Abstract
Hepatitis C virus subverts liver-specific microRNA, miR-122, to upregulate viral RNA abundance in both infected cultured cells and in the liver of infected chimpanzees. These findings have identified miR-122 as an attractive antiviral target. Thus, it is imperative to know whether a distinct functional complex exists between miR-122 and the viral RNA versus its normal cellular target mRNAs. Toward this goal, effects on viral RNA abundance of mutated miR-122 duplex molecules, bound at each of the two target sites in the viral genome, were compared to effects on microRNA- or siRNA-mediated regulation of reporter target mRNAs. It was found that miR-122 formed an unusual microRNA complex with the viral RNA that is distinct from miR-122 complexes with reporter mRNAs. Notably, miR-122 forms an oligomeric complex in which one miR-122 molecule binds to the 5′ terminus of the hepatitis C virus (HCV) RNA with 3′ overhanging nucleotides, masking the 5′ terminal sequences of the HCV genome. Furthermore, specific internal nucleotides as well as the 3′ terminal nucleotides in miR-122 were absolutely required for maintaining HCV RNA abundance but not for microRNA function. Both miR-122 molecules utilize similar internal nucleotides to interact with the viral genome, creating a bulge and tail in the miR-122 molecules, revealing tandemly oriented oligomeric RNA complexes. These findings suggest that miR-122 protects the 5′ terminal viral sequences from nucleolytic degradation or from inducing innate immune responses to the RNA terminus. Finally, this remarkable microRNA-mRNA complex could be targeted with compounds that inactivate miR-122 or interfere with this unique RNA structure.
Hepatitis C virus (HCV) infection is a global health problem with an estimated 170 million people infected worldwide (reviewed in ref. 1). HCV-infected individuals typically develop persistent infections that can lead to chronic hepatitis, cirrhosis, and hepatocellular carcinoma (2). Currently, there are no vaccines available, and the clinical efficacies of modern therapeutics are limited. Thus, understanding fundamental aspects of host–virus interactions in HCV infection is important for the discovery of unique anti-HCV treatments. HCV is a hepatotrophic, positive-sense RNA virus that belongs to the family Flaviviridae. The HCV genome contains a single open reading frame encoding the viral polyprotein, which is subsequently cleaved into at least 10 viral proteins by host and viral proteases. The HCV open reading frame is flanked by 5′ and 3′ noncoding regions (NCRs) that contain RNA elements that are important for viral replication and translation (reviewed in ref. 3). Recently, interactions between the liver-specific microRNA, miR-122, with two sites in the HCV 5′NCR (Fig. 1A) have been shown to be essential to maintain HCV RNA abundance during virus infection in cultured cells (4, 5) and in infected chimpanzees (6).
Fig. 1.
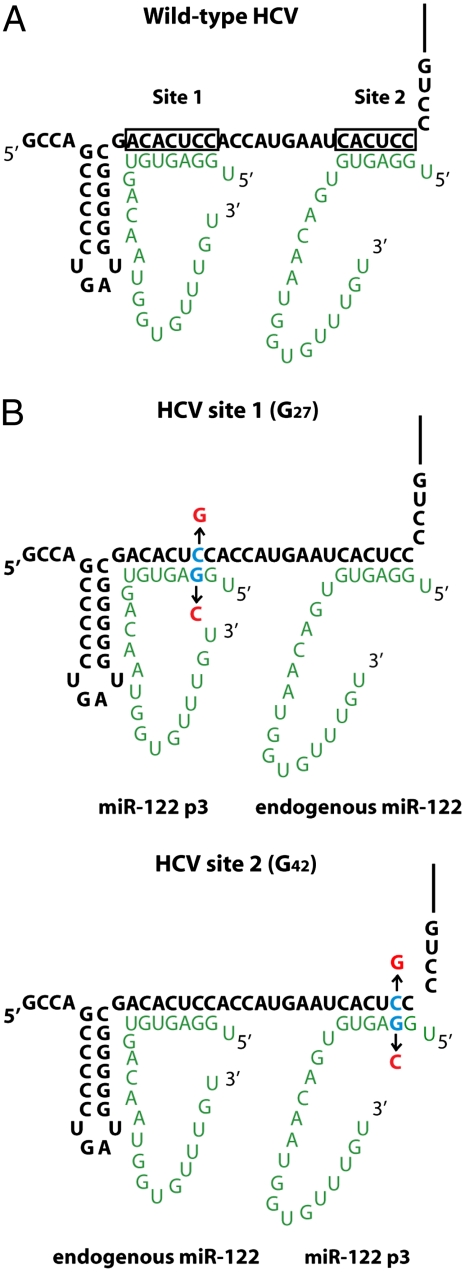
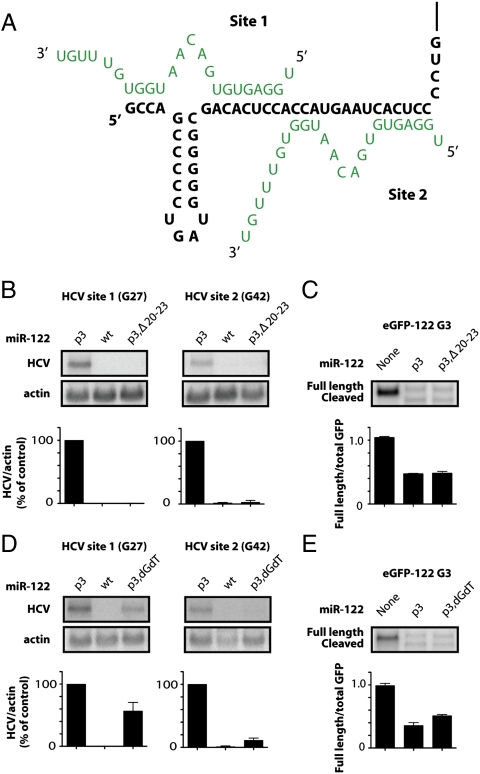
Interactions between miR-122 (green) and HCV (black). (A) Interactions of two miR-122 molecules with HCV RNA. Nucleotides (nts) 2–8 of miR-122 interact at site 1 and nts 2–7 of miR-122 interact at site 2 with the 5′ end of HCV RNA. (B) Location of introduced nucleotide substitutions for stepwise mutational analyses. Mutant miR-122 molecules are directed to HCV site 1 via HCV site 1 (G27), or to HCV site 2 via HCV site 2 (G42). Mutated nucleotides in HCV RNA or miR-122 are shown in red. Endogenous miR-122 binds only to wild-type seed sites in HCV.
MicroRNAs (miRNAs) are small, noncoding RNAs that are predicted to regulate at least one-third of all human mRNAs (7, 8). As a general rule, animal miRNAs function in a sequence-specific manner by binding to imperfectly complementary sites in the 3′NCR of target mRNAs. Such miRNA-target mRNA interactions typically result in decreased target gene expression by mechanisms that involve modulation of translation, mRNA turnover, or both (9–11). Thus, it was surprising that the interaction of miR-122, a liver-specific microRNA, with the HCV RNA genome increases viral RNA abundance. HCV RNA accumulation is dependent upon the miRNA biogenesis pathway, because depletion of Drosha, DGCR8, Dicer, or each of the four Argonaute (Ago) proteins greatly diminished viral RNA abundance in HCV-infected cells (12). Curiously, the interaction of miR-122 with the HCV genome has a minimal effect (1.4- to 3-fold) on viral translation (4, 13, 14) and on the rate of viral RNA synthesis (15), arguing that HCV RNA turnover may be prevented by miR-122.
The predicted stabilization of HCV RNA by miR-122 led us to investigate the contributions of specific miR-122 nucleotides in HCV RNA accumulation. By mutational analysis of miR-122 molecules, we identified nucleotides in miR-122 that are important for HCV RNA accumulation but are dispensable for miRNA- or siRNA-mediated regulation of target mRNAs by canonical microRNA-mRNA interactions. Our results suggest a model for an oligomeric miR-122-HCV complex in which both miR-122 molecules have extensive interactions with the HCV genome. Remarkably, one miR-122 molecule binds to the 5′ terminus of the HCV RNA with 3′ overhanging nucleotides masking the 5′ terminal sequences of the HCV genome. This model suggests that miR-122 may protect HCV RNA from nucleolytic degradation or inhibit the activation of enzymes that induce innate immune responses.
Results
Effects of Stepwise Mutation of miR-122 Molecules on HCV RNA Abundance.
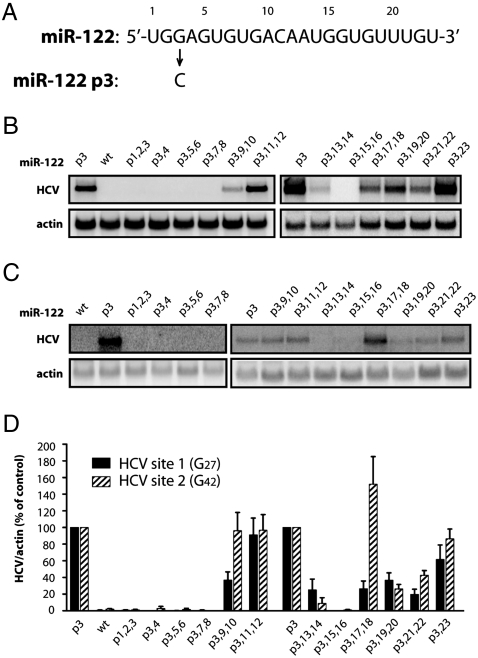
To examine whether miR-122 molecules bound at sites 1 and 2 had interactions outside their seed sequences, i.e., nucleotides 2–8, with HCV RNA, we devised a system to introduce mutations to the miR-122 molecules bound only at site 1 or site 2. Thus, cytidines at HCV nucleotides 27 or 42 were mutated to guanosines (Fig. 1B). Both HCV site 1 (G27) and HCV site 2 (G42) are predicted to bind miR-122 in which the guanosine at nucleotide position 3 (p3) is mutated to a cytidine (miR-122 p3; Figs. 1B and 2A). If both sites need to be occupied by miR-122 with complementary seed sequences, and endogenous wild-type miR-122 can function in the presence of transfected mutant miR-122 p3 duplexes (Fig. 1B), then miR-122 molecules bound to site 1 or 2 will be independently manipulatable. Indeed, HCV site 1 (G27) RNA and HCV site 2 (G42) RNA accumulated only in the presence of duplexes containing a p3 mutation (Fig. 2 B and C, p3 and WT). Therefore, the sequence requirements in miR-122 bound at site 1 and 2 can be independently investigated.
Fig. 2.
Effects of HCV site 1- or HCV site 2-bound miR-122 molecules on HCV RNA abundance. (A) Sequence of wild-type miR-122 and miR-122 p3. Position 3 of miR-122 was mutated from a guanosine to a cytidine to produce miR-122 p3. (B) HCV site 1 (G27) RNA accumulation was measured by Northern blot analysis 5 d postelectroporation in Huh-7 cells. MiR-122 p3 duplexes, mutated in two nucleotide increments, were transfected into cells 1 d prior to, and 1 and 3 d postelectroporation. Nucleotides were mutated to their Watson–Crick base pair. (C) HCV site 2 (G42) RNA and miR-122 mutants were expressed and analyzed as in B. (D) Quantitation of HCV RNA levels. HCV RNA abundances were normalized to γ-actin levels. Data from cells transfected with miR-122 p3 were set to 100%. The data shown represent at least three independent replicates. Error bars represent standard error of the mean.
To test the impact of mutations outside of the seed sequence on the function of miR-122 bound to HCV site 1 or 2, two additional mutations at a time were introduced into miR-122 p3 duplexes. Nucleotides were mutated to their corresponding Watson–Crick base pair (Table S1). The microRNA duplexes were transfected 1 d prior to electroporation of in vitro transcribed HCV site 1 (G27) or HCV site 2 (G42) RNAs, and 1 and 3 d postelectroporation. Viral RNA abundance was measured at 5 d postelectroporation. The patterns of viral RNA accumulation in cells containing either HCV site 1 (G27) (Fig. 2B) or HCV site 2 (G42) (Fig. 2C) were similar, but they displayed some interesting differences that are quantitated in Fig. 2D. HCV RNA accumulation mediated by miR-122 molecules was sensitive to mutations in miR-122 seed nucleotides 2–8 and also in positions 13 and 14, 15 and 16, 19 and 20, and 21 and 22, when bound at either site in HCV (Fig. 2 B–D). Mutating the nucleotides at positions 9 and 10 caused a more dramatic loss of HCV RNA when miR-122 p3,9,10 was directed to site 1, whereas mutating 17 and 18 had opposing effects at site 1 and site 2. These findings argue that distinct nucleotides in miR-122 have different effects on HCV RNA accumulation when miR-122 is bound at site 1 or site 2 of the viral RNA genome.
Effects of miR-122 Mutations in Conventional miRNA and Short-Interfering (si) RNA Assays.
To test whether transfected miR-122 duplexes used in Fig. 2 were able to form miRNA duplexes and engage functional RNA-induced silencing complexes (RISC), all mutated miR-122 duplexes were tested for their abilities to modulate reporter mRNA expression in a miRNA assay. MiRNA assays were performed with firefly luciferase reporter RNAs that contained two identical stretches of the HCV sequence elements (nucleotides 1–45; Fig. 1A) in their 3′ NCRs, yielding a total of four miR-122 binding sites (5). Each miR-122 binding site was mutated to allow binding of miR-122 p3 molecules. After transfection of the luciferase plasmid and mutated miR-122 p3 duplexes into HeLa cells, which do not express miR-122 (4), luciferase activity was measured 24 h posttransfection. To assess efficiency of repression, miR-122 p3 was used as a positive control, and miR-122 p2-8, in which the entire seed sequence had been mutated, was used as a negative control (Fig. S1A). Mutation of any of the seed sequence nucleotides resulted in a considerable increase in luciferase activity over the positive control. In contrast, mutations outside of the seed sequence had limited effects on the ability of mutated miR-122s to repress luciferase expression, although p3,15,16 caused slightly less repression than other mutants (Fig. S1A). Thus, in conventional miRNA assays, mutations outside the seed sequence of miR-122 had minimal effects on miR-122’s function as a microRNA.
As mutation of nucleotides 15 and 16 had such a dramatic effect on HCV RNA abundance (Fig. 2), we further confirmed the functionality of miR-122 p3,15,16 in a siRNA assay. A plasmid encoding EGFP reporter mRNA with a sequence in its 3′ NCR perfectly complementary to miR-122 (4) was mutated to contain a sequence complementary to miR-122 p3,15,16 (EGFP-122-G3G15G16). Fig. S1B shows that miR-122 p3,15,16 was able to exert an siRNA effect, cleaving the GFP reporter mRNA. Thus, miR-122 duplexes with mutations at nucleotides 15 and 16 functioned both in miRNA and in siRNA assays but did not function when bound at either site 1 or site 2 in HCV RNA to augment viral RNA accumulation.
Both Nucleotides 15 and 16 of miR-122 Are Required for miR-122’s Function at Site 1 and Site 2 in HCV RNA.
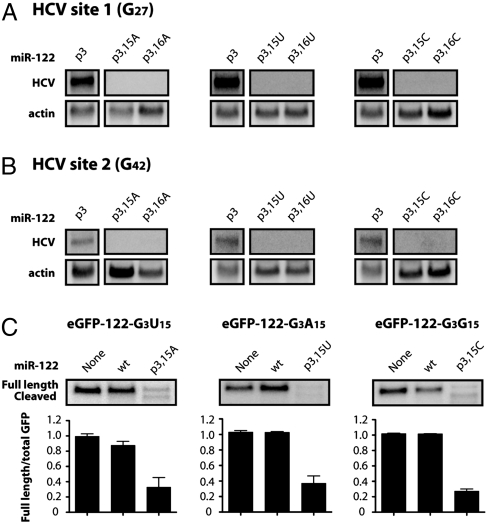
To determine whether the wild-type identity of nucleotide 15, 16, or both was required for HCV RNA accumulation, the guanosines at these positions were individually changed to adenosines, uridines or cytidines. When tested, all of the mutations prevented HCV RNA accumulation, regardless of whether the miR-122 mutants were targeted to site 1 (Fig. 3A) or site 2 (Fig. 3B). However, all six of the individually mutated miR-122 molecules were still able to function in siRNA assays (Fig. 3C and Fig. S2). Therefore, mutation of either position 15 or 16 in miR-122 impedes the function of miR-122 at either site 1 or site 2 in HCV RNA but does not affect the ability of miR-122 to function in RISC-mediated siRNA cleavage.
Fig. 3.
Effects of nucleotide 15 or 16 in miR-122 on HCV RNA abundance (A) and siRNA-mediated cleavage of GFP reporter RNA (B). (A) Position 15 or 16 in miR-122 p3 was mutated to A, U, or C, and the mutants were tested in HCV site 1 (G27) RNA electroporation assays as described in Fig. 2. Northern blots representative of at least three independent replicates are shown. (B) HCV site 2 (G42) RNA and miR-122 mutants were expressed and analyzed as in A. (C) MiR-122 p3 duplexes containing an A, U, or C at position 15 were cotransfected into HeLa cells with a plasmid expressing GFP that contained a perfectly complementary site to the mutant miR-122 molecule in its 3′ noncoding region. GFP mRNA was measured by Northern blot analysis. Full length and cleaved GFP mRNA are indicated. Quantitation is shown as the ratio of full length GFP mRNA to total GFP mRNA and represents at least three independent replicates. Error bars represent standard error of the mean.
Function of miR-122 Nucleotides 15 and 16 at Site 1 and Site 2 in HCV RNA.
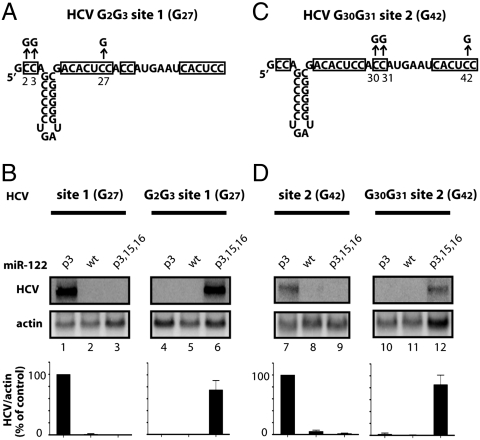
We speculated that the guanosines at positions 15 and 16 of miR-122 might interact directly with HCV RNA. Closer inspection of sequences in the HCV genome surrounding site 1 revealed a highly conserved sequence of two cytidines at the 5′ terminus (nucleotides 2 and 3) (Table S2). To test the possibility that cytidine 2 and cytidine 3 of the HCV genome functionally paired with guanosines 15 and 16 in miR-122, these nucleotides were changed to guanosines in the construct HCV G2G3 site 1 (G27) (Fig. 4A). As is shown in Figs. 2 and 4B, miR-122 p3, but not miR-122 p3,15,16, was able to stabilize HCV site 1 (G27) RNA. However, for HCV G2G3 site 1 (G27) RNA, the three compensatory mutations p3,15,16 were required (Fig. 4B). Thus, positions 15 and 16 in miR-122 molecules bound at site 1 interact directly with nucleotides 2 and 3 of the HCV RNA genome to promote viral RNA accumulation.
Fig. 4.
Interaction of miR-122 nucleotides 15 and 16 with HCV RNA. (A) Diagram of HCV G2G3 site 1 (G27) RNA that was mutated to include additional C to G mutations at nucleotides 2 and 3. (B) HCV site 1 (G27) and HCV G2G3 site 1 G27) RNA abundance in cells transfected with miR-122 p3, WT, or p3,15,16 duplexes, as described in Fig. 2. Northern blot data are representative of at least three independent replicates. Error bars represent standard error of the mean. (C) Diagram of HCV G30G31 site 2 (G42) RNA that was mutated to include additional C to G mutations at nucleotides 30 and 31. (D) HCV site 2 (G42) RNA, HCV G30G31 site 2 (G42), and miR-122 mutants were expressed and analyzed as in Fig. 2.
Inspection of the HCV RNA genome revealed a second conserved motif of two cytidines located between the two seed match sites of miR-122 at positions 30 and 31 of the HCV RNA genome (Table S2 and Fig. 4C). To test whether this region interacted with nucleotides 15 and 16 of miR-122 bound at site 2, the function of miR-122 mutants with HCV site 2 (G42) and HCV G30G31 site 2 (G42) was tested (Fig. 4D). Only miR-122 p3,15,16 was able to rescue HCV RNA accumulation of HCV G30G31 site 2 (G42) RNA (Fig. 4D). These findings argue that miR-122 nucleotides 15 and 16 interact directly with HCV nucleotides 30 and 31 through binding at site 2. As a result, both bound miR-122 molecules contain similar five to six nucleotide bulge sequences and 3′ terminal tails of six to seven nucleotides (Fig. 5A). The proposed pairing shown in the model (Fig. 5A) and the effects of miR-122 nucleotides 19–22 at both site 1 and 2 (Fig. 2 B–D) piqued our curiosity as to the roles of these residues in maintaining HCV RNA abundance.
Fig. 5.
Effects of 3′ end sequences of miR-122 on HCV RNA abundance. (A) Model for miR-122 (green)-HCV (black) interactions at the 5′ end of the HCV genome. (B) Effects of 3′ end deletions in miR-122. A 19-nucleotide-long miR-122 p3 duplex (p3,Δ20–23) was targeted to site 1 or 2 in HCV RNA electroporation assays as described in Fig. 2. (C) siRNA-mediated GFP cleavage assays in HeLa cells transfected with p3,Δ20–23 as described in Fig. 3. (D) Effects of 3′ end nucleotide compositions in miR-122. The last two nucleotides of miR-122 p3 were replaced with deoxynucleotides dG and dT (p3,dGdT) and targeted to site 1 or 2 in HCV RNA electroporation assays as described in Fig. 2. (E) siRNA-mediated GFP cleavage assays in HeLa cells transfected with p3,dGdT as described in Fig. 3.
The 3′ Terminus of miR-122 Is Important to Augment HCV RNA Abundance.
To investigate the importance of the 3′ terminal sequences of miR-122 in HCV RNA abundance further, we tested p3 mutant duplexes in which nucleotides 20–23 were deleted (p3,Δ20–23; Table S1). MiR-122 p3,Δ20–23 duplexes were unable to mediate HCV RNA accumulation when directed to site 1 or site 2 (Fig. 5B). However, p3,Δ20–23 duplexes were able to function in a siRNA assay (Fig. 5C). This finding argues that nucleotides 20–23 are necessary for miR-122’s role in mediating HCV RNA abundance but are dispensable for its incorporation into active RISC complexes that perform siRNA-mediated cleavage.
To further examine the function of the 3′ end sequences of miR-122 in modulating HCV RNA abundance, mutant miR-122 molecules were synthesized that contained two 3′ terminal deoxynucleotides (p3,dGdT, Table S1). Transfection of miR-122 p3,dGdT duplexes rescued HCV site 1 (G27) RNA accumulation to approximately 60% of the positive control when bound at site 1 (Fig. 5D). However, miR-122 p3,dGdT duplexes rescued HCV site 2 (G42) RNA to only 10% when directed to site 2 (Fig. 5D). Importantly, as was observed for the p3,Δ20–23 duplexes, miR-122 p3,dGdT molecules functioned with similar efficiencies in siRNA assays, arguing that they were able to function in active RISC complexes (Fig. 5E). These observations, along with the stepwise mutation analyses (Fig. 2 and Fig. S1A), suggest that the 3′ end sequences of miR-122, i.e., nucleotides 19–23, of miR-122 are important for HCV RNA accumulation and may have distinct roles at site 1 and site 2.
Effects of Ago 1 and 2 on miR-122’s Ability to Modulate HCV RNA Abundance.
Previous results showed that HCV RNA accumulation is dependent upon the intact RNA silencing pathway machinery including Drosha, DGCR8, Dicer, and the four Ago proteins (12). Thus, the RNA silencing pathway is required for the generation of mature miR-122 and/or for positioning miR-122 onto the 5′ end sequences of HCV. The data showing that 3′ deoxynucleotide-containing miR-122 duplexes functioned in the RISC but not in maintaining HCV RNA abundance argue that a canonical RISC may not be needed to maintain HCV RNA abundance. To address this possibility, Ago proteins were depleted by siRNA-mediated gene knockdown. Depleted cells were then electroporated with wild-type HCV RNA and supplemented with wild-type or p3 miR-122 duplexes (Fig. S3 A and B). Depletion of Ago 1 and 2 caused a dramatic decrease in HCV RNA levels compared to mock- and control siRNA-treated cells (Fig. S3A). However, transfection of WT, but not of mutant p3, miR-122 duplexes rescued HCV RNA abundance to similar levels in Ago 1/2 siRNA-treated cells and control siRNA-treated cells (Fig. S3 A and C). These results suggest that exogenously provided mature miR-122 duplexes modulate HCV RNA abundance when Ago 1 and 2 are depleted.
Discussion
Through a series of nucleotide mutations in miR-122 molecules, we have shown that mutation of the seed sequence (nucleotides 2–8) and positions 15 and 16 did not allow HCV RNA accumulation when the mutant miR-122 was bound at either site 1 or site 2 in the HCV RNA genome. Compensatory mutations in the HCV genome at positions 2–3 and 30–31 were able to rescue HCV RNA accumulation when the miR-122 p3,15,16 mutant was directed to site 1 or site 2, respectively. Furthermore, mutation or deletion of the 3′ end sequences in miR-122 decreased HCV RNA accumulation, suggesting that these nucleotides also play a role in mediating HCV RNA abundance. Mutations at positions 9–10, 17–18, and 21–22 revealed differences in the ability of miR-122 molecules to accumulate HCV RNA when directed to either site 1 or site 2. These results suggest a model in which two miR-122 molecules form an oligomeric complex with HCV, in which internal nucleotides in each miR-122 molecule bulge out and the 3′ end of miR-122 engages in base pair interactions with the 5′ terminus of the HCV genome, providing a 3′ overhang (Fig. 5A).
The importance of nucleotides 15 and 16 in miR-122 molecules to maintain HCV RNA abundance was completely unexpected. This finding argues that the miR-122-HCV RNA complex is an unconventional microRNA-target mRNA complex, because miR-122 molecules with mutations at nucleotides 15 and 16 were still able to function in conventional miRNA and siRNA assays. Using genetic complementation assays, we demonstrated that nucleotides 15 and 16 of miR-122 at site 1 interact with nucleotides 2 and 3 of the HCV genome (Figs. 4B and 5A). Additionally, mutation of nucleotides 14 and 17 in miR-122 at site 1 decreased HCV RNA abundance, suggesting that these nucleotides interact with nucleotides 4 and 1 of HCV, respectively. Nucleotides 2 and 3 are highly conserved across all HCV genotypes (Table S2), arguing that the interaction of miR-122 with these nucleotides is conserved among the HCV genotypes. Interestingly, the 5′ terminal guanosine nucleotide in HCV does not seem to be completely conserved among genotypes, and it has been reported that adenosine is the 5′ terminal residue in some HCV genotypes (Table S2). However, nucleotide 17 of miR-122 is a uridine; thus, either a 5′ terminal guanosine or adenosine would be able to interact with miR-122, forming a G-U wobble or a canonical A-U base pair interaction with the first nucleotide of HCV, respectively. In addition, sequence analyses of HCV replicon RNAs recovered from stably selected cell lines revealed that the initial 5′ terminal guanosine residue was replaced by an adenosine residue over time (16, 17). Furthermore, the acquired 5′ terminal adenosine residue was maintained in the HCV genomes, even after numerous rounds of viral RNA replication in cell culture (16). Cai et al. also noted that both guanosine and adenosine residues were present at the 5′ termini of HCV genomes isolated from clinical samples (16). Although this finding was attributed to a preference for the NS5B RNA-dependent RNA polymerase to initiate replication with a purine nucleotide, our results indicate that a 5′ terminal guanosine or adenosine residue may have been selected because it is required to base pair with miR-122.
Our findings also demonstrate that nucleotides 15 and 16 of site 2-bound miR-122 interact with the HCV genome at nucleotides 30 and 31 (Figs. 4D and 5B). The adenosine residues at positions 29 and 32 in the HCV RNA genome are potential binding partners for the uridine residues at positions 17 and 14 of site 2-bound miR-122, respectively. This hypothesis is supported by the data that mutation of nucleotides 13 and 14 of miR-122 at site 2 substantially reduced HCV RNA accumulation (Fig. 2). Although the adenosine residue at position 32 is completely conserved across all HCV genotypes, the adenosine residue at position 29 is a guanosine residue in some genotypes (Table S2). However, the guanosine residue at position 29 would still be predicted to form a G-U wobble pair with miR-122 at this position, providing further evidence for this interaction. Intriguingly, mutation of nucleotides 17 and 18 at site 2 resulted in an increase in HCV RNA abundance (Fig. 2). This finding suggests that pairing of these nucleotides decreases HCV RNA accumulation. This may be due to competition between the uridine at position 1 of site 1-bound miR-122 and the uridine at position 17 of site 2-bound miR-122 with the adenosine at position 29 in HCV (Fig. 5A). Alternatively, mutation of nucleotide 17 could relieve some of the conformational restriction imposed by two miR-122 complexes in such close proximity.
A previous study suggested that nucleotides near the 3′ end of miR-122 were important in maintaining HCV RNA abundance (18). Because our study suggested that the 3′ end sequences of miR-122 form a 3′ overhanging extension with the HCV terminal nucleotides, we investigated their length in maintaining HCV RNA abundance. Although a truncated miR-122 molecule lacking the four 3′ terminal nucleotides was able to incorporate into an active RISC (Fig. 5C), it was unable to stimulate HCV RNA accumulation (Fig. 5B). Similarly, exchanging the terminal two ribonucleotides of miR-122 with deoxynucleotides reduced HCV RNA accumulation (Fig. 5D). The effects of deoxynucleotide substitution were more dramatic when miR-122 molecules were directed to site 2 than to site 1 (Fig. 5D), suggesting that these nucleotides may have distinct roles at site 1 and site 2. The 3′ end of miR-122 sequences may form base pair interactions with other viral or cellular RNA sequence elements or form complexes with proteins. In each case, ribonucleotides may form more stable interactions than deoxyribonucleotides. These observations suggest that the 3′ terminal nucleotides in miR-122 are important for HCV RNA accumulation and may mediate interactions with the viral RNA in unconventional RISCs.
Target mRNA recognition by endogenous microRNAs has been demonstrated to rely on complementarity to nucleotides 2–8 in microRNAs. However, recent results have suggested that nucleotides 13–16 in microRNAs may also play a role in RISC target recognition (19, 20). We discovered that nucleotides 15–16 of miR-122 make important contacts with the HCV genome to mediate effects on HCV RNA abundance. It is therefore possible that Ago proteins play a role in the positioning of miR-122 onto HCV RNA. Although depletion of Ago 1 and 2 resulted in a decrease in HCV RNA abundance, supplementation with exogenous miR-122 was able to stimulate HCV RNA accumulation to a similar extent in Ago 1 and 2 siRNA-treated and control siRNA-treated cells (Fig. S3). Although these results suggest that miR-122 affects HCV RNA abundance in the presence of low abundance of Ago 1 and 2, it is possible that low abundance of Ago 1 and 2 are sufficient to modulate HCV RNA or that Ago 3 and 4 can compensate for the loss of Ago 1 and 2. A more rigorous examination will be required to define the exact role of Ago proteins in miR-122/HCV RNA interactions and to elucidate the precise components of this unique complex at the 5′ end of the HCV genome.
Our finding that miR-122 interacts with the 5′ terminus of the HCV genome and produces 3′ overhanging extensions suggests unique hypotheses regarding its mechanism. Binding of miR-122 to site 1 of the HCV genome may protect HCV RNA from cytoplasmic sensors of viral RNA. For instance, activation of the RNA helicase retinoic acid-inducible gene-I (RIG-I) protein, a cellular sensor of RNA that is up-regulated in chimpanzees persistently infected with HCV (21), is known to be inhibited by 3′ overhangs (22, 23). Thus, the 3′ overhang of miR-122 at the 5′ end of HCV RNA may mask the 5′ terminus from recognition by cytoplasmic RNA sensors. Additionally, binding of miR-122 to the HCV genome may help displace the negative strand during RNA replication to prevent recognition of blunt-ended or long double-stranded RNA intermediates. Another protective role of miR-122’s interaction with the 5′ terminus of the HCV genome could involve protection from 5′ exonucleases. Depletion of XRN-1 mRNA, encoding a 5′-3′ exonuclease that is involved in decay of cellular mRNAs (24, 25), increases the expression of luciferase from tricistronic HCV replicons (26) and the abundance of electroporated HCV replicon RNA. Interestingly, Takahashi et al. reported that the 5′ termini of HCV RNA obtained from HCV replicon cells contains a 5′ monophosphate (17), suggesting that the 5′ terminus of HCV RNA is sensitive to degradation by Xrn-1. However, the chemical composition of the 5′ termini of HCV during infection is unknown. Nonetheless, the 3′ overhang motif created by miR-122 binding could protect the HCV genome from degradation by Xrn-1 or other 5′ exonucleases.
Previous studies of interactions between miR-122 and HCV RNA have yielded conflicting results with regard to the role of miR-122 in the HCV lifecycle. Although initial reports suggested miR-122 had little effect on HCV translation (4), others have reported moderate effects of miR-122 on HCV internal ribosome entry site (IRES)-mediated translation (13, 14). However, the 2- to 3-fold decrease in IRES-directed translation is not sufficient to account for the striking loss of HCV RNA upon mutation of miR-122 binding sites or sequestration of miR-122 (14, 15). These results, together with our findings that miR-122 molecules form complex RNA-RNA interactions with the HCV RNA genome at the viral 5′ terminal RNA sequences, suggest that miR-122 may have multiple roles in the HCV lifecycle. Additionally, the fact that the mutant miR-122 molecules were able to incorporate into RISC and function in conventional miRNA and siRNA assays but not HCV RNA accumulation assays provides further evidence for an unusual oligomeric complex containing miR-122 that functions in a manner unique to the HCV lifecycle. Further elucidation of additional host or viral factors associated with this complex may provide potential antiviral targets to limit HCV replication.
Materials and Methods
Oligonucleotides and DNA Constructs.
All RNA oligonucleotides were synthesized by the Stanford Protein and Nucleic Acids facility. The sequences of microRNAs are as shown in Table S1. Guide strands p3,5,6; p3,15,16; p3,17,18; p3,19,20; p3,21,22; p2-8; and p3,Δ20–23 were annealed to the corresponding mutant passenger strands to maintain base pairing interactions of the duplex. All other guide strands were duplexed with the wild-type passenger strand.
Plasmid eGFP-122 has been previously described (4). To generate the plasmid EGFP-122-G3, mutagenic primers (5′-CAA ACA CCA TTG TCA CAC TGC ATA GAT CAT AAT CAG CCA-3′ and 5′-TGG CTG ATT ATG ATC TAT GCA GTG TGA CAA TGG TGT TTG-3′) were used in site-directed mutagenesis (QuikChange II XL, Stratagene). The same protocol was used to create EGFP-G3G15G16 (primers: 5′-GGC GGC CGC ACA AAA CAG GAT TGT CAC ACT GCA TA-3′ and 5′-TAT GCA GTG TGA CAA TCC TGT TTG TGC GGC CGC C-3′), EGFP-G3U15 (5′-GTA GGC GGC CGC ACA AAC ACT ATT GTC ACA CTG CAT AGA T-3′ and 5′-ATC TAT GCA GTG TGA CAA TAG TGT TTG TGC GGC CGC CTA C-3′), EGFP-G3A15 (5′-GTA GGC GGC CGC ACA AAC ACA ATT GTC ACA CTG CAT AGA T-3′ and 5′-ATC TAT GCA GTG TGA CAA TTG TGT TTG TGC GGC CGC CTA C-3′), and EGFP-G3G15 (5′-GTA GGC GGC CGC ACA AAC ACG ATT GTC ACA CTG CAT AGA T-3′ and 5′-ATC TAT GCA GTG TGA CAA TCG TGT TTG TGC GGC CGC CTA C-3′).
The pH77ΔE1/p7 HCV G2G3 site 1 (G27) and HCV G30G31 site 1 (G42) mutations were created by site-directed mutagenesis (Quikchange II XL, Stratagene). The HCV site 1 (G27) and HCV site 2 (G42) (originally referred to as S1:p3 and S2:p3, respectively) pH77ΔE1/p7 plasmids (5) were mutated according to the manufacturer’s instructions with the following primers: G2G3 site 1 (G27) (5′-CTA ATA CGA CTC ACT ATA GGG AGC CCC CTG ATG GG -3′ and 5′-CCC ATC AGG GGG CTC CCT ATA GTG AGT CGT ATT AG-3′) and HCV G30G31 site 1 (G42) (5′-GGG GCG ACA CTC CAG GAT GAA TCA CTG CCC-3′ and 5′-GGG CAG TGA TTC ATC CTG GAG TGT CGC CCC-3′). The presence of all mutations was confirmed by sequencing.
Cell Culture and Transfection.
Huh-7 cells were cultured in DMEM supplemented with 10% FBS, 1% nonessential amino acids, and 200 μM L-glutamate (GIBCO), and HeLa cells were cultured in DMEM supplemented with 10% FBS and 200 μM L-glutamate.
HCV RNA electroporation assays were carried out in Huh-7 cells as previously described (5). All miR-122 duplexes were transfected into cells at a concentration of 50 nM. For siRNA (GFP cleavage) assays, DNA constructs and oligonucleotides were transfected into HeLa cells in six-well plates using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. GFP plasmids were transfected at 50 ng per well. Twenty-four hours after transfection, cells were lysed and total RNA was extracted in TRIzol reagent (Invitrogen).
RNA Isolation and Northern Analysis.
Total RNA was extracted using TRIzol reagent, according to the manufacturer’s instructions. Ten micrograms of total RNA were separated in 1% agarose gels containing 1× 3-(N-morpholino)propanesulfonic acid (MOPS) buffer and 2.2 M formaldehyde and transferred to Zeta-probe membranes (Bio-Rad). Membranes were hybridized in Express Hyb Hybridization Buffer (ClonTech) to random-primed 32P-labeled DNA probes (RadPrime DNA Labeling System, Invitrogen) complementary to HCV (nucleotides 84-374), GFP (nucleotides 40-814), or γ-actin (nucleotides 685-1171). Autoradiographs were quantitated using ImageQuant (GE Healthcare).
Supplementary Material
Acknowledgments.
We are grateful to Karla Kirkegaard for valuable comments on the manuscript and throughout this study. We thank Teresa Abraham for reagents and many helpful discussions. This study was supported by grants from the National Institutes of Health (AI47365, AI069000). E.S.M acknowledges support from the Paul and Mildred Berg Stanford Graduate Fellowship. S.M.S. acknowledges the National Canadian Research Training Program in Hepatitis C and the Natural Sciences and Engineering Research Council of Canada for postdoctoral fellowships.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.R.C. is a guest editor invited by the Editorial Board.
See Commentary on page 3101.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012464108/-/DCSupplemental.
References
- 1.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. doi: 10.1053/jhep.2002.36227. [DOI] [PubMed] [Google Scholar]
- 3.Tellinghuisen TL, Evans MJ, von Hahn T, You S, Rice CM. Studying hepatitis C virus: Making the best of a bad virus. J Virol. 2007;81:8853–8867. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 5.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanford RE, et al. Therapeutic silencing of MicroRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezikov E, et al. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA. 2006;103:2746–2751. doi: 10.1073/pnas.0511045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;2007(367):re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 11.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 12.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci USA. 2007;104:12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henke JI, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norman KL, Sarnow P. Modulation of hepatitis C virus RNA abundance and the isoprenoid biosynthesis pathway by microRNA miR-122 involves distinct mechanisms. J Virol. 2010;84:666–670. doi: 10.1128/JVI.01156-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai Z, Liang TJ, Luo G. Effects of mutations of the initiation nucleotides on hepatitis C virus RNA replication in the cell. J Virol. 2004;78:3633–3643. doi: 10.1128/JVI.78.7.3633-3643.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi H, et al. Analysis of the 5′ end structure of HCV subgenomic RNA replicated in a Huh7 cell line. Intervirology. 2005;48:104–111. doi: 10.1159/000081736. [DOI] [PubMed] [Google Scholar]
- 18.Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Sym. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- 19.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigger CB, et al. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J Virol. 2004;78:13779–13792. doi: 10.1128/JVI.78.24.13779-13792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nature Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 23.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens A. Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′ leads to a 3′ mode of hydrolysis. J Biol Chem. 1980;255:3080–3085. [PubMed] [Google Scholar]
- 25.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable messenger-RNA encoded by the yeast Mfa2 gene leads to decapping followed by 5′-->3′ digestion of the transcript. Gene Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 26.Jones DM, Domingues P, Targett-Adams P, McLauchlan J. Comparison of U2OS and Huh-7 cells for identifying host factors that affect hepatitis C virus RNA replication. J Gen Virol. 2010;91:2238–2248. doi: 10.1099/vir.0.022210-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.