Abstract
Formation of the complex vertebrate nervous system begins when pluripotent cells of the early embryo are directed to acquire a neural fate. Although cell intrinsic controls play an important role in this process, the molecular nature of this regulation is not well defined. Here we assessed the role for Geminin, a nuclear protein expressed in embryonic cells, during neural fate acquisition from mouse embryonic stem (ES) cells. Whereas Geminin knockdown does not affect the ability of ES cells to maintain or exit pluripotency, we found that it significantly impairs their ability to acquire a neural fate. Conversely, Geminin overexpression promotes neural gene expression, even in the presence of growth factor signaling that antagonizes neural transcriptional responses. These data demonstrate that Geminin's activity contributes to mammalian neural cell fate acquisition. We investigated the mechanistic basis of this phenomenon and found that Geminin maintains a hyperacetylated and open chromatin conformation at neural genes. Interestingly, recombinant Geminin protein also rapidly alters chromatin acetylation and accessibility even when Geminin is combined with nuclear extract and chromatin in vitro. Together, these data support a role for Geminin as a cell intrinsic regulator of neural fate acquisition that promotes expression of neural genes by regulating chromatin accessibility and histone acetylation.
Formation of the cell lineages present in the vertebrate body begins with induction and patterning of the three germ layers during gastrulation. In the mouse embryo, gastrulation involves the movement of uncommitted epiblast cells through the primitive streak to give rise to mesodermal and endodermal derivatives. Anterior epiblast cells that do not enter the primitive streak instead develop as ectoderm, some of which forms the neuroectoderm and gives rise to the vertebrate nervous system. Growth factor signaling contributes to specification and patterning of the germ layers and their derivatives during this period. Transforming growth factor (TGF)-β signaling (BMP and Nodal type) and Wnt signaling promote the formation of mesendodermal cell derivatives, whereas regionalized inhibition of these signaling cues can instead promote neuroectoderm formation (1).
Mouse embryonic stem (ES) cells, derived from the inner cell mass of preimplantation blastocyst embryos (2), provide a useful in vitro model for elucidating early events of germ layer formation at the molecular level. Whereas much of the geometry of the early embryo is absent, formation of germ layer derivatives from ES cells involves many of the same cell extrinsic cues that are used in vivo. Accordingly, Wnt, BMP, and Nodal signaling play important roles in ES-derived mesoderm formation (3–5), whereas neural precursor formation is promoted by antagonism of these extracellular signaling cues (6, 7). These findings suggest that the initial acquisition of neural fate after the exit of ES cells from pluripotency is predominantly regulated by cell intrinsic cues, as in vertebrate animal models (1).
Recent work using ES cells has defined some cell intrinsic transcriptional and epigenetic regulatory mechanisms underlying pluripotency. The pluripotent state of ES cells is maintained by a core transcriptional network involving Oct4, Sox2, and Nanog and characterized by a dynamic chromatin structure with loosely bound histones, which is permissive for the transcriptional machinery (8, 9). Large-scale changes in chromatin structure accompany lineage commitment. These include changes in the composition and activities of chromatin remodeling complexes, alteration of histone modifications, and an overall decrease in chromatin plasticity (10–13). Therefore, proteins that regulate chromatin accessibility by affecting chromatin remodeling or histone modifications can also modulate the cellular plasticity associated with pluripotent and early multipotential cell states. However, cell intrinsic and epigenetic regulatory mechanisms that control neural fate acquisition after pluripotency exit are not well understood. Here, we have assessed the role of the nuclear protein Geminin in this process.
Geminin (Gem or Gmnn) was initially characterized as a dual-function protein that could both expand the neural plate in early Xenopus embryos and inhibit DNA replication origin licensing (14, 15). Gem prevents reinitiation of DNA replication within a single cell cycle by acting as a metazoan-specific inhibitor of the replication licensing protein Cdt1 (16, 17). Gem also interacts with several transcription factors and chromatin regulatory proteins to control transitions from proliferation to differentiation in multiple cell contexts (18–20). Because Gem null mice die by the 32-cell stage, it was not possible to define Gem's role in initial embryonic lineage allocation in mammals (21). Therefore, we developed mouse ES lines for inducible overexpression and knockdown of Geminin to define its roles during the initial aspects of neural lineage commitment.
Our results demonstrate that Geminin plays an important role in promoting neural gene expression during neural fate acquisition in mammals and in antagonizing transcriptional responses to signaling cues that promote nonneural fates. Interestingly, we found that Gem regulates neural commitment by maintaining the chromatin of neural genes in a state of high acetylation and accessibility. Using in vitro experiments with recombinant Geminin, we further determined that Gem can rapidly alter histone acetylation and chromatin accessibility. Together, these results demonstrate that Geminin is a regulator of neural fate acquisition that acts by epigenetic regulation of neural gene expression.
Results
Geminin Activity Regulates Neural Fate Acquisition by Mouse ES Cells.
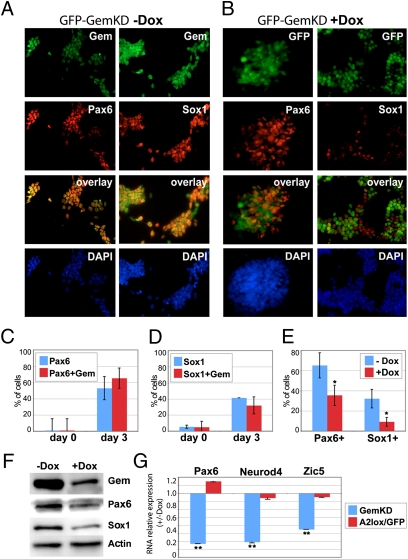
Efforts to examine Geminin's role in mammalian embryonic lineage commitment were hampered by preimplantation lethality (21). Therefore, we instead examined Gem's activities during neural commitment by using mouse ES cells. A2lox ES cells (22) were differentiated on gelatinized dishes in N2B27 medium for 5 d to generate neurectodermal cells expressing Sox1 and Pax6 (6). By 3 d, the mRNA levels of pluripotency markers Nanog and Klf4 decline by >90%, whereas there is a corresponding ∼10-fold increase in Sox1 and Pax6 gene expression (Fig. S1A). Sox1 and Pax6 are also detected at day 3 by Western blotting and in 40–50% of the cells by immunofluorescence (6) (Fig. 1A). Therefore, we focused on defining Gem's activities within this time frame. During the first 3 d of neural commitment in this scheme, Gem protein levels remain relatively constant and Gem colocalizes with markers of both pluripotency (Oct4 and Sox2) and neural fate (Sox1 and Pax6) (Fig. 1 A, C, and D and Fig. S1 B–D). Therefore, Geminin is expressed in ES cells through their acquisition of a neural fate.
Fig. 1.
Knockdown of Geminin decreases neural fate commitment of ES cells. Gem shows significant colocalization with the neural markers Sox1 and Pax6 by day 3 of neural commitment as shown in (A) images and (C and D) quantitated as percentages of total ES cells and Gem immunopositive ES cells also immunopositive for Pax6 or Sox1 (SI Materials and Methods). (B) Dox-induced Gem knockdown (days 0–3) in a GemKD clonal line also expressing GFP. There is decreased Pax6 and Sox1 expression in GFP-expressing Gem knockdown cells (compare A and B, quantitation in E). Gem knockdown also decreases levels of (F) Pax6 and Sox1 protein and (G) Pax6, Neurod4, and Zic5 RNA as detected by immunoblotting or qRTPCR (**P < 0.01, *P < 0.05). Dox treatment of the parental A2lox ES line, which induces GFP overexpression, has no effect on neural gene expression.
To define Geminin's role in neural fate acquisition, we generated clonal ES lines for down-regulating Gem in an acute and doxycycline (Dox)-inducible manner (denoted GemKD). Using the A2lox parental ES line (22), we constructed three independent GemKD lines in which miR30-based short hairpin RNA (shRNAmir) sequences directed against Gem were targeted to a tetracycline-responsive element at the Hprt locus. (Fig. S2 A–C). Dox treatment (500 ng/mL) of any of these GemKD lines for 2 d results in ∼80% Gem knockdown at the protein level (Fig. S2B). Gem knockdown, either in ES culture or during days 0–5 of differentiation in N2B27 medium, does not alter ploidy or cell cycle profiles, as analyzed by FACS (Fig. S3 A–C). Cell viability and proliferation are also unaffected upon Gem knockdown, as measured by TUNEL and MTT assays (SI Materials and Methods) in cells differentiated in N2B27 medium for 5 d (Fig. S3 D and E). The ability of ES cells to proliferate, form alkaline phosphatase-positive colonies (characteristic of pluripotency), and undergo self-renewal in growth medium is also minimally affected by Gem knockdown (Fig. S3 F and G). Therefore, reduction of Geminin levels by ∼80% does not alter the viability, cell cycle characteristics, or chromosomal ploidy of the clones in ES culture or during neural fate commitment.
Although Geminin knockdown does not affect the viability, ploidy, or proliferation rate of these ES clones during commitment, it does significantly impair their ability to acquire a neural fate. Gem knockdown reduces both protein levels and numbers of cells expressing Sox1 (by day 2) and Pax6 (by day 3), as characterized by immunofluorescence (Fig. 1, compare A, B, and E) and immunoblotting (Fig. 1F). Dox-treated Gem knockdown cells also have diminished RNA expression of the neural marker genes Pax6, Neurod4, and Zic5 (Fig. 1G). By contrast, neural gene expression is unchanged following Dox treatment of the parental A2lox ES line (A2lox/GFP), in which Gem levels are unaffected (Fig. 1G). These results demonstrate that Geminin is necessary for cells to efficiently undergo neural fate commitment.
We wanted to determine whether the decrease in neural marker expression upon Geminin knockdown resulted from changes in the ability of ES cells to exit pluripotency and express early markers of pancommitment. To address these questions, we measured the rate at which expression of the pluripotency-associated genes Oct4, Sox2, and Klf4 declined during early neural commitment. Uninduced and GemKD cells show similar levels of Oct4, Sox2, and Klf4 mRNA and Oct4/Sox2 protein through the first 3 d of neural commitment (Fig. S4 A–D). FGF-dependent activation of Erk1/2 kinase activity is required to initiate pancommitment of ES cells, which can be measured by expression of the primitive ectodermal marker Fgf5 and by phosphorylation of Erk1/2 protein (23). By day 2 of neural commitment in control cells, we observe both Erk1/2 phosphorylation and up-regulation of Fgf5 expression (Fig. S4 E and F). Gem knockdown does not affect levels of phospho-Erk1/2 or decrease Fgf5 expression (Fig. S4 E and F). Together, these results suggest that the diminished ability of ES cells to acquire a neural fate following Geminin knockdown is not a consequence of their inability to exit from the pluripotent state.
Geminin Levels Regulate the Choice of Neural Versus Nonneural Lineage Commitment.
To gain additional insight into Geminin's role in neural fate commitment, we used microarray analysis to compare gene expression profiles of ES cells during neural commitment in N2B27 medium with or without Gem knockdown for 2 d. We conducted three independent experiments using Affymetrix microarrays and two clonal GemKD lines that express different microRNA targeting sequences to control for off-target effects (SI Materials and Methods). After normalization, genes that met threshold values with an expression change in the same direction in at least two of the three experiments were further analyzed using the Ingenuity Pathway Analysis (IPA) suite to define prominent biological themes (Fig. S5 A–D). Of the 178 probe sets that show altered expression upon Gem knockdown, the majority (112; 63%) are up-regulated. The top function attributed to these genes is organismal development, whereas top canonical pathways include Wnt and TGF-β signaling, which can antagonize neural fate acquisition. The up-regulated genes include a group of genes with previously described roles in Activin/Nodal signaling-mediated mesoderm formation and/or patterning of the gastrula node (Lefty1, Pitx2, Cited2, Mid1, and Kif3b) (Fig. S5E).
On the basis of these data, we hypothesized that, whereas Geminin knockdown diminishes neural gene expression, it may also elevate expression of nonneural genes. Gem knockdown was insufficient to evoke definitive mesendoderm formation under serum-free conditions (Fig. S5F). However, Gem could potentially counteract signals such as BMP, Wnt3a, and Activin/Nodal that must be antagonized as a prerequisite for neural gene expression (3, 4, 6, 7). We therefore tested the effects on neural gene expression of adding these growth factors during days 1–3 of neural commitment. Adding 5 ng/mL of hBMP4, 10 ng/mL of mWnt3a, or 1 ng/mL of Activin significantly suppressed the expression of a wide range of neural genes (Fig. 2A). Having defined the effects of growth factor addition on neural gene expression under these culture conditions, we next tested whether Gem could modulate these effects. To do this, we used the A2lox ES line to generate clonal cell lines that overexpress FLAG-tagged Gem in a Dox-inducible manner (GemOE) (Fig. S2 A and B). We performed experiments in which N2B27 medium was supplemented with growth factors in the presence or absence of Gem overexpression. Gem antagonized the suppression of neural gene expression that occurred in response to addition of each growth factor in a gene-specific manner (Fig. 2 B–D). These data demonstrate that maintaining high Geminin levels promotes activation of neural gene expression and counteracts the effects of growth factors that can antagonize neural transcriptional responses during commitment.
Fig. 2.
Geminin antagonizes the suppression of neural gene expression by growth factors. GemOE cells were differentiated for 3 d with addition of hBMP4 (5 ng/mL), mWnt3a (10 ng/mL), or Activin A (1 ng/mL) from days 1–3, without or with simultaneous overexpression of Gem (by 500 ng Dox treatment). At day 3, RNA expression levels were determined using qRT-PCR. Y axis of the graphs shows fold changes in neural gene expression (A) with versus without growth factor addition and (B–D) in Dox-treated versus untreated GemOE cells on day 3 of N2B27 differentiation (baseline level without growth factor treatment = 1.0). ***P < 0.001, **P < 0.01, *P < 0.05, +P < 0.1, ns, not significant.
High Levels of Geminin Maintain Histones in a Hyperacetylated State and Increase Chromatin Accessibility.
We found that Geminin overexpression was sufficient to increase expression of many neural genes (Fig. S6A). By day 1 of neural commitment, Gem overexpression caused precocious up-regulation of expression of several genes (Neurod1, Ebf2, and Nestin) that are not normally up-regulated until days 3–5 of neural commitment (Fig. S6 B and E). Previous studies in neural progenitor cells have demonstrated that specific genes, including Neurod1 and Nestin, are also rapidly induced by the addition of hyperacetylating agents such as trichostatin A (TSA) or valproic acid (VPA) (24, 25). Likewise, we found that the same genes that were rapidly induced in response to Gem overexpression by days 1–2 of N2B27 culture were also induced by the addition of hyperacetylating agents such as VPA (2 mM for 8 h) (Fig. S6C). Conversely, genes whose expression decreased upon Geminin knockdown (Sox1, Pax6, and Zic5) were likewise diminished following a brief treatment (4 h) of cells with garcinol, a histone acetyltransferase (HAT) inhibitor (Fig. S6D).
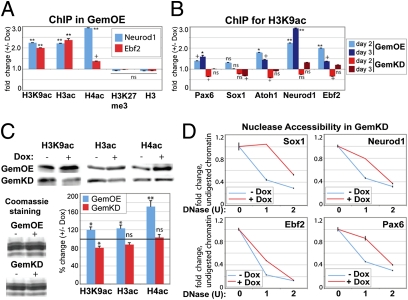
On the basis of the results above, we hypothesized that Geminin's ability to regulate expression of neural genes could involve changes in histone acetylation. To test this hypothesis, we used quantitative chromatin immunoprecipitation (qChIP) to measure the effect of Geminin overexpression on enrichment of panacetylated histone H3 (H3ac), H3 acetylated at lysine 9 (H3K9ac), panacetylated H4, and total H3 (as a control) around the promoter region of two neural genes (Neurod1 and Ebf2) at day 2 of N2B27 culture. We also considered the possibility that the increase in neural gene expression resulted from the loss of repressive activities that prevent the aberrant expression of these genes in ES cells. To test this hypothesis, we assayed changes in the levels of trimethylated lysine 27 of H3 (H3K27me3), a Polycomb-mediated histone modification that correlates with a transcriptionally repressive state of developmental genes. We found that the increases in Neurod1 and Ebf2 expression upon Gem overexpression correlated with increased H3 and H4 acetylation at these genes, while not affecting H3K27me3 (Fig. 3A). Previous studies have observed high H3K9 acetylation at neural genes upon neural commitment (26). We extended our hypothesis that Gem affected histone acetylation to additional neural genes during days 2–3 of neural commitment and found that Gem overexpression consistently elevated H3K9 acetylation, whereas Gem knockdown diminished H3K9 acetylation at these genes (Fig. 3B). We also observed an overall increase in the level of acetylation at core histones upon Gem overexpression from days 1–2 of N2B27 commitment, as detected by immunoblotting total histones for panacetylated H3 and H4 and acetylated H3K9 (Fig. 3C). Conversely, Gem knockdown for 2 d resulted in reduction in acetylation of core histones (Fig. 3C).
Fig. 3.
Geminin regulates histone acetylation and chromatin accessibility at neural genes. (A and B) qChIP detects changes in histone modifications at the indicated genes shown during day 2 (A and B) or 3 (B) of differentiation under conditions of Gem overexpression (A and B) or knockdown (B). The ChIPed material was analyzed using gene-specific primers (Table S3 and SI Materials and Methods). Graphs represent fold changes in histone modifications under +Dox versus −Dox conditions. (C) ES cells were differentiated for 2 d, with or without Gem knockdown or overexpression, followed by total histone extraction. Equal quantities of total histones were resolved by 15% SDS/PAGE and immunoblotted with antibodies against panacetylated H3, H4, and acetyl H3K9 or stained with Coomassie G-250 (loading control). Images were photographed (Biorad Gel Doc XR) and relative intensities determined with Quantity One software. (D) Nuclei were extracted from GemKD cells differentiated for 2 d, with or without Dox induced-Gem knockdown, followed by DNase I digestion with the indicated units. Total DNA was then extracted and quantitated by qPCR with gene-specific primers (same primer sets used for ChIP). Graphs show the percentage of DNA digested (y axis) under increasing DNase I (x axis), ±Gem knockdown (normalized to the 0 unit condition).
Histone acetylation correlates with accessible chromatin and gene transcription (27). To test whether manipulating Geminin levels could also affect chromatin structure at neural genes, we conducted in vivo nuclease accessibility assays under conditions of Gem knockdown during neural commitment. Nuclei were isolated from GemKD cells after 2 d in N2B27 medium with or without Gem knockdown and digested with increasing concentrations of DNase I (Materials and Methods). Total genomic DNA was extracted and levels of chromatin digested by DNase I were quantitated with gene-specific primers using qPCR to determine chromatin accessibility at individual genes. Under conditions of Gem knockdown, neural genes showed significantly less DNase I digestion (Fig. 3D), suggesting that the chromatin at these loci was less accessible. These results were congruent with our other findings and suggest that Geminin levels correlate positively with a hyperacetylated and open chromatin conformation.
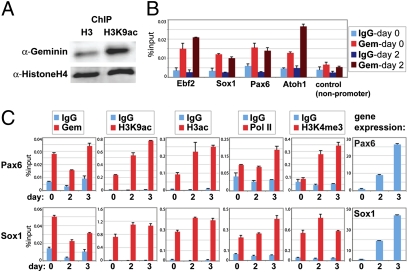
This led us to question whether these effects involve Geminin's direct association with chromatin. We first tested whether Gem preferentially associated with hyperacetylated chromatin. Chromatin extracted after 2 d of neural commitment was immunoprecipitated with antibodies recognizing acetylated histone H3K9 and total histone H3. Immunoprecipitated samples were immunoblotted with Gem antibody (and with an H4 antibody to control for equal loading). We found that Gem is significantly enriched on hyperacetylated chromatin compared with enrichment in total chromatin (Fig. 4A). We conducted additional quantitative ChIP to more specifically assess whether Gem is present at neural genes. Endogenous Gem is significantly enriched at the promoter regions of four candidate neural genes, compared with the IgG control and a nonpromoter region (Fig. 4B). Interestingly, Gem was already enriched at these neural genes in ES cells (by day 0) and remained enriched at day 2. These data demonstrated that Geminin directly associates with neural genes in ES cells through neural fate acquisition.
Fig. 4.
Geminin associates with neural genes before transcriptional activity. (A) Chromatin extracted from ES cells differentiated for 2 d was immunoprecipitated with histone H3 or H3K9ac antibodies. Crosslinks were reversed and equal amounts of total proteins (estimated using Biorad protein assay) were immunoblotted with anti-Gem (N18) antibody or anti-histone H4 antibody. (B) Chromatin from ES cells differentiated for 2 d was immunoprecipitated with anti-Gem or control IgG antibodies. DNA retrieved from ChIP was analyzed using gene-specific ChIP primers (Table S3). (C) qChIP for the proteins/histone modifications indicated was performed at the Sox1 and Pax6 genes on days 0, 2, and 3 and represented as % input (of total input material).
Because Geminin was already enriched at maximal levels at neural promoters in ES cells, we hypothesized that Gem enrichment prefigures their transcriptional activation. To test this, we measured changes in Gem enrichment and compared this to changes in histone acetylation (H3K9ac/H3ac) and components associated with transcriptional activity [binding of RNA polymerase (Pol II) and trimethylation of lysine 4 of histone H3 (H3K4me3)] at the promoters of early neural marker genes, Pax6 and Sox1, through days 0–3 of neural commitment (Fig. 4C). Correlating with increased Pax6 and Sox1 expression by day 2, we observed increased histone acetylation and Pol II and H3K4me3 at these promoters at days 2–3, compared with levels in ES cells (Fig. 4C). These results demonstrate that Geminin enrichment at neural genes prefigures their expression, and could facilitate an early transcriptional competence that enhances later neural gene transcription.
Geminin Increases Histone Acetylation and Chromatin Accessibility in Vitro.
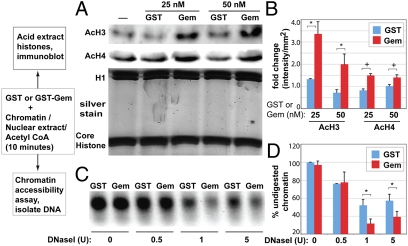
To investigate whether the observed Geminin-mediated changes in histone acetylation and chromatin accessibility were a direct or indirect effect, we tested whether bacterially expressed Gem can regulate histone acetylation in vitro using chromatin and nuclear extracts (NE) purified from ES cells. We purified bacterially expressed GST-Gem or GST and prepared NE and chromatin from ES nuclei. We used these to conduct in vitro HAT reactions by incubating purified chromatin with NE and equimolar amounts of either GST or GST-Gem (0, 25, and 50 nM) for 10 min at 37 °C (Materials and Methods and Fig. 5 A and B). Total histones were then acid extracted and changes in histone acetylation were analyzed by immunoblotting, using antibodies against panacetylated histones H3 and H4. Addition of GST-Gem to the in vitro reaction resulted in a significant increase in acetylation of histones H3 and H4, whereas addition of GST alone did not (Fig. 5 A and B). This Gem-mediated increase in histone acetylation in the in vitro reaction required the presence of the NE. These results indicate that recombinant Geminin can regulate histone acetylation in vitro.
Fig. 5.
In vitro regulation of histone acetylation and chromatin accessibility by Geminin. Schematic at left illustrates the assays used to test in vitro acetylation and chromatin accessibility. (A) nuclear extract (NE, 5 μg), chromatin (2.5 μg), Acetyl CoA (50 μM), and the indicated concentrations of either Gem-GST or GST (0–50 nM) were coincubated for 10 min at 37 °C and histone acetylation was assayed by immunoblotting of acid extracted histones (using panacetylated histone H3, H4 antibodies) or stained with Silver Stain Plus (Biorad). (B) Quantitation of changes in histone acetylation. Images were collected (Biorad; Gel Doc XR) and relative intensities quantitated with Quantity One software. (C) Chromatin remodeling reactions contained 50 nM of purified Gem-GST or GST and NE (5 μg), chromatin (2.5 μg), Acetyl CoA (50 μM), and increasing concentrations of DNase I. DNA was extracted and digestion assessed on a 1.5% agarose gel, imaged using the Biorad Gel Doc XR and (D) relative intensities defined with Quantity One software.
To determine whether Geminin could alter the balance of histone acetylation by enhancing HAT activity or by inhibiting histone deacetylase (HDAC) activity, we tested whether Gem could modulate these activities, using in vitro HAT and HDAC assays (Fig. S7 A–D). First, we assessed whether the addition of GST-Gem could alter the HDAC activity present in nuclear extracts or that of purified HDAC I (Cayman), using an acetylated fluorimetric histone peptide as the substrate (HDAC assay kit; Cayman) (Fig. S7 A and B). We observed no change in the activity of HDACs either in the nuclear extract or for purified HDAC I under increasing concentrations (0–100 nM) of GST-Gem or GST. We likewise tested whether the addition of GST-Gem or GST (0–100 nM) could alter HAT activity levels in nuclear extracts or that of purified p300/CBP-associated factor (PCAF) (Cayman), using a deacetylated histone peptide substrate (HAT assay kit; Cayman) (Fig. S7 C and D). We observed that addition of GST-Gem or GST did not alter the activity of either PCAF or HAT activity present in nuclear extracts. However, it should be noted that these assays use peptide-based histone substrates to assess HAT and HDAC enzymatic activities. Thus, Gem was able to increase histone acetylation only when chromatin was used as a substrate. We hypothesized that Gem might influence histone acetylation levels by regulating chromatin structure and accessibility, perhaps in a manner similar to chromatin remodeling activities. We tested this hypothesis by conducting an in vitro chromatin-remodeling assay, to quantitatively assess changes in chromatin accessibility in the presence of Gem. Chromatin was incubated with NE, Gem-GST, or GST in the presence of increasing concentrations of DNase I (0, 0.5, 1, and 5 units; Materials and Methods and Fig. 5 C and D). DNA was extracted and the levels of chromatin digested by DNase I were quantitated to determine chromatin accessibility. We observed that addition of Gem-GST to the reaction increased the degree of chromatin digestion relative to the control baseline (addition of GST) (Fig. 5 C and D). Gem's ability to increase chromatin accessibility in this assay also required the presence of nuclear extract, resembling the requirement for Gem to hyperacetylate chromatin substrate in vitro. Together, these results further support our hypothesis that maintaining high levels of Geminin facilitates increases chromatin accessibility to maintain chromatin in a hyperacetylated state.
Discussion
Geminin Regulates Neural Fate Acquisition.
In this report, we have defined the role of Geminin in the initial acquisition of neural fate after pluripotency exit. Gem loss in differentiating ES cells significantly impairs their ability to acquire a neural fate. Conversely, Gem overexpression promotes neural gene expression, even in the presence of growth factor signaling that antagonizes neural transcriptional responses. Interestingly, this activity resembles the initial activity for Gem described in the context of Xenopus early development. Gem was identified by functional screening in Xenopus early embryos on the basis of its ability to increase the size of the neural plate at the expense of nonneural cell types such as epidermis (14). Gem was likewise shown to be highly expressed in pluripotent Xenopus ectoderm and to be restricted to the neural precursor territory, while being down-regulated in neural cells before primary neuron differentiation (14, 20, 28). Although the mechanistic basis of Geminin's ability to promote neural gene expression in Xenopus embryos has not been defined, our data suggest that this activity may be conserved between lower vertebrates and mammals.
In addition to its activities as a developmental regulator, Geminin prevents reinitiation of DNA replication within a single cell cycle by binding to and inhibiting Cdt1 (15–17). Cdt1 is the major metazoan regulator of replication origin licensing and is also negatively regulated by Gem-independent proteolysis (29). Although loss of Gem in some cell types can result in genome overreplication due to excessive Cdt1 activity, we did not observe alterations in cell cycle progression or a change in ploidy upon Gem knockdown during neural cell fate acquisition. This is consistent with other reports demonstrating that the cell cycle is unaffected by Gem knockdown in many normal cell types, whereas some cancer and noncancer cell lines are preferentially sensitive to overreplication (30). This suggests that the chromatin- and fate-related phenomenology we observed here upon Geminin loss is not secondary to or resulting from a disrupted cell cycle, and rather reflects a greater sensitivity of these processes to Gem's reduction.
Regulation of Chromatin Acetylation and Accessibility by Geminin.
An interesting observation from this study is the ability of Geminin to increase both histone acetylation and chromatin accessibility. Activities that remodel chromatin and acetylate histones are frequently coordinated to influence chromatin structure and transcription (10, 27, 31). Gem could not alter HAT/HDAC catalytic activity in the in vitro assays using peptide substrates (Fig. S7 A–D) and did not affect HAT/HDAC expression levels, among those we assayed (Fig. S7 E and F). However, Gem did alter both acetylation and accessibility in vitro when chromatin was used as a substrate and ES nuclear extract was included (Fig. 5 A–D). These data suggest that Gem's influence on histone acetylation is not the result of Gem-induced changes in the expression levels or catalytic activity of HATs or HDACs. Rather, it is more likely that Gem affects the chromatin state through interactions with nuclear factors or complexes that affect chromatin remodeling and/or acetylation. Furthermore, Gem is enriched on chromatin at the promoter of neural genes in ES cells, before increased enrichment for histone modifications associated with transcriptional activation. Therefore, Geminin may alter the chromatin structure of neural genes toward a state of high accessibility and acetylation that is conducive to their transcriptional activation.
During Xenopus neuronal differentiation, Geminin can interact with the catalytic subunit of the SWItch/Sucrose NonFermentable (SWI/SNF) remodeling complex (Brg1) and antagonize the ability of SWI/SNF to transactivate target genes involved in regulating neuronal differentiation (20). However, we observed that overexpression of Gem variants containing either intact or mutated Brg1-binding sites could increase neural gene expression to similar levels, suggesting that this activity does not depend on Gem–Brg1 interaction (Fig. S7G). This suggests that additional or alternate Gem interactions with transcription factors or chromatin regulatory proteins may account for its ability to promote chromatin accessibility and neural gene expression. Current efforts are underway to identify the protein complexes through which Geminin mediates structural changes in chromatin and consequently histone acetylation.
Epigenetic Regulation of Early Neural Fate Acquisition.
The cell intrinsic regulators that promote neural gene expression after pluripotency exit remain largely unknown. On the basis of our findings, we hypothesize that the ability of Geminin to rapidly promote the acquisition of an accessible and acetylated chromatin state is required for the initial up-regulation of neural-related gene expression while resulting in a barrier to mesodermal gene expression. Brief histone hyperacetylation induced by TSA treatment induced nestin expression in P19 EC cells and enhanced proneural and Sox2 neural gene expression when performed at the neural progenitor stage (24). In other contexts, hyperacetylation of chromatin enhances cellular plasticity and facilitates somatic cell reprogramming (32, 33). Together, these findings suggest that open and acetylated chromatin is conducive to promoting early and multipotential cell states including neural precursors. Chromatin acetylation must be tightly regulated in early embryonic cells, as this plays an important role in controlling the expression of developmental genes. We found that Gem was enriched at the promoter regions of the neural genes assessed in our study, and could enhance acetylation and accessibility of chromatin at these genes during neural commitment, promoting their expression. It will be very informative to further define the effects of Geminin on chromatin at a genomewide level to better comprehend the global effects of this protein during early lineage commitment.
Materials and Methods
Cell Culture.
The A2lox mouse ES cell line and clonal derivatives were propagated in growth medium and were induced to undergo neural commitment in N2B27 medium as previously described (6) (details in SI Materials and Methods).
qRT-PCR.
Total RNA was extracted and used for cDNA synthesis (details in SI Materials and Methods) followed by analysis using gene-specific primer sequences shown in Table S1.
Immunofluorescence.
Immunofluorescence was as described (6) and see SI Materials and Methods, with quantitation of at least 500 nuclei from three independent experiments using ImageJ. For antibodies, see Table S2.
FACS Analysis.
Methanol-fixed ES cells were stained with propidium iodide and analyzed using the Becton Dickinson FACSCaliber and CELLquest software.
Cell Viability Assay.
Cell viability was determined using Thiazolyl Blue Tetrazolium Bromide (MTT; Sigma) as previously described (34) and see SI Materials and Methods.
Quantitative Chromatin Immunoprecipitation (qChIP).
qChIP was done with modifications to a standard protocol (Upstate; SI Materials and Methods). ChIP antibodies and primers are in Tables S2 and S3.
In Vivo DNase Accessibility Assay.
Nuclei were isolated from 1.5 × 106 ES cells (SI Materials and Methods), washed in 1× nuclei wash buffer (composition in SI Materials and Methods) and resuspended in 300 μL of DNase digestion buffer (1× nuclei wash buffer, 1 mM MgCl2, 0.5 mM CaCl2). Three equal aliquots of 100 μL of DNase digestion buffer containing 0.5 × 106 nuclei each were supplemented with indicated units of DNase I (Roche) and incubated at 37 °C for 5 min. The reaction was stopped with an equal volume of 2× DNase stop buffer (Tris pH 8.0, 20 mM, 4 mM EDTA, 2 mM EGTA) and DNA was extracted and analyzed using the same primers as for ChIP.
In Vitro HAT Assay.
Five micrograms of purified nuclear extract (NE)/reaction was incubated with indicated concentrations of Gem-GST or GST and 50 μM Acetyl CoA (Sigma) in 1× HAT assay buffer (50 mM Tris·HCl, pH 8.0, 10% glycerol, 0.1 mM EDTA, and 1 mM DTT) at room temperature for 10 min. A total of 2.5 μg of chromatin was then added and the reaction was incubated at 37 °C for 10 min. Reaction was stopped and histones were acid extracted (SI Materials and Methods) and analyzed by immunoblotting with anti-acetyl H3 and anti-acetyl H4 antibodies. For further details, see SI Materials and Methods.
In Vitro Chromatin Remodeling Assay.
A total of 2.5 μg of ES-derived chromatin/reaction was incubated with 5 μg of purified nuclear extract, 50 nM Gem-GST or GST, and 50 μM Acetyl CoA (Sigma) in 1× DNase digestion buffer (In Vivo DNase Accessibility Assay) at room temperature for 10 min. Indicated units of DNase I (Roche) were then added and the reaction incubated for an additional 10 min. The reaction was stopped by addition of equal volume of 2× DNase stop buffer.
Supplementary Material
Acknowledgments
We thank Jackie Hughes for help with FACS analysis. Microarray analysis was performed by the Siteman Cancer Center Multiplexed Gene Analysis Core Facility. We are grateful to A. Crossland and J. Enright for assistance with FACS analysis and self-renewal assays. We thank J. Lim, L. Dieden, and J. Witt for technical assistance and E. Caronna for experimental support. We thank Julie Baker, Cathy Collins, Eric George, Asmita Kumar, John Bradley, Kate Beebe, and James Huettner for critical reading of the manuscript and valuable discussions. This work was supported by grants from the National Institutes of Health (GM66815), the March of Dimes, and the American Cancer Society (to K.L.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The Affymetrix microarray datasets reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE25737).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012053108/-/DCSupplemental.
References
- 1.Gaspard N, Vanderhaeghen P. Mechanisms of neural specification from embryonic stem cells. Curr Opin Neurobiol. 2010;20:37–43. doi: 10.1016/j.conb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 3.Lindsley RC, Gill JG, Kyba M, Murphy TL, Murphy KM. Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development. 2006;133:3787–3796. doi: 10.1242/dev.02551. [DOI] [PubMed] [Google Scholar]
- 4.Nostro MC, Cheng X, Keller GM, Gadue P. Wnt, activin, and BMP signaling regulate distinct stages in the developmental pathway from embryonic stem cells to blood. Cell Stem Cell. 2008;2:60–71. doi: 10.1016/j.stem.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfendler KC, Catuar CS, Meneses JJ, Pedersen RA. Overexpression of Nodal promotes differentiation of mouse embryonic stem cells into mesoderm and endoderm at the expense of neuroectoderm formation. Stem Cells Dev. 2005;14:162–172. doi: 10.1089/scd.2005.14.162. [DOI] [PubMed] [Google Scholar]
- 6.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 7.Tropepe V, et al. Direct neural fate specification from embryonic stem cells: A primitive mammalian neural stem cell stage acquired through a default mechanism. Neuron. 2001;30:65–78. doi: 10.1016/s0896-6273(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 8.Kashyap V, et al. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18:1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol. 2009;219:1–7. doi: 10.1002/jcp.21654. [DOI] [PubMed] [Google Scholar]
- 11.Yellajoshyula D, Brown DT. Global modulation of chromatin dynamics mediated by dephosphorylation of linker histone H1 is necessary for erythroid differentiation. Proc Natl Acad Sci USA. 2006;103:18568–18573. doi: 10.1073/pnas.0606478103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemberger M, Dean W, Reik W. Epigenetic dynamics of stem cells and cell lineage commitment: Digging Waddington's canal. Nat Rev Mol Cell Biol. 2009;10:526–537. doi: 10.1038/nrm2727. [DOI] [PubMed] [Google Scholar]
- 14.Kroll KL, Salic AN, Evans LM, Kirschner MW. Geminin, a neuralizing molecule that demarcates the future neural plate at the onset of gastrulation. Development. 1998;125:3247–3258. doi: 10.1242/dev.125.16.3247. [DOI] [PubMed] [Google Scholar]
- 15.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- 16.Tada S, Li A, Maiorano D, Méchali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wohlschlegel JA, et al. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- 18.Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- 19.Del Bene F, Tessmar-Raible K, Wittbrodt J. Direct interaction of geminin and Six3 in eye development. Nature. 2004;427:745–749. doi: 10.1038/nature02292. [DOI] [PubMed] [Google Scholar]
- 20.Seo S, et al. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19:1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez MA, et al. Geminin is essential to prevent endoreduplication and to form pluripotent cells during mammalian development. Genes Dev. 2006;20:1880–1884. doi: 10.1101/gad.379706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyba M, Perlingeiro RC, Daley GQ. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 2002;109:29–37. doi: 10.1016/s0092-8674(02)00680-3. [DOI] [PubMed] [Google Scholar]
- 23.Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134:2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 24.Han DW, et al. Epigenetic hierarchy governing Nestin expression. Stem Cells. 2009;27:1088–1097. doi: 10.1002/stem.43. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams RR, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 27.Neely KE, Workman JL. Histone acetylation and chromatin remodeling: Which comes first? Mol Genet Metab. 2002;76:1–5. doi: 10.1016/s1096-7192(02)00014-8. [DOI] [PubMed] [Google Scholar]
- 28.Spella M, et al. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience. 2007;147:373–387. doi: 10.1016/j.neuroscience.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 29.Nishitani H, et al. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 2006;25:1126–1136. doi: 10.1038/sj.emboj.7601002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu W, Depamphilis ML. Selective killing of cancer cells by suppression of geminin activity. Cancer Res. 2009;69:4870–4877. doi: 10.1158/0008-5472.CAN-08-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 32.Huangfu D, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karantzali E, et al. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: Transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.