Abstract
The pore domain of voltage-gated potassium (Kv) channels consists of transmembrane helices S5 and S6, the turret, the pore helix, the selectivity filter, and the loop preceding S6, with a tertiary reentrant structure between S5 and S6. Using biogenic intermediates, mass tagging (pegylation), and a molecular tape measure, we explored the possibility that the first stages of pore formation occur prior to oligomerization of the transmembrane core. Pegylation of introduced cysteines shows that the pore helix, but not the turret, forms a compact secondary structure in the terminal 20 Å of the ribosomal tunnel. We assessed the tertiary fold of the pore loop in monomeric constructs by determining the relative accessibilities of select cysteines using the kinetics of pegylation. Turret residues are accessible at the extracellular surface. In contrast, pore helix residues are less accessible. All-atom molecular dynamics simulations of a single Kv monomer in a solvated lipid membrane indicate that secondary and tertiary folds are stable over 650 ns. These results are consistent with acquisition of a tertiary reentrant pore architecture at the monomer stage of Kv biogenesis and begin to define a plausible sequence of folding events in the formation of Kv channels.
Keywords: potassium channel biogenesis, permeation pore, protein folding, nascent peptide
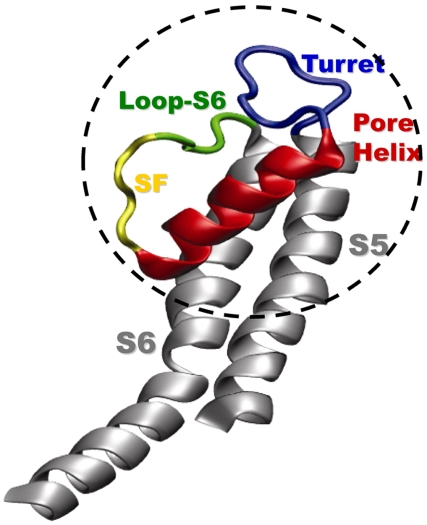
The pore of a selective potassium channel has evolved to be highly selective and rapidly conducting. These permeation properties derive from the exquisitely precise architecture of the tetrameric channel protein (1). Voltage-gated potassium (Kv) channels have six transmembrane segments, S1–S6 and a cytosolic T1 tetramerization domain. The pore region is composed of S5, S6, and an intervening pore loop consisting of a turret, the pore helix, the selectivity filter, and a pre-S6 loop (Fig. 1). Reentry of the pore loop between S5 and S6 is necessary to position the selectivity filter facing both intra- and extracellular aqueous vestibules. The pore loop is structurally highly conserved among all ion channels. Folding defects in this region will lead to channelopathies. For example, mutations in this region cause long QT syndrome and can be fatal (2, 3). Folding of Kv channels, as with all proteins, begins as early as the birth of the nascent peptide attached to the ribosome and continues during its tenure in the endoplasmic reticulum (ER) (4–7). Yet, we know little regarding the sequence of events responsible for the critical folding of the pore itself, e.g., when are the secondary and tertiary folds that define the structure of the reentrant pore loop acquired?
Fig. 1.
The Kv channel pore architecture. Monomer of the Kv1.2 pore region, reproduced from Long et al. (13). The turret is depicted in blue, the pore helix in red, the selectivity filter in yellow, the loop preceding S6 (loop-S6) in green, and transmembrane segments S5 and S6 in gray. The dashed circle highlights the secondary and tertiary architecture investigated in this paper.
Here, we explore the possibility that pore structure forms early in biogenesis, in the monomer Kv1.3 channel protein. Fig. 1 (dashed circle) delineates the secondary and tertiary architecture probed in this study. Using a cysteine scan of the pore helix and turret region in a nascent (attached to the ribosome) Kv1.3 and a mass-tagging strategy, we demonstrate that compaction of the pore helix occurs in the distal portion of the ribosomal tunnel near the exit port. Using the kinetics of modification of cysteines engineered into the turret, the pore helix, and the loop preceding S6 (loop-S6) of Kv1.3, we show that tertiary folding of the pore region occurs in the Kv monomer embedded in the ER membrane. This finding is consistent with all-atom molecular dynamics simulations (650 ns) of a single Kv monomer pore domain in a fully solvated lipid bilayer. These simulations suggest that the monomeric pore domain remains stable in a conformation similar to that observed in the crystal structure of the tetramer in the plasma membrane. Therefore, critical folding events that underlie the conserved pore structure of mature Kv channels occur in the very earliest steps of protein biosynthesis.
Results
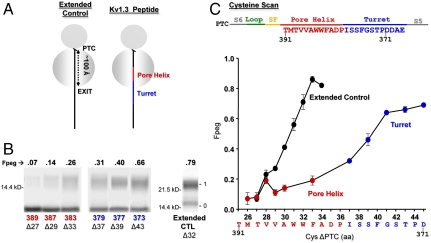
To investigate structure acquisition of the Kv pore region, we focused on two questions. First, does the pore helix form a compact structure within the monomer inside the ribosome, and hence early in biogenesis? Second, does the reentrant loop of the pore acquire its tertiary folded structure in the monomer? We began by addressing the first question. Transmembrane and linker segments of Kv1.3 acquire helicity while exiting the ribosomal tunnel (5, 7). With these precedents, it is entirely reasonable to hypothesize that the pore helix also compacts at this stage. To test this hypothesis, we used a cell-free rabbit reticulocyte lysate translation system that contains all the ingredients necessary to translate mRNA into a peptide. To study the Kv peptide at a defined stage of translation in which the pore helix is in the ribosomal exit tunnel, we created a biogenic intermediate of Kv1.3 that represents an arrested snapshot of the elongating Kv peptide. Such intermediates are made from DNA truncated with a restriction enzyme so that the resulting mRNA lacks a stop codon and thus arrests on the ribosome. We refer to this peptide as a nascent peptide, still attached to tRNA and the ribosome at the peptidyl transferase center (PTC). An SgrAI-cut DNA yields a nascent peptide with transmembrane segment S5 (plus the C terminus of the S4–S5 linker), the turret, the pore helix, the selectivity filter, loop-S6, and the N-terminal half of transmembrane segment S6 (Fig. 2A, right cartoon).
Fig. 2.
Probing secondary structure formation in the ribosomal exit tunnel. (A) Cartoons of two different nascent peptide–ribosome complexes. The gray circles represent the two subunits of the ribosome. The PTC, at the cleft between the two subunits, is approximately 100 Å from the exit port of the tunnel. An all-extended peptide (molecular tape measure) is represented by a solid black line (Left) and a nascent fragment of Kv1.3 containing the pore helix (red) and turret (blue) is depicted schematically as a solid line (Right). The native Kv1.3 peptide is attached to the PTC at residue A415 (SgrAI restriction enzyme). The complex is not drawn to scale. (B) Pegylation of the pore helix and turret. Nascent peptides were translated from mRNA engineered from a DNA that lacked a stop codon, pegylated (1 mM PEG-MAL for 4–6 h), and fractionated using SDS-PAGE as described in the SI Appendix. The number directly under each lane indicates the residue in Kv1.3 mutated to cysteine (pore helix residues in red, turret residues in blue); the number with a Δ indicates the number of amino acids from the PTC to (and including) the cysteine; the numbers to the left of the gel correspond to molecular weight standards; numbers to the right of the gel indicate unpegylated (0) and singly pegylated (1) protein; the number directly above each lane indicates the calculated fraction of nascent peptide pegylated (Fpeg). The rightmost lane is the all-extended control (Extended CTL). (C) Cysteine scan of the pore helix and turret of nascent Kv1.3 peptide. The x axis is the number of amino acids from the PTC to (and including) the labeled cysteine. Pegylation of a known all-extended nascent peptide, is shown by the black circles and represents data taken from Lu and Deutsch (5). The final extent of pegylation of individual residues in the pore helix and turret is represented by the red and blue circles, respectively. Symbols are mean ± SEM (n≥3).
We engineered cysteines, one at a time, in this construct and determined the extent to which each cysteine could be covalently modified by PEG maleimide (PEG-MAL, 5 kD). The adduct is detected as a shift in mobility of the peptide on a protein gel (5, 8). We refer to this mass-tagging method as pegylation. The results are calibrated against a cysteine scan of an all-extended molecular tape measure (Fig. 2A, left cartoon), for which the extent of cysteine labeling depends on the distance from the PTC to each individual cysteine (5). The tape measure is a 95-residue nascent chain composed of the N terminus of Kv1.3. This N terminus includes a portion of the T1 sequence that is known to be all-extended and has been engineered to reside inside the ribosomal exit tunnel. Tape-measure cysteines that are 26 amino acids from the PTC are relatively inaccessible to PEG-MAL. As the target cysteine is moved increasingly further than 26 residues from the PTC, labeling increases monotonically. Those residues that are ≥33 amino acids from the PTC are ≥80% labeled with PEG-MAL (rightmost gel, Fig. 2B; black symbols, Fig. 2C). For an all-extended peptide, 33 residues corresponds to a distance of approximately 99–112 Å (using 3–3.4 Å/amino acid) between the PTC and the exit port, in excellent agreement with anatomical dimensions of the tunnel derived from structural studies (9).
A cysteine scan of selected pore helix residues positioned in the lower portion of the exit tunnel, e.g., residues 389, 387, and 383, shows relatively little pegylation even for a residue that is 30 amino acids from the PTC (Fig. 2B, three leftmost lanes: band 1 vs band 0). In contrast, turret residues, 379, 377, and 373 (center three lanes), show relatively more pegylation, e.g., 66% for residue 373 near the top of the turret. A comparison of the fraction pegylated (Fpeg) for the pore helix (red symbols, Fig. 2C) with the all-extended tape measure reveals a shallower slope and right-shifted curve, whereas the turret (blue symbols) has a steeper slope compared to the pore helix. However, it is somewhat shallower than the tape measure, which is likely due to some contamination of the region probed with the N terminus of the pore helix. Nonetheless, the pore helix is relatively more compact than the turret, as expected from the mature structure.
Tertiary Structure of the Pore.
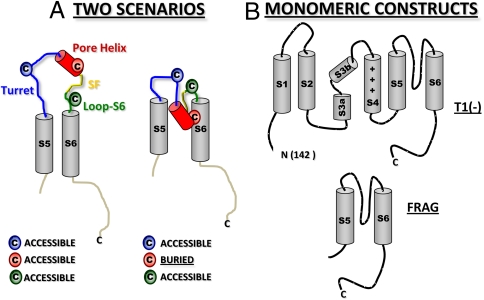
Having determined that the pore helix may already be compact as it exits the ribosomal tunnel, we considered two extreme scenarios for tertiary folding of the pore (Fig. 3A). The cartoon on the left depicts the S5 and S6 segments (gray), which are normally anchored in the membrane, and the intervening pore loop as unstructured and accessible in the aqueous compartment of the ER lumen (topologically equivalent to the extracellular compartment). The second scenario (Fig. 3A, Right) is derived from the crystal structure of the tetrameric Kv1.2/2.1 chimera (10) and manifests a reentrant pore. The two scenarios make different predictions regarding the accessibility of engineered cysteines. In the first scenario, all the indicated cysteines would be accessible to the aqueous compartment; in the second scenario, only those in the exposed turret and loop-S6 would be accessible. The cysteine at the bottom of the pore helix would be buried at protein or lipid interfaces. We used the kinetics of pegylation to explore these predictions. An accessible residue will have a relatively fast modification rate, whereas a buried residue will have a relatively slow modification rate. For these experiments, we chose two Kv1.3 constructs, both shown to be monomeric in the ER membrane (8): a T1-deletion [T1(-)] and an S5-P-S6C-terminus fragment (FRAG; Fig. 3B). The T1(-) peptide is monomeric under the conditions of our experiments, but can produce tetramers under high-RNA conditions and long times (8, 11). In contrast, FRAG never forms tetramers (8) but inserts into the ER membrane with correct orientation (12). A further advantage of FRAG is that the folding phenotype we discover will reflect folding of the pore region itself in the monomer independent of a voltage sensor domain (S1–S4) and therefore be relevant for folding of two-transmembrane-segment K+ channels, e.g., KcsA and Kir.
Fig. 3.
Strategy for assessing tertiary folding. (A) Two scenarios. Depicted are predictions for relative pegylation kinetics for individually engineered cysteines in three component segments of the pore loop between S5 and S6 for an unfolded tertiary pore loop (Left) and a reentrant pore loop (Right). (B) Monomeric constructs for the accessibility assay include Kv1.3 T1(-) (Upper), which lacks only the N-terminal residues 1–141, and a Kv1.3 S5-P-S6C-terminus fragment (FRAG; residues 336–523) (Lower).
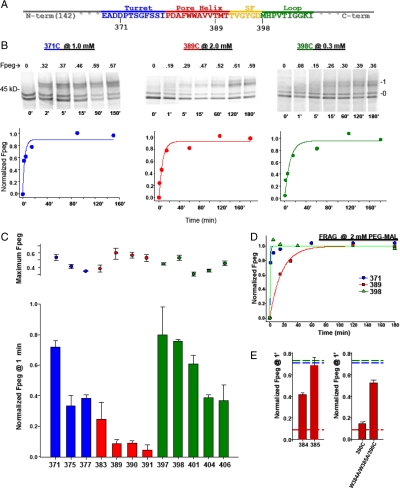
We began with the T1(-) construct, translating it in the presence of microsomal membranes. We solubilized the isolated membranes in dodecylmaltoside (C12M, 0.1%), typically a nondenaturing detergent (SI Appendix), and assessed the accessibility of specific cysteines in the turret, pore helix, and loop-S6. The kinetics of pegylation of residues 371, 389, and 398 in the turret (blue), pore helix (red), and loop-S6 (green), respectively, are shown in Fig. 4B. Note that the concentration of PEG-MAL used in these modification reactions was 1, 2, and 0.3 mM, respectively, in order to capture well-resolved time courses despite the difference in modification reaction rates. For each gel, the lower doublet represents the unpegylated peptide (“0” band), comprised of both unglycosylated (lower band of the doublet) and core (ER) glycosylated (upper band of the doublet) peptide. The upper band(s) represents the pegylated peptide (band “1”). For each indicated time, Fpeg was calculated, normalized to the final data point, and plotted in Fig. 4B below each series of gels. The data show monotonically increasing values that saturate and could be fit with a single exponential to give modification rate constants of 20.2, 1.3, and 6.3 M-1 s-1, respectively. The turret residue, 371, and the loop-S6 residue, 398, are each modified much faster than the pore helix residue, 389. The latter is less accessible than the former two, consistent with a reentrant architecture of the pore. Replicate experiments for each of these cysteines gave mean rate constants of 15.5 ± 2.4 (n = 3, ± SEM), 1.0 ± 0.3 (n = 2, ± average deviation), and 6.6 ± 0.4 (n = 3, ± SEM) M-1 s-1, for 371, 389, and 398, respectively. The rate constant in the pore helix is significantly lower than that in either the turret (P = 0.02) or the loop-S6 (P = 0.002). Residue 390, also at the bottom of the pore helix, gives a mean rate constant of 0.4 ± 0.1 (n = 3, ± SEM) M-1 s-1, which is significantly different from the turret and loop-S6 (both P ≤ 0.003). These results (also see SI Appendix) suggest that residues in the C-terminal pore helix are more hindered and the pore region is folded in the T1(-) Kv1.3 monomer.
Fig. 4.
Relative pegylation kinetics of select cysteines in Kv1.3T1(-) and FRAG. Cysteines were engineered, one at a time, in each of three segments: the turret, the pore helix, and loop-S6, and pegylated. (A) The primary sequence of the pore region. (B) Pegylation kinetics of D371C (turret, blue), T389C (pore helix, red), and M398C (loop-S6, green) were determined at 1, 2, and 0.3 mM PEG-MAL, respectively. Each lane represents a sample quenched at the indicated time (below each lane, in minutes) and the number directly above each lane indicates the calculated Fpeg. Right and left numbers as in Fig. 2B. For each residue, a time course of normalized Fpeg is plotted below each set of gels and fit with a single exponential. (C) Normalized Fpeg at one minute in T1(-) for residues in the turret (blue), pore helix (red), and loop-S6 (green). The final PEG-MAL concentration was 2 mM in all reactions shown here. Identical pegylation reactions were quenched at 1 min and at > 3 h, fractionated using SDS-PAGE, and quantified as described in the SI Appendix. Pegylation at 1 min was normalized to the maximum fraction pegylated at > 3 h (maximum Fpeg, dot plot) and shown as Fpeg at 1 min (bar graph). Symbols are mean ± SEM (n≥3) for all residues except 377 and 397, which are mean ± average deviation for duplicate samples. (D) Time course of cysteine modification in FRAG. Cysteines engineered in the turret (371), the pore helix (389), and the loop-S6 (398) were engineered in the background of FRAG and pegylated (2 mM PEG-MAL), as described in Fig. 4B. Results were fit with a single exponential. (E) Cysteines were engineered in the pore helix at positions 384 and 385, each a tryptophan in the native sequence. (Left) W384C and W385C, separately, were pegylated (2 mM PEG-MAL) as described in Fig. 4B. Data are shown as Fpeg at 1 min normalized to the maximum Fpeg at 4 h. Values are mean ± SEM for n = 3. The red dashed line (derived from Fig. 4C) indicates a minimum Fpeg at 1 min for pore helix residues and the blue and green dashed lines (derived from Fig. 4C) indicate a maximum Fpeg value at 1 min for the turret and loop-S6, respectively. (Right) W384C and W385C were simultaneously mutated in the background of M390C and pegylated as described in Fig. 4B. The normalized Fpeg at 1 min for this triple mutant, W384A/W385A/M390C, is compared with M390C alone, and shown as mean ± SEM, n = 3, for M390C, and mean ± average deviation, n = 2, for the triple mutant. Dashed lines as described above for Fig. 4E, Left.
If the folded monomer is stable, then those residues ultimately fated to be at an intersubunit interface in the mature Kv channel will no longer be masked by protein in the ER membrane, whereas those residues fated to be at an intrasubunit or lipid interface will remain masked by the lipid or protein. For example, residue 398, equivalent to residue L81 in KcsA and M380 in Kv1.2, is located at an intersubunit interface according to the models derived from the respective crystal structures (1, 13). Additionally, this residue can be crosslinked to produce dimers and the equivalent mutation in Shaker (M448C) also can be crosslinked to give dimers (14). This residue should therefore be relatively accessible in the T1(-) Kv1.3 monomer, which is indeed the case (see also Fig. 4C).
To probe these regions more completely, we carried out a more extensive cysteine scan of the three regions, using a fixed concentration of PEG-MAL (2 mM). The range of normalized Fpeg at 1 min is approximately 0.4–0.7 for turret residues, approximately 0.1 for the bottom of the pore helix, and approximately 0.4–0.8 for loop-S6 residues (Fig. 4C). A pattern emerges that indicates relatively faster modification rates for the turret and loop-S6 than for the pore helix, consistent with a reentrant pore architecture in the T1(-) monomer. To support this conclusion, we probed FRAG for the same three residues (371, 389, and 398) shown in Fig. 4B. Fig. 4D recapitulates the results obtained for the T1(-) monomer. The modification rate for the 389 residue is much slower, approximately 0.4 M-1 s-1 compared with that for the 371 and 398 residues. We suggest that the pore is reentrant in the monomer and does not require a tetramer to achieve a pore-like architecture. Furthermore, we do not think that the pore helix predominantly lies parallel to the membrane surface, but rather that it adopts a reentrant position. This conclusion derives from three arguments. First, the pore helix itself is not amphipathic and has a mean relative hydrophobic moment of 0.12, which represents its hydrophobic moment relative to a perfectly amphipathic peptide (Eisenberg scale, HydroMCalc; http://www.bbcm.univ.trieste.it/~tossi/HydroCalc/HydroMCalc.html). Second, there are many strong interactions between the pore helix and S5 and S6 within the monomer proper (see calculated van der Waals interaction energies, Fig. 5C), so that the pore helix is stable as a reentrant loop. Third, three adjacent residues, 389, 390, and 391, at the C terminus of the pore helix, each exhibit slow modification rate constants. It is unlikely that all three residues would be slow if the pore helix were lying at the membrane–water interface parallel to the membrane. The most parsimonious explanation for the observed pattern of rate constants is a reentrant loop in the monomer.
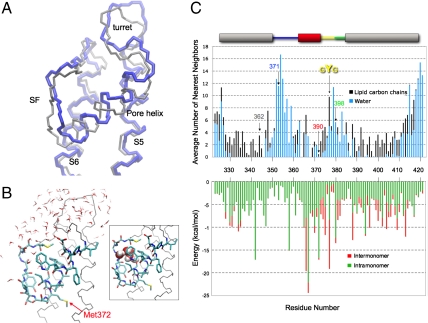
Fig. 5.
Stability of the reentrant pore loop. (A) Overlay of the monomer pore domain from the crystal structure of Kv1.2 (gray) and the pore domain at the 650th nanosecond of the molecular dynamics simulation (blue). (B) Snapshot of Kv1.2 pore domain at 650 ns. The pore helix and the selectivity filter backbone atoms (except O) and side chains, including Met372 (corresponds to Kv1.3 Met390), are shown. Waters within 4 Å of protein at 650 ns are shown. Pictures were prepared in the molecular viewing program VMD. (Inset) Same structure as in B, highlighting five water molecules between the pore helix and selectivity filter as space filling Corey–Pauling–Koltun atoms. (C, Upper) Average number of water and lipid heavy atoms around each residue. Number of water (blue bar) or lipid (black bar) heavy atoms (all atoms except for hydrogen) calculated within 4 Å of each heavy atom in each side chain and averaged throughout the 650-ns simulation. The bar along the top of the plot is color coded as described in Fig. 1 to indicate Kv segments. The numbers along the x axis are residue numbers of native Kv1.2. Tcl command interface of the VMD program were used to perform the analysis. Residues 346, 364, and 368 are probably surrounded only by protein (i.e., neither lipid nor water). The corresponding Kv1.3 residues 362 (S5, gray), 371 (turret, blue), 390 (pore helix, red), and 398 (loop-S6, green) are indicated by arrows, as is the GYG sequence (yellow). (Lower) The van der Waals interaction energies within a Kv1.2 monomer (green bars) and between one monomer and its three partner subunits in the tetramer Kv1.2 (red bars), both calculated from the Kv1.2 crystal structure.
If the monomer is folded, then unfolding the pore should increase the relative accessibility of a residue at the bottom of the pore helix. We undertook two approaches to explore this hypothesis. First, we solubilized the ER membrane-integrated T1(-) peptides in lithium dodecyl sulfate, a denaturing detergent, pegylated and determined the Fpeg. Under these conditions, residues 371, 389, and 398 gave normalized 1-min Fpeg values of 0.97, 0.62, and 0.99, respectively, a marked increase over those shown in Fig. 4C for the folded monomer. Second, we mutated residues in the pore helix that might contribute to folding free energy. We chose the two adjacent tryptophan residues (W384 and W385) in the middle of the pore helix of Kv1.3 because tryptophans have a preference for membrane interfaces (15–17). W384C and W385C each exhibit increased pegylation at 1 min (Fig. 4E, left bar graph), compared to other pore helix residues (dashed red line, derived Fig. 4C). If these results are a consequence of unfolding of the pore tertiary structure due to removal of aromatic residues, then a 390C at the bottom of the pore helix should be more accessible in a construct in which the W384 and W385 have been mutated to nonaromatic residues. The triple mutant, W384A/W385A/M390C, is pegylated approximately fivefold faster than the single M390C mutant containing the WW motif (Fig. 4E, right bar graph), consistent with increased accessibility of 390C. Mutation of either or both tryptophans fails to produce current upon transfection in Xenopus oocytes, consistent with failure of the channel to form a proper pore architecture.
Molecular Dynamics Simulations of a Kv Monomer.
To examine the conclusion that the reentrant pore loop can exist in the Kv monomer, we performed all-atom molecular dynamics simulations. The simulation system was constructed from the crystal structure of the Kv1.2 [Protein Data Bank (PDB) ID 2A79] pore domain (99 residues from the beginning of S5, Ala323, to the end of S6, Thr421) embedded in palmitoyloleoylphosphatidylcholine (POPC) and surrounded by an aqueous solution of 150-mM KCl. After a 50-ns equilibration, the structure was simulated for an additional 650 ns. Fig. 5A shows a backbone structure at 650 ns (blue) superimposed on the backbone of the crystal structure (gray). There are some local structural deviations (rmsd is 3.9 Å for all atoms and 3.0 Å for α-carbon atoms for this snapshot at 650 ns) along the pore loop and transmembrane segments, and the pore helix is shifted about a half turn toward the extracellular side in the 650-ns snapshot. Nevertheless, the tertiary reentrant fold of the pore loop is manifest in both structures. The time-dependence of the rms deviations of the monomer relative to either the crystal structure or the final snapshot of the trajectory (650 ns) indicates that the monomer stabilizes in its final conformation after 350 ns.
In Fig. 5B, a detailed snapshot of the simulated structure at 650 ns is shown depicting side chains and water molecules located within 4 Å of the protein. The inset (box) highlights the presence of a mini-water-droplet, five water molecules, lodged between the pore helix and the selectivity filter, which likely contributes to stabilizing the tertiary fold. The surrounding water molecules, along with surrounding lipid carbons are represented quantitatively in Fig. 5C, Upper. Most of transmembrane segments S5 and S6, especially the middle stretch of residues, is in contact with lipid chains, whereas the turret and loop-S6 are in contact with water molecules. In contrast, for most of the pore helix and selectivity filter, residues are in contact with some lipid and some water (see SI Appendix). The pore helix does not need to reside at the lipid–water interface to be hydrated. This hydration of the reentrant helix may be sufficient for cysteine modification to occur. It is striking that the profile for the water distribution is very similar to that for distribution of normalized Fpeg (Fig. 4C). Because cysteine modification rates are a measure of water accessible volume surrounding the cysteine side chain, the excellent correlation of the two plots is strong evidence in support of the conclusion that the pore domain of a monomer contains a reentrant pore loop.
To better understand the energetics underlying the stability of the monomer reentrant architecture, we calculated the van der Waals interaction energies from the Kv1.2 crystal structure. The van der Waals energy directly reflects the extent of atomic contact between the side chains, which are predominantly nonpolar. Intramonomer energies correspond to the interactions of the side chain of residue i in monomer A with the side chains of all residues of monomer A (excluding the nearest neighbors along the sequence i - 1 and i + 1). Similarly, intermonomer energies correspond to the interactions of the side chain of residue i in monomer A with the side chains of all residues of monomers B, C, and D in the tetramer. As shown in Fig. 5C, Lower, the major contribution (except for residues 366 and 376) is always intramonomer.
Discussion
The pore of a potassium channel was selected by evolution for its high conductance and exquisite selectivity for potassium ions. These properties derive from the precise, and highly conserved, architecture of the tetrameric channel protein, specifically that of a reentrant pore loop. Although the primary sequence of the pore is synthesized vectorially in the ribosome, the stage in Kv biogenesis when the loop between S5 and S6 folds back into the membrane is not known. The pore helix is compact in the more distal regions of the tunnel, near the exit port, whereas the turret is more extended. Because secondary structure acquired in the ribosomal tunnel is retained upon migration through the translocon (7), it is likely that the compact structure of the pore helix sequence is retained and available for tertiary folding when this segment of Kv1.3 exits the translocon. Cooperativity among secondary, tertiary, and quaternary folding events may be pivotal in acquisition of correctly folded mature Kv structure.
Tertiary Structure Formation of the Pore.
Two arguments support the hypothesis that a reentrant pore architecture may be established early in biogenesis and does not require tetramerization of Kv subunits. First, there is precedent for a structurally homologous helix/fold/helix motif among soluble proteins. Using DaliLite version 3 (http://ekhidna.biocenter.helsinki.fi/dali_server) to search a protein structure database, several examples of monomeric soluble proteins were identified that are similar (rmsd values of ≤ 3 Å; see SI Appendix) to the Kv pore segment (residues 333–398). Second, we previously showed that S5, the pore loop, and S6C-terminus likely exist as a cotranslationally folded biogenic unit (12).
Although pore architecture can arise early in biogenesis of the monomer and does not require formation of a Kv tetramer, the tertiary fold of the monomer likely differs from that in the mature tetramer because additional folding may accompany tetramerization of the pore domain. Such fine tuning would serve to optimize ion conductance and selectivity of the functional tetrameric channel. Moreover, formation of a reentrant loop might facilitate tetramerization of the transmembrane segments due to additional intersubunit interactions (1). Although the monomer—by itself—has enough van der Waals stabilization energy to maintain the conformation observed in the tetramer, tetramerization may be necessary to help catalyze the folding process, the subunits acting collectively as chaperones for one another. Regardless, the pegylation results suggest that the monomer can fold on its own.
What Drives Reentry of the Loop in the Monomer?
Our results suggest that van der Waals interactions within the monomer are sufficient to stabilize a reentrant configuration of the pore loop. Moreover, the crystal structure of the Kv1.2/Kv2.1 chimera reveals a network of interactions within 3.8 Å, consistent with extensive intrasubunit hydrogen bond formation (see SI Appendix for examples), albeit these interactions exist in the monomer within the context of the mature tetramer and may not be as extensive at an earlier biogenic stage. Mutation of some analogous hydrogen-bonding residues in Kv1.3 prevents functional expression in Xenopus oocytes. In addition, the molecular dynamics simulations reveal a droplet of five water molecules that simultaneously bridges the pore helix and selectivity filter in the tertiary folded conformation and may contribute to stabilizing the reentrant topology of the monomer.
An additional consideration is the role of aromatic pore residues in promoting reentry. Mutation of W384 and W385 to alanine produced a more accessible T1(-) M390C. The pair of tryptophans in the pore helix is highly conserved across K+ channels. Tryptophans preferentially reside at membrane interfaces (17–19). The complex properties known as aromaticity underlie this preference (17). In addition, aromatic side chains are key determinants for insertion of transmembrane segments into the lipid bilayer and topogenic events (18). We therefore suggest that this preference of aromatic residues for the lipid–water interface may also contribute to reentry of the pore sequence into the membrane. In Kv1.3, mutation of W384 or W385 to A, E, L, or P, prevents functional expression in Xenopus oocytes. In KcsA, mutations W67A or W68A in the pore helix yield a peptide that fails to assemble correctly (20). These scenarios implicate a role for aromatic pore helix residues in tertiary folding.
The sequential coordination of folding events is undoubtedly critical for acquisition of a mature structure, and our results support the plausibility that some aspects of tertiary folding, notably the formation of a reentrant pore loop, precede oligomerization. Folding of the monomer might help create an appropriate interaction interface that energetically favors tetramer formation. An alternative possibility is that tetramer formation provides a scaffold that favors the entry of an extracellular pore loop into its mature residence within the transmembrane domain. Our results indicate that this prerequisite is not necessary. Defining the sequence of folding events is key to understanding not only Kv biogenesis and its efficiency, but also subsequent trafficking mechanisms, because retention, export, and degradation signals are modulated by how the protein is folded and packed.
Methods
Constructs, in Vitro Translation, Pegylation.
All constructs were created in a pSP64 plasmid. The pSP64/full-length Kv1.3, pSP64/T1(-)Kv1.3, and pSP64/S5-P-S6C-terminus Kv1.3 (FRAG) were constructed as described previously and are cysteine free (8). Engineered cysteines and restriction enzyme sites were introduced using Stratagene™ QuikChange Kit. For details of molecular biology methods and in vitro translation see SI Appendix and ref. 8.
Isolated ribosome-nascent peptide-membrane pellets were resuspended on ice with PBS* (CaCl2- and MgCl2-free Dulbecco’s PBS, pH 7.4, supplemented with 4 mM MgCl2) containing 50 μM DTT. An equal volume of methyl PEG-MAL (Mr 5000, > 99%, SunBio, Inc.; prepared in PBS*) was added (final PEG-MAL concentration of 1–2 mM) and incubated on ice from 0 to 6 h. The pegylation reaction was quenched with 100-fold excess DTT at ambient temperature for 10 min and then 0 °C for 10 min and fractionated as described in the SI Appendix.
Pegylation Measurements for Tertiary Folding.
Pellets containing membrane-embedded peptides were resuspended on ice with 50–100 μL of 0.1% Anatrace™ n-dodecyl-β-d-maltopyranoside, Anagrade® (C12M, prepared in PBS* supplemented with 50–500 μM DTT) and incubated 1 h on ice to dissolve membranes. Samples were centrifuged to pellet undissolved membrane and the supernatant transferred to a fresh tube and an equal volume of PBS* solution containing PEG-MAL (final PEG-MAL concentration was 1–2 mM), incubated on ice from 0 to 3 h, quenched as described above, and analyzed as described in the SI Appendix.
Molecular Dynamics.
The simulation system was constructed from the crystal structure of the Kv1.2 (PDB ID 2A79) pore domain embedded in POPC surrounded by an aqueous solution of 150 mM KCl. See the SI Appendix for details.
Supplementary Material
Acknowledgments.
We thank Dr. Richard Horn for careful reading of the manuscript and helpful discussion, Timothy Pian for technical assistance, and Dr. LiWei Tu for constant guidance and helpful discussion. Supported by National Institutes of Health Grant GM 52302 (to C.D.) and GM062342 to (B.R.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017097108/-/DCSupplemental.
References
- 1.Doyle DA, et al. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 1998;280:69–76. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 2.Benson DW, et al. Missense mutation in the pore region of HERG causes familial long QT syndrome. Circulation. 1996;93:1791–1795. doi: 10.1161/01.cir.93.10.1791. [DOI] [PubMed] [Google Scholar]
- 3.Huang FD, Chen J, Lin M, Keating MT, Sanguinetti MC. Long-QT syndrome-associated missense mutations in the pore helix of the HERG potassium channel. Circulation. 2001;104:1071–1075. doi: 10.1161/hc3501.093815. [DOI] [PubMed] [Google Scholar]
- 4.Kosolapov A, Tu L, Wang J, Deutsch C. Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 6.Tu L, Wang J, Deutsch C. Biogenesis of the T1-S1 linker of voltage-gated K+ channels. Biochemistry. 2007;46:8075–8084. doi: 10.1021/bi700319f. [DOI] [PubMed] [Google Scholar]
- 7.Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J Mol Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Deutsch C. Pegylation: A method for assessing topological accessibilities in Kv1.3. Biochemistry. 2001;40:13288–13301. doi: 10.1021/bi0107647. [DOI] [PubMed] [Google Scholar]
- 9.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 10.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 11.Tu L, et al. Voltage-gated K+ channels contain multiple intersubunit association sites. J Biol Chem. 1996;271:18904–18911. doi: 10.1074/jbc.271.31.18904. [DOI] [PubMed] [Google Scholar]
- 12.Tu L, Wang J, Helm A, Skach WR, Deutsch C. Transmembrane biogenesis of Kv1.3. Biochemistry. 2000;39:824–836. doi: 10.1021/bi991740r. [DOI] [PubMed] [Google Scholar]
- 13.Long SB, Campbell EB, MacKinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Jurman ME, Yellen G. Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron. 1996;16:859–867. doi: 10.1016/s0896-6273(00)80106-3. [DOI] [PubMed] [Google Scholar]
- 15.Wimley WC, White SH. Membrane partitioning: Distinguishing bilayer effects from the hydrophobic effect. Biochemistry. 1993;32:6307–6312. doi: 10.1021/bi00076a001. [DOI] [PubMed] [Google Scholar]
- 16.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 17.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 18.Higy M, Gander S, Spiess M. Probing the environment of signal-anchor sequences during topogenesis in the endoplasmic reticulum. Biochemistry. 2005;44:2039–2047. doi: 10.1021/bi047976z. [DOI] [PubMed] [Google Scholar]
- 19.Braun P, von Heijne G. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry. 1999;38:9778–9782. doi: 10.1021/bi990923a. [DOI] [PubMed] [Google Scholar]
- 20.Cordero-Morales JF, et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat Struct Mol Biol. 2006;13:311–318. doi: 10.1038/nsmb1069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.