Abstract
Transneuronal transport of neurotropic viruses is widely used to define the organization of neural circuitry in the mature and developing nervous system. However, interconnectivity within complex circuits limits the ability of viral tracing to define connections specifically linked to a subpopulation of neurons within a network. Here we demonstrate a unique viral tracing technology that highlights connections to defined populations of neurons within a larger labeled network. This technology was accomplished by constructing a replication-competent strain of pseudorabies virus (PRV-263) that changes the profile of fluorescent reporter expression in the presence of Cre recombinase (Cre). The viral genome carries a Brainbow cassette that expresses a default red reporter in infected cells. However, in the presence of Cre, the red reporter gene is excised from the genome and expression of yellow or cyan reporters is enabled. We used PRV-263 in combination with a unique lentivirus vector that produces Cre expression in catecholamine neurons. Projection-specific infection of central circuits containing these Cre-expressing catecholamine neurons with PRV-263 resulted in Cre-mediated recombination of the PRV-263 genome and conditional expression of cyan/yellow reporters. Replication and transneuronal transport of recombined virus produced conditional reporter expression in neurons synaptically linked to the Cre-expressing catecholamine neurons. This unique technology highlights connections specific to phenotypically defined neurons within larger networks infected by retrograde transneuronal transport of virus from a defined projection target. The availability of other technologies that restrict Cre expression to defined populations of neurons indicates that this approach can be widely applied across functionally defined systems.
Keywords: autonomic, preautonomic network, sympathetic, transneuronal tracing
Knowledge of the synaptic organization of neural circuitry is foundational for understanding the way in which the nervous system functions, or malfunctions, in health, disease, and injury. Experimental approaches for circuit definition have long exploited the axonal transport capabilities of neurons (1). A vast literature continues to define systems organization of brain circuitry through localization of “classic” tracers that are transported through axons but do not cross synapses. Such studies define regional associations but, in the absence of transmission electron microscopic analysis, do not define the synaptology of the system under study. The ability of neurotropic viruses to replicate and spread through neural circuits has brought a polysynaptic perspective to circuit analysis (2–6). Recent genetic engineering of such viruses has produced increasingly powerful probes that have provided novel insights into the identity, organization, and activity of neural networks in a variety of systems (7–9).
The core strengths of the viral transneuronal tracing method lie in the ability of neurotropic viruses to cross synapses and generate infectious progeny in each neuron of a circuit. In effect, these viruses represent self-amplifying neural tracers that efficiently label synaptically linked neurons in a time-dependent manner. However, these desirable attributes also limit the ability to define details of synaptic connectivity within complex networks. For example, definition of the brain's neural network, which regulates homeostasis through autonomic outflow, has benefited enormously from temporal analysis of the replication and transneuronal passage of pseudorabies virus (PRV; a DNA swine α-herpesvirus) from peripheral tissues (e.g., refs. 10–12). Functionally distinct neurons influential in the regulation of cardiovascular function (e.g., blood pressure versus heart rate) are distributed throughout this network and their activity is coordinated through local circuits connecting network nodes (13–15). The reciprocity of connections that ensures integrated activity among such functionally related components of preautonomic circuitry also promotes efficient viral spread throughout the network. Thus, although viral transneuronal tracing has proven to be an effective means of defining the full extent of preautonomic circuitry, the efficiency of viral transport through the entire network undermines the fine-scale resolution of synaptic inputs to defined populations of neurons.
Creative approaches for limiting replication of virus to targeted populations of neurons have substantially improved the ability to define the synaptology of neural circuitry. Wickersham et al. developed a novel tracing approach in which replication of rabies virus (an RNA virus) is restricted to targeted populations of neurons and their first-order synaptic partners (16). This approach was achieved by deleting the rabies glycoprotein gene from the viral genome and pseudotyping the virus so that it differentially infects neurons engineered to express the deleted glycoprotein. Because the rabies glycoprotein is necessary for retrograde transneuronal passage of progeny virus, infection is limited to the targeted neurons and their first-order synaptic inputs. DeFalco et al. constructed a PRV recombinant (PRV-2001), whose replication is dependent upon the presence of the bacterial enzyme Cre recombinase (Cre), and used it to define polysynaptic circuits in transgenic mice that express Cre in neuropeptide Y neurons (17). In this approach, Cre mediates recombination of the viral genome to eliminate a floxed stop cassette that prevents transcription of thymidine kinase, a gene essential for viral replication in nonmitotic cells. Once the stop cassette is eliminated, thymidine kinase can be expressed and the resulting viral genome is permanently replication competent and passes retrogradely to infect neurons synaptically linked to the Cre-expressing neurons. In addition to neuropeptide Y-containing neurons (17), this approach has been used to define circuits synaptically linked to gonadotropin- and serotonin-containing neurons (17–20). The considerable power inherent in both of these adaptations of the viral tracing method lies in restricting the ability of the virus to replicate or move transneuronally between neurons. In addition to conferring important advantages to directed analysis of defined neural circuits, these approaches also place limitations upon the way in which the method can be applied. Chief among these limitations is the need for targeted delivery of virus to neurons permissive to viral replication (Table S1).
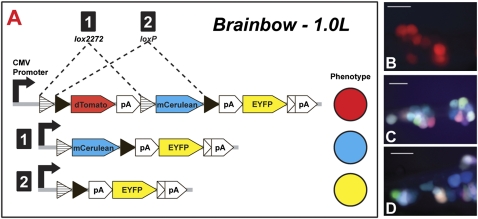
In this study, we describe a viral-tracing technology based on PRV, where the virus is replication-competent in all neurons, but expresses unique reporters in response to Cre-mediated recombination of the viral genome. This technology involves insertion of the Brainbow 1.0L cassette developed by Lichtman and colleagues (21) into the genome of a PRV recombinant (PRV-152) commonly used for viral transneuronal tracing (Fig. 1A). The resulting virus (PRV-263) is replication-competent and reproduces the selective retrograde invasive profile of PRV-152. In the absence of Cre, PRV-263–infected neurons express only a red reporter. However, when Cre is present, Cre-dependent recombination of the Brainbow cassette efficiently, and permanently, removes the red reporter gene and enables expression either of yellow (EYFP) or cyan (mCerulean) fluorescent reporters (22). We further demonstrate that this conditional expression approach can be used to highlight neurons synaptically linked to phenotypically defined neurons embedded within a polysynaptic network retrogradely labeled from a projection target. In our proof-of-principle experiments, selective expression of Cre was achieved in a specific group of rat brainstem catecholamine neurons using a lentivirus vector in which Cre expression is controlled by a synthetic dopamine-β-hydroxylase (DβH) promoter (23, 24). Injection of the vector into the brainstem followed by injection of PRV-263 into the kidney allowed us to distinguish neurons synaptically linked to Cre-expressing catecholamine neurons (yellow and cyan fluorescence) from neurons of the renal preautonomic network infected through neural pathways that did not involve those neurons (only red fluorescence). In this fashion, microcircuit connectivity to targeted neurons was visualized within the context of larger complex circuits labeled by projection-specific transport of virus from a distant target. The availability of increasingly sophisticated approaches for targeted expression of Cre indicates that this unique approach can be applied across systems to provide novel insights into the organization and plasticity of neural circuits of mature and developing nervous systems.
Fig. 1.
To prepare PRV-263, the EGFP cassette in the gG locus of the PRV-152 was replaced with the Brainbow 1.0L cassette (A) developed by Lichtman and colleagues (21), using homologous recombination. Cre-mediated recombination of the viral genome occurs at either paired lox2272 or loxP sites, permanently eliminating the red dTomato reporter and, depending upon the site of recombination, liberating expression of either mCerulean or EYFP. Images B-D illustrate the phenotype of cultured SCG neurons infected with PRV-263 grown in Cre or non-Cre cells. SCG neurons infected by PRV-263 grown in non–Cre-expressing PK15 cells only fluoresce red (B). In contrast, infection of neurons from virus produced by Cre-expressing PK-15 cells revealed expression of the EYFP or mCerulean genes of the Brainbow cassette (C and D). (Scale bars, 50 μm.)
Experimental Objectives and Design
Our goal was to construct a robust, replication-competent strain of PRV that marks neurons in a circuit but changes the expression profile of reporter genes in phenotypically defined populations of neurons within that circuit. There are three essential elements in our strategy. The first element is to maintain the ability of the virus to replicate in all permissive neurons. As a result, it will be possible to introduce the virus to a circuit in a projection-specific fashion: for example, a circuit can be infected on the basis of its target (e.g., kidney) rather than depending on the presence of a molecule essential for viral invasiveness or replication. The second element is to provide a genetic means to change the profile of reporter-gene expression from the virus in a controlled, permanent, and highly reproducible fashion. The Brainbow technology using Cre-promoted, site-specific recombination provides a well-characterized system for achieving this goal. The third element is to provide a method for Cre expression in a targeted population of neurons within the circuit. Lentivirus-mediated gene delivery is well suited for this purpose in that Cre expression can be restricted to phenotypically defined neurons in a regionally defined manner through the use of cell-specific promoters. In addition, lentivirus vectors can infect a wide variety of animals, enabling use of this PRV technology in different species. The details of our experimental approach are provided in SI Materials and Methods.
Results and Discussion
Characterization of PRV-263 Neuroinvasiveness.
We first compared the neuroinvasiveness of PRV-263 to that produced by PRV-152, the parental strain of PRV-263, in animals that were not previously injected with lentivirus vector. PRV-263 was injected into the kidney and the pattern of retrograde transneuronal infection was analyzed 4 to 7 d later using immunoperoxidase localization of viral antigens. Fluorescent reporter expression was analyzed in adjacent sections from each case. The immunoperoxidase and fluorescence analyses demonstrated that the distribution of infected neurons recapitulated our previous findings using PRV-152 (12). Injection of PRV-263 into the kidney produced a predictable course of retrograde transneuronal infection that resulted from first-order infection of sympathetic postganglionic neurons, transneuronal infection of sympathetic preganglionic neurons (SPGs) in the thoracic cord, and retrograde transneuronal infection of the central preautonomic network (Fig. 2). PRV-263 invasion of this circuitry was temporally defined, with initial infection of the brain occurring in the caudal brainstem and progressing to circumscribed populations of neurons in all major subdivisions of the neuraxis. Importantly, the virus remained confined to the preautonomic network in all cases, with the most extensive infection and fluorescence analysis demonstrated that all infected neurons selectively expressed the red dTomato reporter from the Brainbow cassette.
Fig. 2.
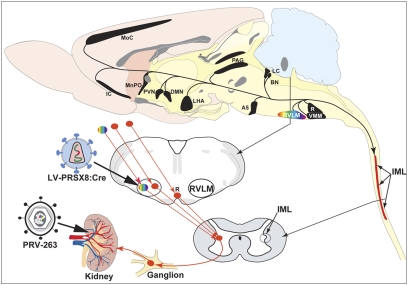
The experimental paradigm used for proof-of-principle studies is illustrated. Selective expression of Cre in C1 neurons of the RVLM was achieved using a replication defective lentivirus vector that expresses Cre under the control of a synthetic DβH promoter. Seven days after vector injection, renal preautonomic circuitry was infected by retrograde transneuronal transport of PRV-263 from the kidney. The sagittal schematic of the rat brain illustrates the major cell groups that comprise the renal preautonomic network (black). Transneuronal passage of PRV-263 to the RVLM infected C1 and non-C1 neurons via their projections to the intermediolateral cell column (IML) of the thoracic spinal cord. Restricted expression of Cre in C1 neurons limited recombination of the PRV-263 genome to this catecholamine population, permanently removing the red dTomato gene and enabling the expression of either EYFP or mCerulean transgenes. Infected neurons in the RVLM, and in parallel pathways that did not contain Cre, did not undergo recombination of the viral genome and remained red. A5, A5 catecholamine cell group; BN, Barrington's nucleus; DMN, dorsomedial hypothalamic nucleus; IC, insular cortex; LC, locus coeruleus; LHA, lateral hypothalamic nucleus; MnPO, median preoptic nucleus; MoC, motor cortex; PAG, periaqueductal gray; PVN, paraventricular hypothalamic nucleus; R, raphe; RVLM, rostroventrolateral medulla; VMM, ventromedial medulla.
Dual Injections of Lentivirus Vector and PRV-263.
The above experiments demonstrated that the RVLM was first infected with PRV-263 4 d after kidney injection and infection spread transneuronally to infect all major components of the preautonomic network by 5 d postinoculation. Thus, we chose these survival intervals for in vivo analysis of the fluorophor profile of infected neurons. Because parametric studies demonstrated that transgene expression from the lentivirus vector is robust by 7 d and stable through 64 d, we injected the vector unilaterally into the left RVLM 7 d before injection of PRV-263 into the left kidney (Fig. 2). Thus, we had two informative survival intervals to assess Cre-mediated recombination in RVLM neurons and transneuronal passage to afferent neurons. Our analysis of tissue from each survival interval was done in two steps. We first conducted a detailed quantitative analysis of the distribution of infected neurons in coronal sections using immunoperoxidase localization of viral antigens. These data served as the template for the second phase of analysis, in which the fluorophor profiles of infected neurons were documented. Sections adjacent to those processed for the immunoperoxidase analysis were systematically examined for fluorophor expression using fluorescence microscopy.
The distribution of infected neurons revealed in immunoperoxidase localizations of each case was mapped in 48 coronal sections that provided a detailed sampling of the renal preautonomic network (Figs. S2 and S3). In addition, we analyzed the extent of infection in horizontal sections of the entire spinal cord processed for immunocytochemical localization of viral antigens. PRV-263 invaded the circuitry linked to the kidney with kinetics and a pattern of infection that recapitulated infection when PRV-263 was injected alone (detailed above). At the early survival interval (4 d), infection in the spinal cord was largely confined to the sympathetic preganglionic neurons of the thoracic spinal cord and infected brainstem neurons were differentially concentrated in cell groups that constitute the baroreceptor reflex (RVLM, caudal ventrolateral medulla, and nucleus of the solitary tract) and in the A5 catecholamine cell group. It is well established that the dense descending projections of A5 and the RVLM account for the early infection of this brainstem circuitry (12). The paraventricular nucleus (PVN) of the hypothalamus also projects densely to SPG in the thoracic intermediolateral cell column (IML) and a subset of animals at the short survival interval exhibited a small number of infected PVN neurons (Fig. S2). The extent of infection within the preautonomic network was substantially greater 5 d postinoculation. Prominent among the expanded circuitry infected 5 d after injection of PRV-263 into the kidney, was the ventromedial medulla (VMM) surrounding the pyramidal tracts, the locus coeruleus (LC), and hypothalamic cells groups that play essential roles in coordinating behavioral state and arousal with adaptive changes in physiology.
Immunofluorescence analysis revealed differential expression of the three fluorescent reporters from the Brainbow cassette based upon the location of the neurons within the preautonomic circuitry. Neurons in the sympathetic ganglia and the IML only expressed the red dTomato reporter (Fig. S4). This expression profile is consistent with selective retrograde transport of PRV-263 from the kidney and the absence of Cre from this component of the circuitry (Fig. 2). The left RVLM, which was injected with the lentivirus vector, demonstrated Cre-mediated recombination of the PRV-263 genome in all animals. It is important to note that the overlapping wavelengths for detection of EGFP (the reporter of lentivirus transgene expression) and EYFP (one of the reporters of the Brainbow cassette) do not permit their differential localization in the fluorescence analysis. Nevertheless, our parametric control studies demonstrated that YFP and CFP reporters are only expressed in the presence of Cre and that lentivirus-mediated transgene expression is confined to the C1 catecholamine neurons in the RVLM. Thus, although we cannot unambiguously distinguish EYFP and EGFP in the fluorescence analysis, we can assert with confidence that mCerulean and EYFP expression in the preautonomic network are the result of Cre-mediated recombination of the PRV-263 genome in C1 neurons.
Within the left RVLM we observed neurons with mixed and pure color profiles with a distribution consistent with the known circuit organization of the RVLM (Fig. 3 B and C). At early survival intervals, the majority of these neurons were concentrated in the rostral portion of the RVLM that is the principal source of descending C1 projections to SPGs in the IML of the thoracic spinal cord (27, 28). A day later, neurons throughout the RVLM exhibited yellow, green, and cyan fluorescence, either alone or in combination. Importantly, a population of RVLM neurons also exhibited only red fluorescence, even in animals with the most extensive infection of CNS circuitry (Fig. 3 B and C). Although it is well documented that C1 and non-C1 neurons both contribute to the descending reticulospinal projections of the RVLM to the IML (29–31), little is known regarding the synaptic interaction between neurons giving rise to these pathways within the RVLM. The fact that both of these populations (red only and cyan/yellow) remain stable in cases with the most extensive infection raises the possibility that projections from the RVLM to the spinal cord are organized in parallel. Nevertheless, a more detailed analysis incorporating longer surviving animals is necessary to rigorously test this hypothesis.
Fig. 3.
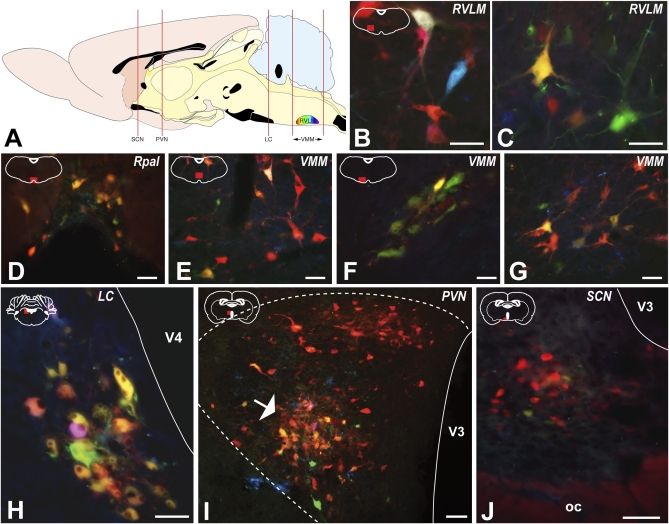
Fluorophor profiles of neurons in different regions of the renal preautonomic network in animals infected with PRV-263 7 d after injection of the Cre-vector into the RVLM are illustrated. The red lines in the sagittal schematic diagram A define the relative location of the regions included in the analysis. The red boxes in the coronal schematics included in the upper left portions of B to J illustrate the location of the cell group for each photographed area. Neurons exhibiting red, yellow, and cyan fluorescence were prominent within the RVLM injected with the Cre-expressing vector (B and C). Neurons in all major subdivisions of the VMM exhibited neurons expressing the cyan and yellow reporters among neurons only expressing the red reporter. These included neurons in the rostral portion of the raphe pallidus (D) and both the medial (E) and lateral (F and G) subdivisions of the VMM. Neurons in the ventral tier of the LC were infected by retrograde transneuronal passage of PRV-263 (H). The presence of yellow and cyan reporters in essentially all of these neurons indicates that the LC exerts its influence upon cardiovascular function through synaptic contacts with the C1 catecholamine neurons of the RVLM. Cyan and yellow fluorophors were differentially concentrated within a distinct subfield of the medial parvicellular subdivision of the PVN (arrow in I) and within the SCN (J) of animals with the most extensive infection of the preautonomic circuit. See text for discussion of these data. (Scale bars, 50 μm.)
We sought further evidence of the utility of the method for deciphering the synaptic organization of microcircuits within complex neural networks by examining the fluorescence profiles of neurons known to project to the RVLM. Toward this end, we documented the fluorophor profile of infected neurons throughout the renal preautonomic network. That analysis revealed neurons expressing the conditional reporters were a clear and reproducible subset of the neurons within the network. Areas expressing cyan/yellow reporters included the contralateral RVLM, VMM, LC, PVN, and the suprachiasmatic nucleus (SCN) (Fig. 3 A–J). Data derived from each of these regions validated the experimental approach and also provided unique insights into the way in which the RVLM functions within the larger renal preautonomic network. In interpreting these findings it is important to note that, although we know Cre-expression was restricted to C1 neurons, it is likely that the lentivirus injection only produced Cre-expression in a subset of the C1 population. Thus, it is also likely that reticulospinal C1 neurons are among the RVLM neurons expressing only the default dTomato reporter. In any event, the presence of conditional reporters in infected neurons that project to the RVLM provides unique insights into the components of the renal preautonomic network synaptically linked to the C1 cell group.
Prominent replication of the recombined genome of PRV-263 (expression of cyan/yellow reporters) following retrograde transneuronal transport of virus from Cre-expressing C1 neurons was consistently observed within distinct subdivisions of the VMM (Fig. 3 D–G). These subdivisions included neurons within the raphe pallidus (Fig. 3D), the parapyramidal region that contains the raphe magnus (Fig. 3E), and a compact group of neurons in the area of the lateral paragigantocellular cell group (Fig. 3 F and G). The distribution of these neurons within these regions was consistent among all of the animals in the 5-d survival group and, importantly, constituted a subset of the total population of infected neurons. Neurons expressing the red dTomato reporter were observed throughout the rostrocaudal extent of the raphe pallidus, but cyan and yellow fluorescing neurons were differentially concentrated in the rostral two thirds of this serotonergic cell group. Serotonergic neurons of the raphe magnus are a prominent component of this region and, although we were not able to characterize the neurotransmitter phenotype, the morphology and distribution of the cyan/yellow-labeled neurons suggest that at least a portion of these neurons express serotonin. CFP/YFP-expressing neurons in the lateral paragigantocellular region also formed a distinctive grouping immediately lateral to the pyramids that was present bilaterally, but most extensive ipsilateral to Cre-expressing C1 neurons (Fig. 3 F and G). The VMM is a well-documented component of the preautonomic network labeled by viral transport from a variety of peripheral tissues (e.g., refs. 11, 12). Functional roles for VMM neurons in the control of thermogenesis, cardiovascular function, and nociception are well established (32–36). In addition, Mason (37) has provided evidence that neurons in this region play an important role in multimodal sensory processing, some of which is relevant to cardiovascular homeostasis. Our data bring further clarity to the identity of the subset of neurons within this region that influence arterial blood pressure through their connections with the C1 catecholamine neurons.
A considerable amount of literature has demonstrated that the LC plays an important role in coordinating behavioral state and arousal with adaptive changes in physiology (38, 39). Activity in the LC mirrors behavioral state, with increased arousal and task-related behavior correlating with an increased firing rate of LC neurons (40). Our data are consistent with the prior demonstrations that neurons in the ventral tier of the LC project upon the RVLM and other nodes within the preautonomic network (Fig. 3H). The data further demonstrate that the vast majority of neurons infected by retrograde transneuronal transport of PRV-263 from the kidney expressed yellow and cyan reporters (Fig. 3H). This finding demonstrates that ventral tier LC neurons exert their influence upon the RVLM through synaptic connections with C1 catecholamine neurons.
The PVN exerts prominent influences over endocrine and autonomic regulation, particularly as it relates to cardiovascular homeostasis and responses to stress (41, 42). Retrograde transneuronal passage of virus from peripheral tissues infects PVN neurons in all of the preautonomic subdivisions of this complex cell group, but the precise pathways through which the PVN controls autonomic outflow in an organ-specific fashion are not known. Retrograde transneuronal infection of the PVN by injection of PRV-263 into the kidney recapitulated the findings of prior viral-tracing studies. We observed infected neurons in the dorsal and medial parvicellular PVN subdivisions, but also documented a differential pattern of reporter-gene expression in the medial and dorsal parvicellular subfields of the PVN (Fig. 3I). Whereas neurons exhibiting red reporter-gene expression were present in both of these subfields, neurons expressing cyan and yellow reporters were largely confined to a distinct region within the medial parvicellular PVN (arrow in Fig. 3I). These data indicate that the influence of the PVN upon RVLM C1 neurons arises from neurons in the medial parvicellular PVN, and that dorsal parvicellular PVN neurons are acting through a pathway independent of the C1 population.
We also observed neurons in the SCN in the animals with the most extensive viral transport (e.g., see cases 10 and 11 in Fig. S2). These neurons were present in both the dorsomedial and ventrolateral subdivisions of the SCN and were of mixed phenotypes (Fig. 3J). The majority of the neurons expressed the red fluorophor but a subset expressed the cyan and yellow fluorophors. It is well known that cardiovascular homeostasis exhibits a circadian rhythm that is subject to the control of the SCN (43, 44). In addition, a recent study demonstrated that a mutation in the Per2 clock protein, an integral component of the molecular machinery in SCN neurons responsible for the generation of rhythmicity, interferes with the circadian expression of rhythms of heart rate and blood pressure (45). Prior studies have demonstrated retrograde transneuronal infection of SCN neurons after injection of PRV into the adrenal gland (46), pineal gland (47), liver (48), and autonomic ganglia (49). On the basis of these data it has been postulated that the SCN orchestrates the rhythmic functional activity of autonomic targets. Furthermore, dual infection studies with viruses expressing unique reporters have provided evidence that there is functional parcellation of the SCN with respect to the peripheral organ systems that the clock regulates (46). Our data are in accord with this conclusion in that the neurons expressing cyan and yellow reporters are a subset of the total population of infected neurons in the SCN. The fact that these neurons are only present in the cases with the most extensive infection further supports the conclusion that the influence of the SCN upon the RVLM C1 population occurs through a relay. Further directed analysis of this circuitry provides a means of defining this relay.
Summary and Conclusions
Our experimental approach using PRV Brainbow recombinants provides a powerful beginning to defining the identity and organization of circuits synaptically linked to phenotypically identified populations of neurons. In the context of the current experimental design, the power of the PRV Brainbow virus derives from localized expression of Cre within components of a circuit and reveals unique insights into the distributed circuit of neurons synaptically linked to reticulospinal C1 neurons in the renal preautonomic network. In preliminary studies presented in Fig. S5, we demonstrate that lentivirus-mediated Cre expression under the control of a universal promoter can be effectively applied to define routes of viral transport through a node within a circuit independent of the neurotransmitter phenotype of the resident neurons. This ability and the increasing availability of transgenic animals engineered to express Cre in phenotypically defined neurons substantially expand the experimental scope of the approach.
Use of the Brainbow cassette in the current investigation derives from in vitro studies with PRV-263 that demonstrated a limited number of genomes are expressed in infected cells (22). We also used this cassette to optimize the possibility that a recombination event will occur in Cre-expressing neurons because of the two pairs of lox sites for Cre-mediated recombination. This feature may have contributed to the remarkably bright and diverse hues produced by combinatorial expression of different fluorescent proteins observed in our analysis. This feature permitted unambiguous identification of neurons replicating recombined genomes, and also offers the possibility of circuit analysis at the single-cell level. However, it should be noted that Cre-mediated expression of a single novel reporter (e.g., cyan) would further expand the utility of the conditional approach by allowing phenotypic characterization of neurons expressing the conditional reporter in dual labeling immunofluorescence localizations.
This conditional reporter approach for viral transneuronal circuit analysis also holds substantial promise for production of more powerful probes of circuit organization that will increase the versatility of the viral-tracing method. For example, construction of viruses that conditionally express membrane-tethered fluorescent proteins can expand the power of the approach by densely labeling the axons of identified infected neurons. Considered collectively, these unique tools have the potential to substantially improve the resolution of viral tracing by revealing details of synaptology of identified populations of neurons within the context of functionally related partners in complex networks.
Materials and Methods
Details regarding the experimental animals and procedures used in the analysis are provided in SI Materials and Methods. SI Materials and Methods also describes the in vitro studies conducted to validate Cre-mediated recombination of the PRV-263 genome. All experimental procedures were approved by University of Pittsburgh and Princeton University regulatory bodies.
Supplementary Material
Acknowledgments
We thank Jeff Lichtman for the Brainbow plasmids and acknowledge members of the L.W.E. and J.P.C. laboratories for advice, encouragement, and technical assistance. O.K. is funded by the International Human Frontier Science Program. This research was supported by National Institute of Health Grants 1RC1NS068414, 1R01HL093134, and P40 RR018604, and National Science Foundation Grant 0918867.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015033108/-/DCSupplemental.
References
- 1.Zaborszky L, Wouterlood FG, Lanciego JL. Neuroanatomical Tract-Tracing 3: Molecules, Neurons, and Systems. Springer, New York; 2006. [Google Scholar]
- 2.Song CK, Enquist LW, Bartness TJ. New developments in tracing neural circuits with herpesviruses. Virus Res. 2005;111:235–249. doi: 10.1016/j.virusres.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 3.Ekstrand MI, Enquist LW, Pomeranz LE. The alpha-herpesviruses: Molecular pathfinders in nervous system circuits. Trends Mol Med. 2008;14:134–140. doi: 10.1016/j.molmed.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldogköi Z, et al. Novel tracing paradigms—genetically engineered herpesviruses as tools for mapping functional circuits within the CNS: Present status and future prospects. Prog Neurobiol. 2004;72:417–445. doi: 10.1016/j.pneurobio.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Card JP. Pseudorabies virus neuroinvasiveness: A window into the functional organization of the brain. Adv Virus Res. 2001;56:39–71. doi: 10.1016/s0065-3527(01)56004-2. [DOI] [PubMed] [Google Scholar]
- 6.Loewy AD. Pseudorabies virus: A transneuronal tracer for neuroanatomical studies. In: Kaplitt MG, Loewy AD, editors. Viral Vectors. Gene Therapy and Neuroscience Applications. San Diego: Academic Press; 1995. pp. 349–366. [Google Scholar]
- 7.Boldogkoi Z, et al. Genetically timed, activity-sensor and rainbow transsynaptic viral tools. Nat Methods. 2009;6:127–130. doi: 10.1038/nmeth.1292. [DOI] [PubMed] [Google Scholar]
- 8.Granstedt AE, Szpara ML, Kuhn B, Wang S-H, Enquist LW. Fluorescence-based monitoring of in vivo neural activity using a circut-tracing pseudorabies virus. PLos One. 2009;4 doi: 10.1371/journal.pone.0006923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothermel M, et al. Advanced tracing tools: Functional neuronal expression of virally encoded fluorescent calcium indicator proteins. J Neurovirol. 2009;15:458–464. doi: 10.3109/13550280903473460. [DOI] [PubMed] [Google Scholar]
- 10.Jansen ASP, Van Nguyen X, Karpitskiy V, Mettenleiter TC, Loewy AD. Central command neurons of the sympathetic nervous system: Basis of the fight-or-flight response. Science. 1995;270:253–260. doi: 10.1126/science.270.5236.644. [DOI] [PubMed] [Google Scholar]
- 11.Strack AM, Sawyer WB, Hughes JH, Platt KB, Loewy AD. A general pattern of CNS innervation of the sympathetic outflow demonstrated by transneuronal pseudorabies viral infections. Brain Res. 1989;491:156–162. doi: 10.1016/0006-8993(89)90098-x. [DOI] [PubMed] [Google Scholar]
- 12.Cano G, Card JP, Sved AF. Dual viral transneuronal tracing of central autonomic circuits involved in the innervation of the two kidneys in rat. J Comp Neurol. 2004;471:462–481. doi: 10.1002/cne.20040. [DOI] [PubMed] [Google Scholar]
- 13.Guyenet PG. The sympathetic control of blood pressure. Nat Rev Neurosci. 2006;7:335–346. doi: 10.1038/nrn1902. [DOI] [PubMed] [Google Scholar]
- 14.Sved AF, Ito S, Sved JC. Brainstem mechanisms of hypertension: Role of the rostral ventrolateral medulla. Curr Hypertens Rep. 2003;5:262–268. doi: 10.1007/s11906-003-0030-0. [DOI] [PubMed] [Google Scholar]
- 15.Spyer KM. Centeral nervous mechansims contributing to cardiovascular control. J Physiol. 1994;474:1–19. doi: 10.1113/jphysiol.1994.sp019997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickersham IR, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeFalco J, et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- 18.Braz JM, Enquist LW, Basbaum AI. Inputs to serotonergic neurons revealed by conditional viral transneuronal tracing. J Comp Neurol. 2009;514:145–160. doi: 10.1002/cne.22003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell RE, Herbison AE. Definition of brainstem afferents to gonadotropin-releasing hormone neurons in the mouse using conditional viral tract tracing. Endocrinology. 2007;148:5884–5890. doi: 10.1210/en.2007-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell RE. Defining the gonadotrophin-releasing hormone neuronal network: Transgenic approaches to understanding neurocircuitry. J Neuroendocrinol. 2007;19:561–573. doi: 10.1111/j.1365-2826.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- 21.Livet J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- 22.Kobiler O, Lipman Y, Therkelsen K, Daubechies I, Enquist LW. Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes. Nat. Commun. 2010;1:146. doi: 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Card JP, et al. Efferent projections of rat rostroventrolateral medulla C1 catecholamine neurons: Implications for the central control of cardiovascular regulation. J Comp Neurol. 2006;499:840–859. doi: 10.1002/cne.21140. [DOI] [PubMed] [Google Scholar]
- 24.Hwang D-Y, Carlezon WA, Jr., Isacson O, Kim K-S. A high-efficiency synthetic promoter that drives transgene expression selectively in noradrenergic neurons. Hum Gene Ther. 2001;12:1731–1740. doi: 10.1089/104303401750476230. [DOI] [PubMed] [Google Scholar]
- 25.Card JP, Lois J, Sved AF. Distribution and phenotype of Phox2a-containing neurons in the adult sprague-dawley rat. J Comp Neurol. 2010;518:2202–2220. doi: 10.1002/cne.22327. [DOI] [PubMed] [Google Scholar]
- 26.Kang BJ, et al. Central nervous system distribution of the transcription factor Phox2b in the adult rat. J Comp Neurol. 2007;503:627–641. doi: 10.1002/cne.21409. [DOI] [PubMed] [Google Scholar]
- 27.Tucker DC, Saper CB, Ruggiero DA, Reis DJ. Organization of central adrenergic pathways: I. Relationships of ventrolateral medullary projections to the hypothalamus and spinal cord. J Comp Neurol. 1987;259:591–603. doi: 10.1002/cne.902590408. [DOI] [PubMed] [Google Scholar]
- 28.Schreihofer AM, Guyenet PG. Sympathetic reflexes after depletion of bulbospinal catecholaminergic neurons with anti-DbetaH-saporin. Am J Physiol Regul Integr Comp Physiol. 2000;279:R729–R742. doi: 10.1152/ajpregu.2000.279.2.R729. [DOI] [PubMed] [Google Scholar]
- 29.Jeske I, McKenna KE. Quantitative analysis of bulbospinal projections from the rostral ventrolateral medulla: contribution of C1-adrenergic and nonadrenergic neurons. J Comp Neurol. 1992;324:1–13. doi: 10.1002/cne.903240102. [DOI] [PubMed] [Google Scholar]
- 30.Schreihofer AM, Guyenet PG. Identification of C1 presympathetic neurons in rat rostral ventrolateral medulla by juxtacellular labeling in vivo. J Comp Neurol. 1997;387:524–536. doi: 10.1002/(sici)1096-9861(19971103)387:4<524::aid-cne4>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Sartor DM, Verberne AJ. Phenotypic identification of rat rostroventrolateral medullary presympathetic vasomotor neurons inhibited by exogenous cholecystokinin. J Comp Neurol. 2003;465:467–479. doi: 10.1002/cne.10840. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura K, Matsumura K, Kobayashi S, Kaneko T. Sympathetic premotor neurons mediating thermoregulatory functions. Neurosci Res. 2005;51:1–8. doi: 10.1016/j.neures.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Babic T, Ciriello J. Medullary and spinal cord projections from cardiovascular responsive sites in the rostral ventromedial medulla. J Comp Neurol. 2004;469:391–412. doi: 10.1002/cne.11024. [DOI] [PubMed] [Google Scholar]
- 34.Nason MW, Jr., Mason P. Modulation of sympathetic and somatomotor function by the ventromedial medulla. J Neurophysiol. 2004;92:510–522. doi: 10.1152/jn.00089.2004. [DOI] [PubMed] [Google Scholar]
- 35.Basbaum AI, Fields HL. Endogenous pain control mechanisms: Review and hypothesis. Ann Neurol. 1978;4:451–462. doi: 10.1002/ana.410040511. [DOI] [PubMed] [Google Scholar]
- 36.Basbaum AI, Fields HL. Endogenous pain control systems: brainstem spinal pathways and endorphin circuitry. Annu Rev Neurosci. 1984;7:309–338. doi: 10.1146/annurev.ne.07.030184.001521. [DOI] [PubMed] [Google Scholar]
- 37.Mason P. Ventromedial medulla: Pain modulation and beyond. J Comp Neurol. 2005;493:2–8. doi: 10.1002/cne.20751. [DOI] [PubMed] [Google Scholar]
- 38.Svensson TH. Peripheral, autonomic regulation of locus coeruleus noradrenergic neurons in brain: Putative implications for psychiatry and psychopharmacology. Psychopharmacology (Berl) 1987;92:1–7. doi: 10.1007/BF00215471. [DOI] [PubMed] [Google Scholar]
- 39.Samuels ER, Szabadi E. Functional neuroanatomy of the noradrenergic locus coeruleus: Its roles in the regulation of arousal and autonomic function part II: Physiological and pharmacological manipulations and pathological alterations of locus coeruleus activity in humans. Curr Neuropharmacol. 2008;6:254–285. doi: 10.2174/157015908785777193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aston-Jones G, Cohen JD. An integrative theory of locus coeruleus-norepinephrine function: Adaptive gain and optimal performance. Annu Rev Neurosci. 2005;28:403–450. doi: 10.1146/annurev.neuro.28.061604.135709. [DOI] [PubMed] [Google Scholar]
- 41.Simmons DM, Swanson LW. Comparison of the spatial distribution of seven types of neuroendocrine neurons in the rat paraventricular nucleus: Toward a global 3D model. J Comp Neurol. 2009;516:423–441. doi: 10.1002/cne.22126. [DOI] [PubMed] [Google Scholar]
- 42.Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci. 2009;10:397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssen BJ, Tyssen CM, Duindam H, Rietveld WJ. Suprachiasmatic lesions eliminate 24-h blood pressure variability in rats. Physiol Behav. 1994;55:307–311. doi: 10.1016/0031-9384(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 44.Saleh MA, Winget CM. Effect of suprachiasmatic lesions on diurnal heart rate rhythm in the rat. Physiol Behav. 1977;19:561–564. doi: 10.1016/0031-9384(77)90234-7. [DOI] [PubMed] [Google Scholar]
- 45.Vukolic A, et al. Role of mutation of the circadian clock gene Per2 in cardivascular circadian rhythms. Am J Physiol Regul Integr Comp Physiol. 2010;298:R627–R634. doi: 10.1152/ajpregu.00404.2009. [DOI] [PubMed] [Google Scholar]
- 46.Buijs RM, et al. The suprachiasmatic nucleus balances sympathetic and parasympathetic output to peripheral organs through separate preautonomic neurons. J Comp Neurol. 2003;464:36–48. doi: 10.1002/cne.10765. [DOI] [PubMed] [Google Scholar]
- 47.Larsen PJ, Enquist LW, Card JP. Characterization of the multisynaptic neuronal control of the rat pineal gland using viral transneuronal tracing. Eur J Neurosci. 1998;10:128–145. doi: 10.1046/j.1460-9568.1998.00003.x. [DOI] [PubMed] [Google Scholar]
- 48.la Fleur SE, Kalsbeek A, Wortel J, Buijs RM. Polysynaptic neural pathways between the hypothalamus, including the suprachiasmatic nucleus, and the liver. Brain Res. 2000;871:50–56. doi: 10.1016/s0006-8993(00)02423-9. [DOI] [PubMed] [Google Scholar]
- 49.Ueyama T, et al. Suprachiasmatic nucleus: A central autonomic clock. Nat Neurosci. 1999;2:1051–1053. doi: 10.1038/15973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.