these genes have been clearly identified as positive regulators of ABA signaling such as MYB2, PP2C (HAI1) (Abe et al. 2003; Fujita et al. 2009) or as downstream targets of ABA-signaling pathways such as COR15A, KIN1, RD29B, AIA1, HIS1-3 (Fujita et al. 2005). Therefore, their up-regulation in ABP9 overexpressing transgenic plants under normal growth and/or salt stress conditions (Fig. 6) provides further molecular validation that ABP9 may act as an upstream positive regulator in ABA and stress signaling. Previous studies have reported that enhanced expression of stress-responsive genes such as COR15A, KIN1, RD29B, AIA1, HIS1-3 or overexpression of transcription factor genes such as MYB2, CBF1, CBF2, AZF2 imparts tolerance to low temperature and water stress in plants (Abe et al. 2003; Gilmour et al. 2004; Sakamoto et al. 2000). In addition, a role of heat shock transcription factor genes, such as HSFA6A and HSFA4A, in stress tolerance and ROS signaling has also been reported (Chao et al. 2007; Miller and Mittler 2006). Thus, the up-regulation of these stress tolerance marker genes and signaling molecules or transcription factors may collectively set the molecular basis for ABP9-mediated enhancement of abiotic stress tolerance. Future studies will be aimed to unravel the direct target genes of ABP9 in maize and to explore its utilization in engineering transgenic crops with enhanced tolerance to multiple abiotic stresses.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Professor Shihuang Zhang for providing maize Qi319 seeds, Professor Baolu Zhao, Dr. Yangcang Xu, and Yongsheng Chen for technical assistance in ESR assay. We are also grateful to Dr. Haiyang Wang for helpful discussion and critical reading on the manuscript. This work was supported by the National Basic Research Program of China (grant no. 2006CB100102) and by the National High Technology Research and Development Program of China (grant no. 2006AA10Z116).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bartels D, Sunkar R. Drought and salt tolerance in plants. Crit Rev Plant Sci. 2005;24:23–58. doi: 10.1080/07352680590910410. [DOI] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie Life Sci. 1993;316:1194–1199. [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu J-K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Mol Biol. 1998;37:425–435. doi: 10.1023/A:1006058700720. [DOI] [PubMed] [Google Scholar]
- Capani F, Loidl CF, Aguirre F, Piehl L, Facorro G, Hager A, De Paoli T, Farach H, Pecci-Saavedra J. Changes in reactive oxygen species (ROS) production in rat brain during global perinatal asphyxia: an ESR study. Brain Res. 2001;914:204–207. doi: 10.1016/S0006-8993(01)02781-0. [DOI] [PubMed] [Google Scholar]
- Chao X, Wang D-p, Gong Z-h, Liang Y, Zhao J. Functional analysis of Arabidopsis transcriptional factor in heat stress tolerance. Acta Bot Boreal Occident Sin. 2007;27:1305–1310. [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Schlauch K, Coutu J, Mittler R. The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Reynolds A, Hancock JT, Neill SJ. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem J. 1998;330:115–120. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Mackerness SAH, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127:159–172. doi: 10.1104/pp.127.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–610. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala SSL, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14:S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR. Imaging of photo-oxidative stress responses in leaves. J Exp Bot. 2002;53:1249–1254. doi: 10.1093/jexbot/53.372.1249. [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR. Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 2003;33:691–705. doi: 10.1046/j.1365-313X.2003.01656.x. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–3488. doi: 10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y, Nakashima K, Yoshida T, Katagiri T, Kidokoro S, Kanamori N, Umezawa T, Fujita M, Maruyama K, Ishiyama K, Kobayashi M, Nakasone S, Yamada K, Ito T, Shinozaki K, Yamaguchi-Shinozaki K. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009;50:2123–2132. doi: 10.1093/pcp/pcp147. [DOI] [PubMed] [Google Scholar]
- Garlick PB, Davies MJ, Hearse DJ, Slater TF. Direct detection of free radicals in the reperfused rat heart using electron spin resonance spectroscopy. Circ Res. 1987;61:757–760. doi: 10.1161/01.res.61.5.757. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Guan L, Scandalios JG. Effects of the plant growth regulator abscisic acid and high osmoticum on the developmental expression of the maize catalase genes. Physiol Plant. 1998;104:413–422. doi: 10.1034/j.1399-3054.1998.1040317.x. [DOI] [Google Scholar]
- Guan LM, Zhao J, Scandalios JG. Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J. 2000;22:87–95. doi: 10.1046/j.1365-313x.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- Halliwell B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006;141:312–322. doi: 10.1104/pp.106.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang MY, Zhang JH. Water stress-induced abscisic acid accumulation triggers the increased generation of reactive oxygen species and up-regulates the activities of antioxidant enzymes in maize leaves. J Exp Bot. 2002;53:2401–2410. doi: 10.1093/jxb/erf090. [DOI] [PubMed] [Google Scholar]
- Kim SY. The role of ABF family bZIP class transcription factors in stress response. Physiol Plant. 2006;126:519–527. [Google Scholar]
- Kim M, Ahn J-W, Jin U-H, Choi D, Paek KH, Pai HS. Activation of the programmed cell death pathway by inhibition of proteasome function in plants. J Biol Chem. 2003;278:19406–19415. doi: 10.1074/jbc.M210539200. [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle J, Van Montagu M, Inze D. Oxidative stress tolerance and longevity in Arabidopsis: the late-flowering mutant gigantea is tolerant to paraquat. Plant J. 1998;14:759–764. doi: 10.1046/j.1365-313x.1998.00168.x. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 2003;22:2623–2633. doi: 10.1093/emboj/cdg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A, Tenhaken R, Dixon R, Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;18:350–382. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2T−ΔΔC Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Miller G, Mittler R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot. 2006;9:279–288. doi: 10.1093/aob/mcl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mittler R, Kim Y, Song L, Coutu J, Coutu A, Ciftci-Yilmaz S, Lee H, Stevenson B, Zhu J-K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006;580:6537–6542. doi: 10.1016/j.febslet.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige Y, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Ono A, Izawa T, Chua NH, Shimamoto K. The rab16B promoter of rice contains two distinct abscisic acid-responsive elements. Plant Physiol. 1996;112:483–491. doi: 10.1104/pp.112.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei ZM, Kuchitsu K, Ward JM, Schwarz M, Schroeder JI. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 Mutants. Plant Cell. 1997;9:409–423. doi: 10.1105/tpc.9.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Z-M, Murata Y, Benning G, Thomine S, Klusener B, Allen GJ, Grill E, Schroeder JI. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR. Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell. 1994;6:65–74. doi: 10.1105/tpc.6.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Hallak-Herr E, Van Breusegem F, Rachmilevitch S, Barr JE, Rodermel S, Inz D, Mittler R. Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J. 2002;32:329–342. doi: 10.1046/j.1365-313X.2002.01427.x. [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Davletova S, Liang H, Mittler R. The zinc finger protein Zat12 is required for cytosolic ascorbate peroxidase 1 expression during oxidative stress in Arabidopsis. J Biol Chem. 2004;279:11736–11743. doi: 10.1074/jbc.M313350200. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Araki T, Meshi T, Iwabuchi M. Expression of a subset of the Arabidopsis Cys2/His2-type zinc-finger protein gene family under water stress. Gene. 2000;248:23–32. doi: 10.1016/S0378-1119(00)00133-5. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Maruyama K, Sakuma Y, Meshi T, Iwabuchi M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis Cys2/His2-Type zinc-finger proteins function as transcription repressors under drought, cold, and high-salinity stress conditions. Plant Physiol. 2004;136:2734–2746. doi: 10.1104/pp.104.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thalianaCBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF. Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:571–599. doi: 10.1146/annurev.arplant.50.1.571. [DOI] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- von Koskull-Döing P, Scharf K-D, Nover L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007;12:452–457. doi: 10.1016/j.tplants.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Wang L, Zhao J, Fan YL. Gene cloning and function analysis of ABP9 protein which specifically binds to ABRE2 motif of maize Cat1 gene. Chinese Sci Bull. 2002;47:1871–1875. doi: 10.1360/02tb9409. [DOI] [Google Scholar]
- Xin Z, Zhao Y, Zheng ZL. Transcriptome analysis reveals specific modulation of abscisic acid signaling by ROP10 small GTPase in Arabidopsis. Plant Physiol. 2005;139:1350–1365. doi: 10.1104/pp.105.068064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song C-P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001;126:1438–1448. doi: 10.1104/pp.126.4.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wollenweber B, Jiang D, Liu F, Zhao J. Water deficits and heat shock effects on photosynthesis of a transgenic Arabidopsis thaliana constitutively expressing ABP9, a bZIP transcription factor. J Exp Bot. 2008;59:839–848. doi: 10.1093/jxb/erm364. [DOI] [PubMed] [Google Scholar]
- Zhu D, Scandalios JG. Differential accumulation of manganese-superoxide dismutase transcripts in maize in response to abscisic acid and high osmoticum. Plant Physiol. 1994;106:173–178. doi: 10.1104/pp.106.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.