Abstract
Wild-type strains of the protozoan ciliate Euplotes collected from different locations on the coasts of Antarctica, Tierra del Fuego and the Arctic were taxonomically identified as the morpho-species Euplotes nobilii, based on morphometric and phylogenetic analyses. Subsequent studies of their sexual interactions revealed that mating combinations of Antarctic and Arctic strains form stable pairs of conjugant cells. These conjugant pairs were isolated and shown to complete mutual gene exchange and cross-fertilization. The biological significance of this finding was further substantiated by demonstrating that close homology exists among the three-dimensional structures determined by NMR of the water-borne signaling pheromones that are constitutively secreted into the extracellular space by these interbreeding strains, in which these molecules trigger the switch between the growth stage and the sexual stage of the life cycle. The fact that Antarctic and Arctic E. nobilii populations share the same gene pool and belong to the same biological species provides new support to the biogeographic model of global distribution of eukaryotic microorganisms, which had so far been based exclusively on studies of morphological and phylogenetic taxonomy.
Keywords: ciliate mating types and sexual interactions, microbial ecology, NMR structures, polar biology, signaling by water-borne pheromones
Microorganisms thrive in ocean waters and knowledge of their biology and ecology is essential to better understanding how ocean life evolves and responds to a more and more rapidly changing environment (1, 2). Through sampling of the most remote ecosystems of our planet and the application of modern molecular procedures, new archea, bacteria and protists are discovered, and this calls for unequivocal and effective criteria to determine their species status and to investigate if their geographic distribution is either global or local (3–8). The more conventional concept of species is primarily derived from analysis of the phenotype, behavior, and, most importantly, the ability of organisms to interbreed and generate fertile offspring. Whereas these procedures are well suited to more complex life forms, they are less readily applied to microorganisms, because these are for the most part poorly differentiated, both morphologically and behaviorally, and proliferate only through vegetative growth. In this context, the protist group of ciliates represents an exception and offers unique experimental opportunities to investigate species diversity and biogeography when compared to other eukaryotic microbes. In addition to possessing cell-body structures that are much more complex and taxonomically distinctive than in any other microbial group, ciliates are microorganisms characterized by a sexual phenomenon known as conjugation (or mating), in which two functionally hermaphroditic individuals of differing mating types unite temporarily in pairs to carry out a reciprocal gene exchange (9, 10). Therefore, considering the relative ease with which cospecific strains of differing mating types can be collected in the wild, they are ideal microorganisms for breeding analyses of natural microbial populations under laboratory conditions. Among a vast collection of Antarctic, Fuegian, and Arctic strains of the most cosmopolitan and ubiquitous ciliate, Euplotes, we identified a number of cospecific strains of Euplotes nobilii of differing mating types and studied in detail the mating interactions of Antarctic and Arctic strains along with the three-dimensional structures of the water-borne, strain-specific signaling proteins (pheromones) that govern these interactions. The results provided compelling evidence that these strains share the same gene pool and represent a unique biological species.
Results and Discussion
Strain Collection, Morphospecies Identification, and Phylogenetic Relationships.
The initial material for this study consisted of a collection of 18 wild-type strains isolated during several polar expeditions from bottom sediments of shallow coastal waters at Terra Nova Bay (Ross Sea, Antarctica, 7 strains), Tierra del Fuego (sub-Antarctica, 4 strains), Western Greenland (Arctic, 6 strains), and Svalbard Islands (Arctic, 1 strain) (Fig. 1A and Table S1). These strains were never found to form cysts or other inactive cellular stages in response to changes in cultivation conditions.
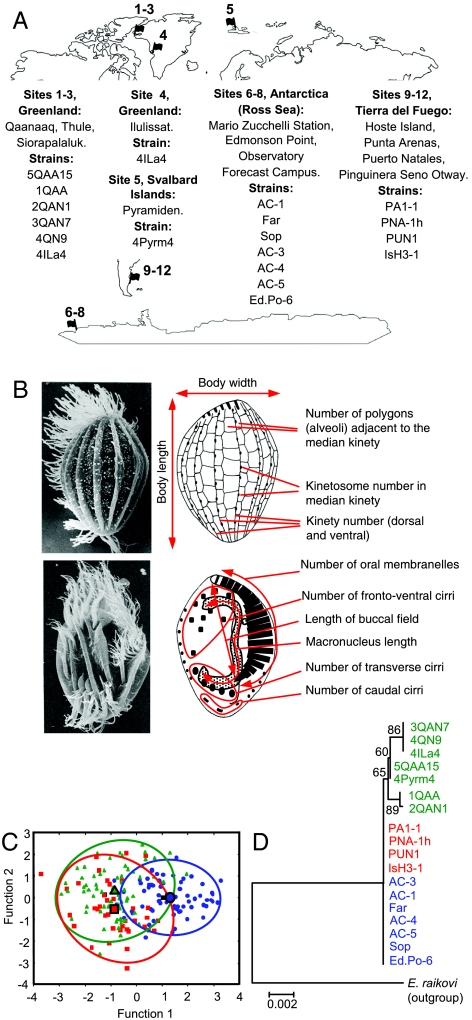
Fig. 1.
Geographic origin, morphometry, and phylogenetic relationships of E. nobilii strains. (A) Strain denominations and map of the collection sites (see Table S1 for more detailed information). (B) Illustration of the 11 phenotypic traits of taxonomic value used in the morphologic analysis of E. nobilii. The true structure of most traits represented schematically in the drawings on the right can be observed in the corresponding SEM micrographs on the left. (Upper) Cell dorsal surface. (Lower) Cell ventral surface. (C) The two axes of the plot represent the first and second functions of a statistical analysis of the analyzed E. nobilii taxonomic traits (data in Table S2) on ten cells of each strain. Antarctic, Fuegian, and Arctic strains are distinguished by blue, red, and green symbols, respectively. Ellipses and larger symbols represent the 95% confidence area and the centroid for each source, respectively. (D) Phylogenetic correlations among Antarctic (blue), Fuegian (red), and Arctic (green) strains derived from multiple alignment of the SSU-rRNA gene sequences (accession numbers in Table S3). The numbers at nodes of the phylogenetic tree are values estimated as percentages from 1,000 bootstrap replicates. E. raikovi, used as outgroup, is the species most closely allied to E. nobilii (15).
For each of the 18 strains we first established its taxonomic status as morpho-species, by measuring or counting all the phenotypic traits (Fig. 1B and Table S2) that are commonly used in the diagnosis of the nearly one hundred species that are classified in the genus Euplotes (11, 12). The data were then subjected to multivariate statistics methods to assess the degree of interstrain variability. The results indicated that all the strains form an overlapping group of units (Fig. 1C), uniformly fitting with the original morphological description of E. nobilii (13).
The 18 strains were then analyzed (Table S3) to determine their phylogenetic relationships on the basis of a comparison of their small subunit (SSU)-rRNA nuclear gene sequences. The Antarctic and Fuegian strains showed complete SSU-rRNA gene sequence identity, whereas the Arctic strains diverged up to a maximum of four nucleotide mutations, and one to three mutations were found to separate the Arctic strains from the Antarctic and Fuegian strains (Fig. 1D). Although a single nucleotide mutation in the SSU-rRNA gene sequence has been proposed to represent a significant interspecies divergence in other ciliate taxa such as Stylonychia (14), more than 60 nucleotide mutations have been found in SSU-rRNA gene sequence comparisons between E. nobilii and Euplotes raikovi, which is a temperate water species that is phylogenetically closely related to E. nobilii (15). In light of this situation, we interpret the one- to four-nucleotide substitutions detected among the SSU-rRNA gene sequences of the E. nobilii strains as reflecting sequence variations that are merely intraspecific, and not indicative of interstrain divergences of phylogenetic significance.
Mating Interactions and Breeding Analysis.
By studying the Antarctic strains, which had been collected before the other strains (Table S1), we discovered that E. nobilii, like other Euplotes species (16), performs conjugation under the genetic control of a mechanism of multiple mating types mediated by diffusible signaling pheromones (17). When the Arctic and Fuegian strains were subsequently isolated, the study of the mating interactions was thus extended to the complete collection of strains. Conjugation was regularly observed to occur at varying levels of intensity in each pairwise combination among the three Arctic strains, 5QAA15, 2QAN1, and 4Pyrm4, and the three Antarctic strains, AC-1, AC-3, and AC-4, thus implying that these six strains shared genetic homogeneity and mating compatibility. This implication was assessed by analyzing these strains in detail with regard to the outcome of their breeding interactions and the three-dimensional structure homology of their pheromones.
When analyzing mating interactions in Euplotes and other ciliates it is important to consider that mixing of two strains of differing mating types (e.g., A and B) does not necessarily generate mating pairs that are exclusively heterotypic (AB) and destined to perform mutual exchange of gamete nuclei and cross-fertilization. Homotypic (or self) pairs (AA, BB) obliged to carry out self-fertilization, or varied assortments of homo- and heterotypic pairs may be generated (9, 16). The genetic procedure conventionally adopted to distinguish between these morphologically alike homo- and heterotypic pairs is the Mendelian analysis of the mating-type inheritance. However, this procedure has disadvantages in the case of polar ciliates because their generation times are at least four-fold those of temperate water ciliates and the so-called period of sexual immaturity of their life cycle (during which cells are unable to conjugate) can extend over several months (18). Therefore, to unequivocally identify homo- and heterotypic pairs as well as to distinguish between heterotypic pairs with or without cross-fertilization, we used the SSU-rRNA gene sequences (which had previously been determined in relation to the phylogenetic analysis) as cell-specific, biparentally inheritable nuclear signatures.
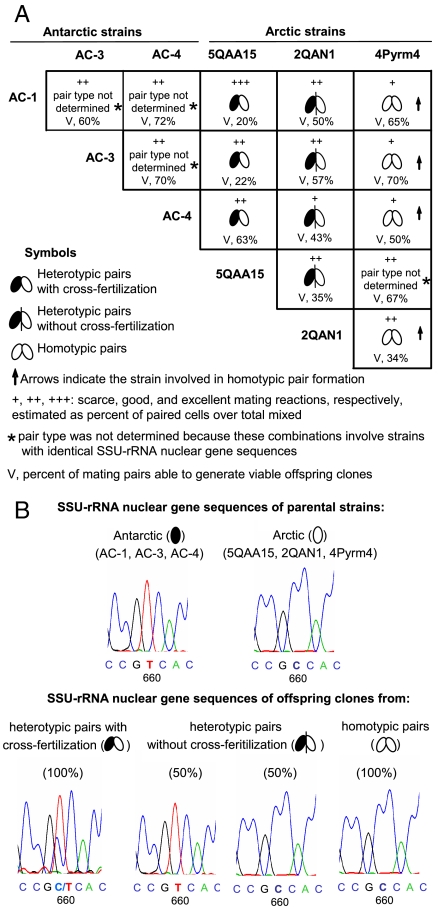
All three possible types of mating pairs were detected in the mating mixtures and each type appeared able to generate viable offspring clones, albeit with different rates of survival (Fig. 2A). Most important was the finding that the mixtures of the Arctic strain 5QAA15 with each one of the three Antarctic strains all formed only heterotypic pairs, which were fully capable of performing mutual gene exchange and cross-fertilization. This was revealed by observing that the offspring clones all carried both the parental SSU-rRNA gene sequences, which were distinguished from one another by the presence of different nucleotides (C or T) in the sequence position 660 (Fig. 2B). Formation of homotypic mating pairs and heterotypic pairs with self-fertilization was instead observed in the mixtures involving the other two Arctic strains, 4Pyrm4 and 2QAN1. The former were identified by detecting identical SSU-rRNA gene sequences among offspring clones as well as between the offspring clones and one parental strain (Fig. 2B), and the latter by detecting a 1∶1 inheritance between the two parental SSU-rRNA gene sequences (Fig. 2B).
Fig. 2.
Mating interactions between Antarctic and Arctic E. nobilii strains. (A) For each two-strain combination the corresponding box indicates the intensity of mating interactions given on a four-step scale, the types of cell mating pairs formed, and the viability rate computed as percentage of ex-conjugant cells that were able to develop a new nuclear apparatus and expand into fully viable progeny clones. (B) Schematic presentation of the utilization of the strain-specific SSU-rRNA gene sequences to distinguish the different types of mating pairs formed by mating mixtures between Antarctic and Arctic strains. The numbers below the sequences indicate the positions showing nucleotide variations that are distinctive between the Antarctic and Arctic strains. The heterotypic pairs with cross-fertilization are unique in generating offspring clones with hybrid sequences characterized by a double C/T peak at the position 660, whereas the heterotypic pairs without cross-fertilization and the homotypic pairs generate offspring clones with unchanged sequences with respect to the parental strains.
Pheromone Structure and Activity.
As pointed out above, the mating-type mechanism of E. nobilii involves the expression of diffusible signaling pheromones mediating the cell-cell recognition phenomena that trigger the switch between the growth and mating stages of the cell life cycle. The polypeptide sequences of three Antarctic and four Arctic E. nobilii pheromones (the former designated En-1, En-2 and En-6, and the latter designated En-A1, En-A2, En-A3, and En-A4) have previously been shown to contain 52–63 amino acids, with only the eight Cys residues and one Ala residue carried in conserved positions (19) (Fig. 3A). However, despite this low extent of amino acid sequence identity, close homology among the three-dimensional NMR solution structures of the three Antarctic pheromones was previously established (20–22). We have now investigated that this structural homology extends to the pheromones of the Arctic strains, by purifying the pheromone En-A1 from the strain 4Pyrm4 (which can be grown in large cultures more easily than any of the other Arctic strains) and by determining its NMR structure in solution (Table S4).
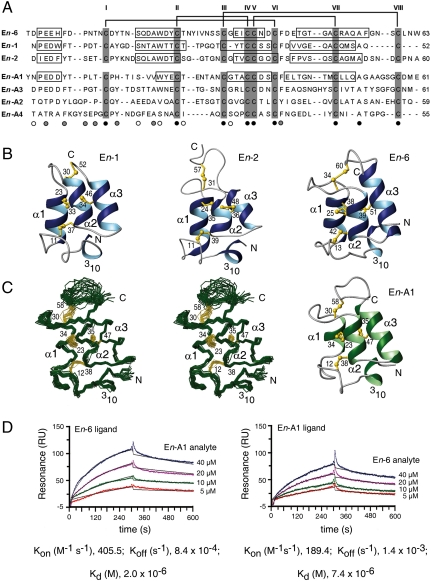
Fig. 3.
Structure and activity of E. nobilii pheromones. (A) Multiple amino acid sequence alignment between the sequences of E. nobilii pheromones of Antarctic (top three columns) and Arctic (bottom four columns) strains. The sequences were deduced from the respective coding nucleotide sequences (19), and their alignment was optimized by deliberate insertion of gaps. Positions that in this alignment are occupied by only one, two, or three different residue types are indicated below the En-A4 sequence by black, gray, and empty circles, respectively. The cysteines are shadowed and identified by Roman numerals progressing from the amino to the carboxyl terminus. The horizontal lines at the top indicate the disulfide bonds, which are common to both groups of pheromones. The boxes identify the locations of helical secondary structures in the four pheromones for which three-dimensional structures are available (20–22). (B) Ribbon presentations of the previously determined En-1, En-2 and En-6 pheromone solution NMR structures from the Antartic E. nobilii strains AC-1 and AC-4 (20, 21). The helical regions are highlighted in blue, regions of nonregular secondary structure in gray, and the disulfide bonds in yellow. The amino and carboxyl chain ends are indicated by N and C, respectively, the four helical structures are marked with 310, α1, α2, and α3, and the cysteine positions are numbered. (C) Newly determined NMR solution structure of the pheromone En-A1 [Protein Data Bank (PDB) entry 2KK2] isolated from culture filtrates of the Arctic strain 4Pyrm4 at natural isotope abundance. On the left, there is a stereo presentation of the polypeptide backbone (green) and the disulfide bonds (yellow) of a bundle of 20 energy-minimized DYANA conformers superimposed for minimal backbone rmsd of the residues 2–53. On the right, there is a ribbon presentation, characterized as in (B), of one of these En-A1 conformers. (D) Surface plasmon resonance measurements at 4 °C of protein–protein interactions between Antarctic and Arctic pheromones. In the sensorgram on the left, the pheromones En-6 (Antarctic) and En-A1 (Arctic) were used as ligand and analyte, respectively, whereas in the sensorgram on the right the roles were reversed. Four different concentrations of one pheromone were exposed to a sensor chip surface previously coated with the other pheromone during a 300-sec association phase and then removed by running buffer during a 300-sec flow dissociation phase. The two plots show the intensity of the signal in resonance units (RU) as a function of time. The curves obtained from the different injections of the analyte were superimposed using the BIA-Evaluation 4.1 software. Their fit was evaluated according to the Langmuir 1∶1 binding model, and for each analyte concentration the experimental and calculated curves are represented by colored and black lines, respectively. The plotted data represent differences in the RU signal between the flow cell (with the immobilized ligand) and the reference flow cell (without the ligand) and reflect specific ligand–analyte complexes, characterized by the kinetic and thermodynamic parameters reported at the bottom.
The Fig. 3 B and C document extensive three-dimensional structure homology between the Arctic pheromone En-A1 and the three Antarctic pheromones En-1, En-2 and En-6. The En-A1 molecular architecture includes an up-down-up three-helix core anchored by the disulfide bonds II-IV and V-VII, and extended regions of nonregular secondary structure. These are a well-defined 14-residue amino terminal segment that includes a 310 turn and is anchored to the molecular core by the disulfide bond I-IV, another well-defined 7-residue segment that connects the helices α-1 and α-2, and a structurally disordered carboxy-terminal 11-residue segment linked to the molecular core by the disulfide bond III-VIII (Fig. 3C). Therefore, the three-dimensional structure homology of the Arctic En-A1 pheromone with the Antarctic pheromones involves not only the approximate sequence locations of the regular helical structures and the conservation of the disulfide bonds, but also the long segments of nonregular secondary structure preceding helix α-1 and linking the helices α-1 and α-2. These nonregular structural features have no counterparts in the NMR structures of pheromones from the temperate water species Euplotes raikovi (23), and they appear to be specific traits of the cold-adapted E. nobilii pheromones. Variable length and variable three-dimensional structure of the carboxy-terminal peptide segment following helix α-3 have previously been observed also among different E. raikovi pheromones (23), indicating that this structural variability confers species specificity.
Purified En-A1 preparations were used in combination with preparations of the Antarctic pheromone En-6 to mimic in vitro the protein–protein interactions that pheromones are expected to carry out with their receptors to elicit cell mating activity. Results from studies on E. raikovi (24) suggest that the pheromone receptors are membrane-bound pheromone isoforms, characterized in each cell type by an extracellular domain that is a structural counterpart to the secreted pheromone capable of selective, competitive binding to multiple closely homologous pheromone molecules (25). Mutual binding reactions between En-6 and En-A1 pheromone preparations were analyzed with Surface Plasmon Resonance experiments, by linking one preparation to a sensor chip to act as a ligand and injecting the second preparation into the flow chamber of the experimental apparatus to act as the analyte (Fig. 3D). The equilibrium constants (Kd) were 2 × 10-6 M for En-A1 binding to immobilized En-6 and 7.4 × 10-6 M for En-6 binding to immobilized En-A1. These values imply that the two structurally homologous pheromones of Antarctic and Arctic origin specifically interact with each other.
Conclusions
This paper provides evidence from structural biology of protein pheromones and interstrain breeding analyses that, in addition to taxonomic implications derived from morphological and phylogenetic observations, demonstrates that Antarctic and Arctic populations of E. nobilii are genetically homogeneous and interfertile, and thus form a unique biological species with a bipolar distribution. Genetic exchange among high-latitude populations was previously proposed to occur, on the basis of SSU-rDNA sequence comparisons, in three distinct morphospecies of planktonic foraminifers collected along transects between Iceland and Greenland, and between the Falkland Islands and the Antarctic Peninsula (26). Similar to these foraminiferal species and other polar microorganisms (27), the Antarctic and Arctic E. nobilii populations thus seem to “tunnel” their geographic separation by relying on the dispersal of swarming individuals in the stably cold (4–5 °C) deep currents of the oceanic waters. An additional potential force that preserves the status of cospecific populations has been shown here to be represented by the pheromone-mediated cell-cell communication that Euplotes species evolved in association with their mating-type mechanism (28). Because of their activity in promoting not only mating interactions but also cell-vegetative proliferation through interactions of autocrine type with the same cells from which they are secreted (29), Euplotes pheromones can be regarded, like their structurally related species-specific attractins of the mollusk Aplysia (30), as key signaling molecules in directing phenomena of speciation and environmental radiation of natural populations. By determining the molecular structures of Antarctic and Arctic E. nobilii pheromones, we provided direct evidence that these water-borne signaling molecules, in addition to being structurally homologous and cross-reactive, possess common specific cold-adaptive modifications while maintaining the compactness of “disulfide-rich small proteins” (31) that can ensure a long-lasting activity and enable wide-range dispersion in variable environments.
Materials and Methods
Strain cultivation.
Antarctic and Arctic strains were maintained as stable laboratory cultures in cold rooms at 2–4 °C, while the Fuegian strains were maintained at 6–8 °C. Nutrients were provided by the green algae Dunaliella tertiolecta grown in natural sea water supplemented with Walne medium.
Morphology.
Morphology was studied on silver- and Feulgen-stained specimens prepared according to standard protocols (32). The determination of the phenotypic traits was carried out on ten selected specimens of each preparation at 1000× magnification on a Leica DMR microscope, using the dedicated Leica IM1000 version 1.0 software STATISTICA (StatSoft, Inc.).
Phylogenetic Analysis.
The nuclear SSU-rRNA gene sequences were amplified by polymerase chain reaction (PCR), using the 18S universal eukaryotic forward primer F9 [5′-CTGGTTGATCCTGCCAG-3′] and the 18S Hypo reverse primer R1513 [5′-TGATCCTTCYGCAGGTTC-3′] (33, 34). The sequences were aligned using the program ClustalX and the alignment was adjusted interactively with the program BioEdit Sequence Alignment Editor to optimize the base-pairing scheme of the rRNA gene molecule secondary structure (35). The phylogenetic analysis was performed in PAUP v4.0b10 (36), applying a neighbor-joining method and using Modeltest 3.06 (37) to select the appropriate model of substitution. The Akaike information criterion indicated that the Tamura-Nei (TrN) model (38), which considers unequal base frequencies, was most appropriate (-lnL = 2872.9131). The settings for the TrN model produced the following frequencies: 0.2673 for A, 0.2030 for C, 0.2436 for G, and 0.2861 for T. The reliability of the internal branches of the phylogenetic tree was assessed using the bootstrap method (39).
Mating.
Mating mixtures were prepared from cultures resuspended in fresh sea water for 1–2 d at an adjusted concentration of about 3 × 103 cells/mL. The intensity of mating reactions was assessed from 2–3 d after cell mixing as ratio of paired to single cells. Mating pairs were individually isolated in a few drops of filtrate of their original mixtures, and left to separate into the two exconjugant cells and generate offspring clones. Ten pairs of sister progeny clones were expanded in stable cultures and used to determine the type of parental mating pairs.
Pheromone Protein Preparation.
Homogeneous pheromone preparations were obtained from culture filtrates according to a standard procedure originally devised to purify E. raikovi pheromones (40). On average, 200–300 μg of homogenous protein were extracted from 10 L of culture filtrate. The purified material was stored at -20 °C after lyophilization, and no significant loss of activity was observed, as verified by mating induction assays (41).
NMR Structure Determination of Pheromone En-A1.
All NMR measurements were performed at 25 °C with a 1 mM aqueous En-A1 pheromone solution containing 10% D2O and 20 mM sodium phosphate buffer at pH 6.0. The following spectra were acquired: 2D 2QF [1H,1H]-COSY; 2D [1H,1H]-TOCSY with 60 ms mixing time; 2D [13C,1H]-HSQC (all at 600 MHz); 2D [1H,1H]-NOESY with 120 ms mixing time (at 800 MHz). NMR assignments were obtained following standard techniques for proteins at natural isotope abundance (42). The En-A1 molecular structure was calculated following a standard protocol consisting of seven automated cycles of the ATNOS/CANDID/DYANA programs (43–45), and subsequent energy-minimization in explicit water with the program OPALp (46, 47). Statistics of the structure determination are reported in Table S4.
Surface Plasmon Resonance Analysis.
In Surface Plasmon resonance experiments carried out on a BIAcore-X instrument, pheromones were immobilized at 25 °C on the activated surface of CM5 sensor chips by amine coupling of their primary amino groups, as indicated by the manufacturer. All binding experiments were carried out at 4 °C with a flow rate of 5 μL/ min, using pheromones dissolved at different concentrations in 10 mM acetate buffer at pH 5.5 containing 150 mM NaCl and 0.005 % surfactant P-20 (BIAcore).
Supplementary Material
Acknowledgments.
This study was funded by the National Program for Antarctic Research (PNRA) to the P.L. and F.D. laboratories, and by the ETH Zürich through research support to K.W. Support for B.P. provided by the Schweizerischer Nationalfonds Fellowship PA00A/104097-1. K.W. is the Cecil H. and Ida M. Green Professor of Structural Biology at the Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2KK2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019432108/-/DCSupplemental.
References
- 1.Green JL, Bohannan BJM, Whitaker RJ. Microbial biogeography: From taxonomy to traits. Science. 2008;320:1039–1043. doi: 10.1126/science.1153475. [DOI] [PubMed] [Google Scholar]
- 2.Fraser C, et al. The bacterial species challenge: Making sense of genetic and ecological diversity. Science. 2009;323:741–746. doi: 10.1126/science.1159388. [DOI] [PubMed] [Google Scholar]
- 3.Finlay BJ, Esteban GF, Fenchel T. Global diversity and body size. Nature. 1996;383:132–133. [Google Scholar]
- 4.Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- 5.Fenchel T, Finlay BJ. The ubiquity of small species: patterns of local and global diversity. Bioscience. 2004;54:777–784. [Google Scholar]
- 6.Fenchel T. Cosmopolitan microbes and their “cryptic” species. Aquat Microb Ecol. 2005;41:49–54. [Google Scholar]
- 7.Foissner W. Biogeography and dispersal of microorganisms: A review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 8.Caron DA. Protistan biogeography: Why all the fuss? J Eukaryot Microbiol. 2009;56:105–112. doi: 10.1111/j.1550-7408.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- 9.Nanney DL. Experimental Ciliatology. New York: Wiley; 1980. [Google Scholar]
- 10.Lynn DH. The Ciliate Protozoa. Berlin: Springer; 2008. [Google Scholar]
- 11.Gates MA. Analysis of positional information applied to cirral patterns of the ciliate Euplotes. Nature. 1977;268:362–364. [Google Scholar]
- 12.Borror AC, Hill BH. The order Euplotida (Ciliophora): Taxonomy, with division of Euplotes into several genera. J Eukaryot Microbiol. 1995;42:457–466. [Google Scholar]
- 13.Valbonesi A, Luporini P. Description of two new species of Euplotes and Euplotes rariseta from Antarctica. Polar Biol. 1990;11:47–53. [Google Scholar]
- 14.Bernhard D, et al. Phylogenetic relationships within the class Spirotrichea (Ciliophora) inferred from small subunit rRNA gene sequences. Mol Phylogenet Evol. 2001;21:86–92. doi: 10.1006/mpev.2001.0997. [DOI] [PubMed] [Google Scholar]
- 15.Vallesi A, Di Giuseppe G, Dini F, Luporini P. Pheromone evolution in the protozoan ciliate, Euplotes: The ability to synthesize diffusibile forms is ancestral and secondarily lost. Mol Phylogenet Evol. 2008;47:439–442. doi: 10.1016/j.ympev.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Dini F, Nyberg D. In: Advances in Microbial Ecology. Jones JG, editor. Vol. 13. New York: Plenum; 1993. pp. 85–153. [Google Scholar]
- 17.Felici A, Alimenti C, Ortenzi C, Luporini P. Purification and initial characterization of two pheromones from the marine Antarctic ciliate, Euplotes nobilii. Ital J Zool. 1999;66:355–360. [Google Scholar]
- 18.Valbonesi A, Luporini P. Biology of Euplotes focardii, an Antarctic ciliate. Polar Biol. 1993;13:489–493. [Google Scholar]
- 19.Vallesi A, et al. Characterization of the pheromone gene family of an Antarctic and Arctic protozoan ciliate, Euplotes nobilii. Mar Gen. 2009;2:27–32. doi: 10.1016/j.margen.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 20.Placzek WJ, et al. Cold-adapted signal proteins: NMR structures of pheromones from the Antarctic ciliate Euplotes nobilii. IUBMB Life. 2007;59:578–585. doi: 10.1080/15216540701258165. [DOI] [PubMed] [Google Scholar]
- 21.Pedrini B, et al. Cold-adapted in sea-water-borne signal proteins: sequence and NMR structure of the pheromone En-6 from the Antarctic ciliate Euplotes nobilii. J Mol Biol. 2007;372:277–286. doi: 10.1016/j.jmb.2007.06.046. [DOI] [PubMed] [Google Scholar]
- 22.Alimenti C, et al. Molecular cold-adaptation: comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate Euplotes. IUBMB Life. 2009;61:838–845. doi: 10.1002/iub.228. [DOI] [PubMed] [Google Scholar]
- 23.Luginbühl P, Ottiger M, Mronga S, Wüthrich K. Structure comparison of the pheromones Er-1, Er-10, and Er-2 from Euplotes raikovi. Protein Sci. 1994;3:1537–1546. doi: 10.1002/pro.5560030919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortenzi C, et al. The autocrine mitogenic loop of the ciliate Euplotes raikovi: the pheromone membrane-bound forms are the cell binding sites and potential signalling receptors of soluble pheromones. Mol Biol Cell. 2000;11:1445–1455. doi: 10.1091/mbc.11.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallesi A, et al. Autocrine, mitogenic pheromone receptor loop of the ciliate Euplotes raikovi: pheromone-induced receptor internalization. Eukaryot Cell. 2005;4:1221–1227. doi: 10.1128/EC.4.7.1221-1227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darling KF, et al. Molecular evidence for genetic mixing of Artic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. [DOI] [PubMed] [Google Scholar]
- 27.Montresor M, et al. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 2003;26:186–194. [Google Scholar]
- 28.Luporini P, Alimenti C, Ortenzi C, Vallesi A. Ciliate mating types and their specific protein pheromomes. Acta Protozool. 2005;44:89–101. [Google Scholar]
- 29.Vallesi A, Giuli G, Bradshaw RA, Luporini P. Autocrine mitogenic activity of pheromones produced by the protozoan ciliate Euplotes raikovi. Nature. 1995;376:522–524. doi: 10.1038/376522a0. [DOI] [PubMed] [Google Scholar]
- 30.Painter SD, et al. Structural and functional analysis of Aplysia attractins, a family of water-borne protein pheromones with interspecific attractiveness. Proc Natl Acad Sci USA. 2004;101:6929–6933. doi: 10.1073/pnas.0306339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson J. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 32.Foissner W, Berger H, Schaumburg J. Identification and Ecology of Limnetic Plankton Ciliates. Munich: Bavarian State Office for Water Management; 1999. [Google Scholar]
- 33.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 34.Petroni G, Dini F, Verni F, Rosati G. A molecular approach to the tangled intrageneric relationships underlying phylogeny in Euplotes (Ciliophora, Spirotrichea) Mol Phylogenet Evol. 2002;22:118–130. doi: 10.1006/mpev.2001.1030. [DOI] [PubMed] [Google Scholar]
- 35.Van de Peer Y, De Rijk P, Winkelmans J, De Watcher R. The European small subunit ribosomal RNA database. Nucleic Acids Res. 2000;28:175–176. doi: 10.1093/nar/28.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 37.Posada D, Crandall KA. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 39.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 40.Luporini P, Raffioni S, Concetti A, Miceli C. The ciliate Euplotes raikovi heterozygous at the mat genetic locus co-releases two individual species of mating pheromone: Genetic and biochemical evidence. Proc Natl Acad Sci USA. 1986;83:2889–2893. doi: 10.1073/pnas.83.9.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyake A, Beyer J. Blepharmone: A conjugation-inducing glycoprotein in the ciliate Blepharisma. Science. 1974;185:621–623. doi: 10.1126/science.185.4151.621. [DOI] [PubMed] [Google Scholar]
- 42.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 43.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 44.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 45.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 46.Luginbühl P, Güntert P, Billeter M, Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J Biomol NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 47.Luginbühl P, Güntert P, Billeter M, Wüthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J Biomol NMR. 1997;9:212. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.