Abstract
Elongation factor 4 (EF4) is one of the most conserved proteins present in bacteria as well as in mitochondria and chloroplasts of eukaryotes. Although EF4 has the unique ability to catalyze the back-translocation reaction on posttranslocation state ribosomes, the physiological role of EF4 remains unclear. Here we demonstrate that EF4 is stored at the membrane of Escherichia coli cells and released into the cytoplasm upon conditions of high ionic strength or low temperature. Under such conditions, wild-type E. coli cells overgrow mutant cells lacking the EF4 gene within 5–10 generations. Elevated intracellular Mg2+ concentrations or low temperature retard bacterial growth and inhibit protein synthesis, probably because of formation of aberrant elongating ribosomal states. We suggest that EF4 binds to these stuck ribosomes and remobilizes them, consistent with the EF4-dependent enhancement (fivefold) in protein synthesis observed under these unfavorable conditions. The strong selective advantage conferred by the presence of EF4 at high intracellular ionic strength or low temperatures explains the ubiquitous distribution and high conservation of EF4.
Keywords: bacterial membrane, protein synthesis under stress
The prolongation of the nascent polypeptide chain on ribosomes is driven by two universal elongation factors (EF): EF-Tu (EF1A in Archaea and eukarya), which transports aminoacyl-tRNAs to the ribosomal A site and EF-G (EF2), which translocates the ribosomal tRNA2•mRNA complex by one codon length moving the peptidyl-tRNA and deacylated-tRNA from the A and P sites to the P and E sites, respectively (1).
Recently a unique elongation factor was detected in bacteria, termed elongation factor 4 (EF4).† In Escherichia coli EF4 was originally called LepA, because the lepA gene is located in the Lep operon upstream of the Lep protein, a universal peptidase that cleaves the signal peptide from the N terminus of proteins after translocation through the membrane (4). EF4 was considered to be a membrane protein (5), although a knockout of the gene did not exhibit any observable protein transport deficiencies. Moreover, the growth rate of the knockout strain was not significantly affected (6), which greatly reduced the general interest in this gene. However, the picture began to change when Kusters and coworkers in Amsterdam reported a systematic knockout analysis in Helicobacter pylori (7), a medically important pathogen responsible for the widespread disease of stomach ulcera. Among 10 genes identified to be essential for the survival of H. pylori in the hostile low-pH milieu of the stomach, one of them was the lepA gene (7).
A systematic analysis of distribution of the lepA gene revealed that EF4 is one of the best conserved proteins present in practically all bacteria and the organelles mitochondria and chloroplasts. Functional studies indicate that EF4 is characterized by the following features: (i) a ribosome-stimulated GTPase activity (2, 8), (ii) the ability to back-translocate posttranslocational complexes, i.e., EF4 catalyzes the movement of the tRNA2•mRNA complex from E and P sites to P and A sites, respectively (2, 8); and (iii) increases the active fraction of newly translated proteins (2). The structure of EF4 is related to that of EF-G, comprising equivalents to all EF-G domains except the 130-aa-long domain IV. In addition, EF4 contains both a unique C-terminal domain that is structurally unrelated to known proteins and a short V′ subdomain (9, 10). On the ribosome, domain IV of EF-G occupies the decoding center at the A site, which has been suggested to act as a “doorstop” preventing back-translocation as long as EF-G is bound. The lack of the domain IV in EF4, coupled with the additional C-terminal domain that interacts with the A-tRNA (tRNA at the A site) (10) are consistent with the observation that EF4 can actively back-translocate the tRNA2•mRNA complex (8, 10).
The suspicion that EF4 prevents misincorporation of amino acids during translation (2) cannot be easily reconciled with the fact that misincorporations are not often detrimental to the stability or function of a protein. This plasticity is because of the fact that near-cognate amino acids, the prime candidates for misincorporations, are of the same chemical character as the cognate one. For example, near-cognate amino acids are hydrophobic if the cognate is hydrophobic. The dangerous noncognate amino acids, which are often of a different chemical character as the cognate amino acids, are carefully excluded from the ribosomal selection process because of coevolution of the decoding mechanisms and the organization of the genetic code (for definition of near-cognate and noncognate, see ref. 11; for review, see ref. 12).
Therefore we set out to study the physiological importance of the back-translocation function of EF4. Here we demonstrate that with increasing intracellular ionic strength the cellular localization of EF4 is shifted from the membrane to the cytoplasm, changing the cytoplasm∶membrane ratio from 0.25∶1 to 5∶1. This ratio suggests that the membrane functions as a storage place for EF4, releasing the protein when needed. Accordingly, we find that the presence of the lepA gene only provides a viability advantage for the cell under unfavorable growth conditions. At high intracellular ionic strength, the fraction of unscheduled stalled ribosomes is dramatically increased and impairs synthesis rate and cotranslational folding of proteins. We find that EF4 does not prevent misincorporation, in agreement with ref. 13, but rather remobilizes the stalled ribosomes and thus restores the rate of protein synthesis and supports domain folding of the nascent peptide chain.
Results
Lack of EF4 Impairs Growth Under Stress Conditions.
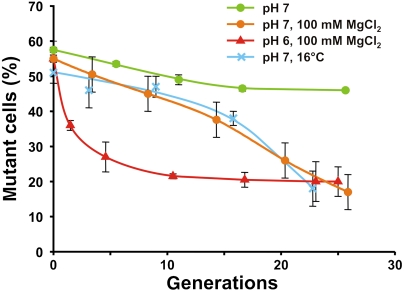
A knockout mutant of the lepA gene (ΔlepA) in E. coli showed almost no phenotype when grown in rich LB medium (6). However, the enormous conservation and wide distribution of the EF4 factor (2) prompted us to explore the phenotype of the ΔlepA strain in more detail. For this purpose, we first prepared an isogenic ΔlepA strain where a kanamycin-resistant gene replaces the wild-type lepA gene. Growth competition experiments were performed between the wild-type E. coli and ΔlepA strains under various stress conditions. For example, low-pH media was tested, having in mind the phenotype of the ΔlepA mutant of H. pylori reported by ref. 7. Equal amount of wild-type and mutant cells were mixed and cocultured for 24 h and the population distribution was determined every 4 h. At pH 7 in minimal media, the growth of the ΔlepA strain was slightly impaired compared to the wild type, decreasing from 58 to 48% after approximately 15 generations. Addition of 100 mM MgCl2 (Fig. 1) or 200 mM KCl (14) to the growth medium led to a dramatic decrease of ΔlepA cells (to 20% after 25 generations). A similar reduction was observed at 16 °C (Fig. 1). Furthermore, the same decrease in ΔlepA population was observed after only 10–15 generations when the pH of the medium was reduced to 6, both in the absence (14) and presence of 100 mM MgCl2. These results demonstrate clearly that EF4 plays an important role for bacterial growth under high ionic conditions as well as at low temperature. The leveling off in ΔlepA cells at ∼20% and eventually obtaining the growth rate of wild-type cells is probably because of the second phase of osmoregulation in bacteria (see Discussion).
Fig. 1.
Growth competition. Equal mixtures of E. coli wild-type cells and ΔlepA cells were grown under various conditions as indicated and the fraction of ΔlepA cells was determined (mutant cells in %), as described in the text and Materials and Methods.
EF4 Moves Between Cytoplasm and Membrane.
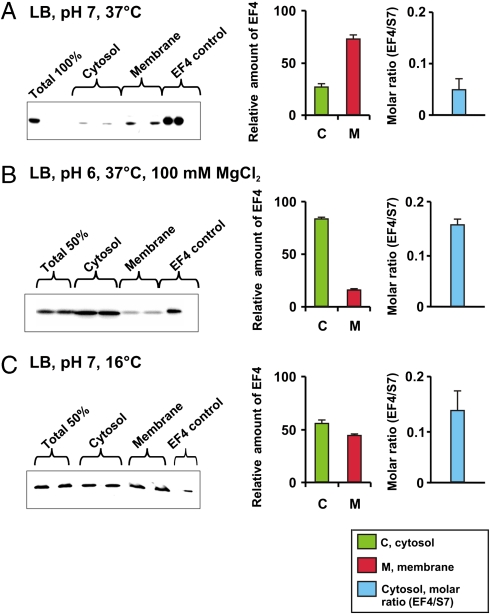
The observation that EF4 is a translational G protein that interacts with the ribosome (2, 10) is hard to reconcile with the observation that EF4 is a membrane protein (4), because the majority of ribosomes in bacteria are cytoplasmic, rather than membrane bound. Therefore, we readdressed this issue by determining the distribution of EF4 between cytosol and membrane fractions under various growth conditions, using Western blot analysis with a polyclonal antibody against EF4 (Fig. 2). Under optimal conditions (pH 7, 37 °C, LB medium) the cytosol∶membrane distribution of EF4 was 0.25∶1, i.e., as might be expected for a membrane protein. Surprisingly, the situation is reversed in cells grown at pH 6 in media containing 100 mM MgCl2, such that the cytosol∶membrane ratio of 5∶1 was determined. Similarly, at pH 7 but with growth at low temperature (16 °C), EF4 is preferentially present in the cytosol. Thus, we suggest that EF4 is not a bona fide membrane protein, but rather that the membrane acts as a storage vessel for EF4 and that EF4 is released into the cytoplasm as required under specific stress conditions. Indeed, the EF4∶70S molar ratio in the cytoplasm, which was determined with the help of antibodies against the ribosomal protein S7 as a ribosome marker, revealed that the ratio increased by up to threefold under various stress conditions, i.e., from 0.06 (pH 7, 37 °C, LB medium) to 0.14–0.17 (Fig. 2, Right).
Fig. 2.
EF4 distribution between cytosol and membranes. (Left) Immunoblotting using polyclonal anti-EF4 antibody as described in Materials and Methods. Total, the total amount of EF4 in a cell sample corresponding to the cell fraction used for EF4 determination in the cytosol and membranes, respectively. (Center) Estimates of the relative amounts (%) of EF4 in membrane and cytosol fractions based on the intensities of the corresponding immunoblotting bands. (Right) Molar ratio between EF4 and ribosomal protein S7 in the cytosol fraction based on intensities of corresponding immunoblotting bands (14), using polyclonal anti-EF4 and anti-S7 antibodies; band intensities were calculated as described in Materials and Methods.
EF4 Increases the Rate of Protein Synthesis.
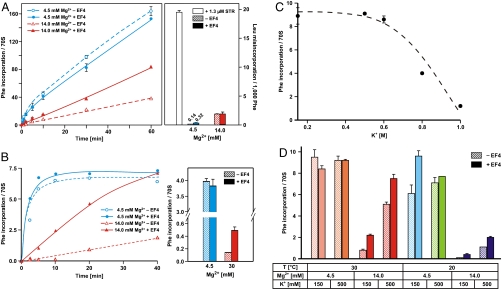
An optimized poly(Phe) synthesis system (15) was used to monitor the effects of EF4 on the rate and fidelity of translation. In this system, > 150 Phe can be incorporated into the nascent polypeptide chain per ribosome. Because this activity assumes ∼100% active ribosomes (Fig. 3A, Left), which is usually not the case, the incorporation per active ribosome will be even higher. The misincorporation error in this system was monitored by incorporation of near-cognate Leu: At 4.5 mM Mg2+ ∼0.14 Leu misincorporations per 1,000 Phe could be detected (Fig. 3A, Right), indicating the sensitivity of the system. The misincorporation of Leu could be increased 150-fold (20 Leu/1,000 Phe) by the addition of streptomycin (STR, open bar), an aminoglycoside well known for its ability to induce translational misreading (16). In contrast, the addition of EF4 had little if any effect on the misincorporation rate. The effect of EF4 in the system was more evident when the rate of protein synthesis was monitored at higher Mg2+ concentration: EF4 increased the rate about twofold when the Mg2+ concentration was raised from 4.5 to 14 mM (Fig. 3A, Left). Despite the strong EF4-dependent stimulation of the poly(Phe) synthesis at the high Mg2+ concentration, EF4 had no noticeable effect on the misincorporation frequency.
Fig. 3.
EF4 effects on rate and accuracy of in vitro protein synthesis. (A, Left) Kinetics at 30 °C of poly(Phe) synthesis under optimal (4.5 mM) and high Mg2+ concentrations (14 mM) using S100 enzymes, which already contain EF4. (A, Right) Leu misincorporation per 1,000 incorporated Phe under the same conditions. (B, Left) Kinetics of oligo(Phe) in a pure system containing precharged 10 Phe-tRNAs per ribosome and the factors EF-Tu, EF-Ts, EF-G, and ± EF4. (B, Right) Oligo(Phe) incorporation in the pure system after 20 min at optimal and unfavorable Mg2+ concentrations (4.5 and 30 mM Mg2+, respectively). (C) Oligo(Phe) synthesis in the pure system incubated 30 °C for 60 min at various K(acetate) concentrations. (D) Oligo(Phe) synthesis in the pure system at various ionic concentrations either at 30 or 20 °C for 20 min as indicated.
The EF4-dependent stimulation of poly(Phe)-synthesis probably represents a minimal value, because the standard poly(Phe)-synthesis assay utilizes S100 fractions that already contain some EF4. To assess more precisely the stimulation factor of EF4, we analyzed the effect of EF4 in a purified system containing only EF-Tu, EF-Ts, and EF-G, together with precharged Phe-tRNA and poly(U) mRNA. In this system, we observed a maximal incorporation of about seven Phe/70S, because only 10 precharged Phe-tRNA/70S were present in the assay. Again no significant effect of EF4 was seen under optimal conditions (4.5 mM Mg2+ plus polyamines), whereas at 14 mM Mg2+ (plus polyamines) EF4 increased the rate approximately fivefold (Fig. 3B, Left). At 30 mM Mg2+ protein synthesis is practically blocked, yet under these extremely restricted conditions EF4 allows for slow but significant protein synthesis, indicating the mobilizing effect of EF4 on the ribosome (Fig. 3B, Right).
In vivo the Mg2+ concentration can increase two- to fivefold in bacteria and mitochondria (17–19), and the K+ concentration as much as 7- to 10-fold, reaching values approaching 1 M in E. coli depending on the salt conditions of the media (17, 20). In vitro elevated K+ concentrations above 0.5 M increasingly block protein synthesis (incubation temperature of 30 °C) (Fig. 3C), whereas dramatic impairments of protein synthesis are already seen at 14 mM Mg2+ (150 mM K+) (Fig. 3B and D, see also ref. 15). At 30 °C, high Mg2+ is the only ionic condition where EF4 improves protein synthesis at low and high K+. In contrast, at lower temperature (20 °C) EF4 already exerts a beneficial effect at moderate salt conditions (Fig. 3D, light-blue bars).
Discussion
High concentrations (10–15 mM) of Mg2+ influence the structure of the ribosome, impairing the accuracy and reducing the rate of protein synthesis. Error-inducing aminoglycoside antibiotics such as streptomycin also impair both accuracy and rate of protein synthesis, but stimulates poly(Phe) synthesis (15). However, the underlying mechanisms of error induction must be quite different in the two cases because EF4 reverses only the salt-induced effects rather than those induced by aminoglycosides (2). One proposed explanation was that aminoglycosides induce a localized change of the decoding center, whereas high salt and particularly high Mg2+ is likely to cause a more general conformational change in the ribosome; these conformational changes might occasionally impair EF-G-dependent translocation in such a way that the stalled ribosomes display the codon improperly at the decoding center in the A site, thus increasing the misincorporations (2). In this model, EF4 would recognize such defective translocated ribosomes and trigger a back-translocation, thereby preventing a possible misincorporation and providing a second chance for correct translocation. Substoichiometric amounts of EF4 relative to ribosomes were shown to be sufficient for increasing the active fraction. The specificity for impaired ribosomes was lost at high EF4 levels, resulting in inhibition of protein synthesis via futile cycles of translocation and back-translocation, consistent with the toxic effect of EF4 overexpression (2).
Here we demonstrate that EF4 does not in fact prevent misincorporations, consistent with observations in vivo (13), but rather EF4 stimulates protein synthesis by up to five times in conditions of high (14 mM) Mg2+ (Fig. 3 A and B) or by about 50% at lower temperature (20 °C) and moderate salt conditions (Fig. 3D, light-blue columns). This surprising observation was not anticipated considering the back-translocation function of the factor and calls for another explanation of the role of EF4 that has to be reconciled with the in vivo data presented here.
Under hyperosmotic conditions in the media, the growth rate of ΔlepA cells initially fell rapidly compared to that of wild-type cells, but after 10–20 generations, the growth rate of both cell types equalized and remained so for many generations (Fig. 1). This observation is consistent with the known two-phase response of bacterial osmoregulation. During the first phase, intracellular Mg2+ and K+ concentrations (normally maintained at 1–4 and 100–150 mM) increase, reaching three- to sevenfold higher concentrations in bacteria, respectively (17–20). In the second phase osmolytes such as disaccharide trehalose and others are synthesized, resulting in a reduction of the K+ concentration back to its normal level (21). The first phase can be further subdivided into a fast initial phase that occurs within seconds where the Mg2+ concentration is raised actively, and a second (probably slow) phase, during which K+ and its counterion glutamate are increased (18, 19). Our data indicate that it is preferentially this Mg2+-dominated phase that requires the presence of EF4, whereas at high K+ concentrations only moderate EF4 effects are observed. Interestingly, hyperosmotic conditions generally leave the interior ionic conditions, including Mg2+ and K+ concentrations, of Archaea and eukarya cells largely unchanged (22), which correlates with the lack of EF4 in these evolutionary domains.
The beneficial effect of EF4 seen at lower temperature in vivo (Fig. 1) does not correspond to an increased intracellular ionic strength, because osmolytes rather than K+ accumulate under chill stress (23, 24). This in vivo effect is nicely reflected by our in vitro observation that at lower temperature (20 °C) and moderate salt concentrations EF4 is accelerating the rate of poly(Phe) synthesis by 50% (Fig. 3D, light-blue columns). It is likely that the increased stability of mRNA secondary structures at lower temperatures fosters the tendency of ribosomal stalling and thus requires the presence of EF4.
Under moderate conditions such as pH 7 and 37 °C, EF4 is only present in trace amounts in the cytoplasm, whereas large amounts are found in the membrane fraction, as noted previously (5). Because EF4 is not needed under these mild conditions, little phenotype is observed when the EF4 gene was knocked out (6). In striking contrast, a dramatic shift of EF4 from the membrane to the cytoplasm is observed under specific stress conditions, for example, the EF4 cytosol∶membrane distribution changes from 0.25∶1 to 5∶1 at high Mg2+ (Fig. 2). When the intracellular Mg2+ concentration is raised rapidly in response to high salt concentrations in the media, the bacterial cell might not have time enough to synthesize the 599-aa-long EF4 (indeed, E. coli EF4 is markedly longer than an average bacterial protein with 300–400 amino acids), but rather exploits the membrane as a storage vessel for EF4.
An important in vitro observation was the fact that EF4 induced a rate increase in the poly(Phe) synthesis at higher Mg2+ concentrations (Fig. 3). Similarly, low amounts of EF4 increased the synthesis of GFP at 12 mM Mg2+ in a coupled transcription/translation system (2). At 30 mM Mg2+ most if not all ribosomes are stalled, yet even under these extreme conditions EF4 remobilized the ribosomes allowing low but significant protein synthesis (Fig. 3B, Right). In a purified system, EF4 accelerated protein synthesis about fivefold at 14 mM Mg2+, whereas under optimal 4.5 mM Mg2+ and 30 °C EF4 played no role.
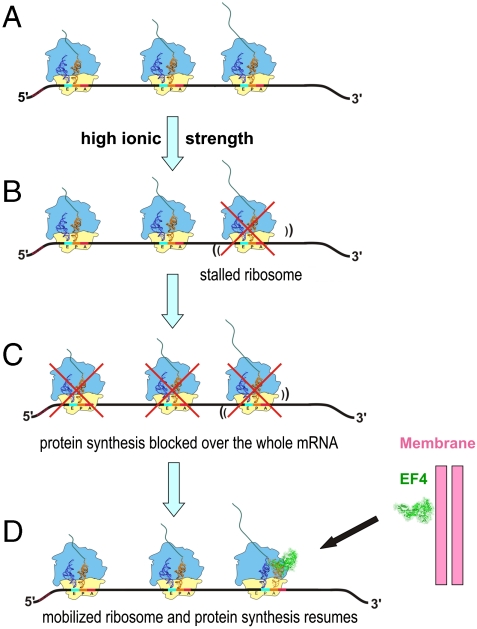
The observed acceleration effect of EF4 on protein synthesis came as a surprise, because the function of EF4 in promoting back-translocation would suggest an inhibitory rather than stimulatory effect. In line with this expectation, the effect of EF4 to increase translational accuracy was thought to be related to the assumed retardation of protein synthesis (25) in accord with the general wisdom “the slower, the better,” or adapted to our case “the slower, the more accurate.” The overall stimulatory effect of EF4 coupled with its back-translocation function suggests that EF4 recognizes stalled ribosomes, remobilizes them, and thus reactivates protein synthesis. On polysomes an EF4-induced mobilization of a single ribosome can amplify the positive effect on protein synthesis, because all the following ribosomes on the same mRNA are also blocked by just a single stalled ribosome (Fig. 4). We conclude that back-translocation and acceleration of protein synthesis are just two sides of the same coin, and that mobilization of stalled ribosomes is the central function of EF4.
Fig. 4.
Model showing EF4 importance at high ionic strength. (A) Normal protein synthesis on polysomes; (B) high ionic strength can lead to a single stalled ribosome on a polysome; (C) upstream ribosomes are therefore blocked stopping protein synthesis on this mRNA. (D) EF4 addition mobilizes and rescues the stalled ribosome by provoking a back-translocation resulting in a pretranslocational state that can be correctly translocated by EF-G and thus allows translation of the preceding ribosomes as well.
Materials and Methods
Bacterial Strains and Culture Conditions.
The ΔlepA strain JW2553 and its parental strain BW25113 were obtained from the Keio collection (26). The lepA∷kan deletion mutation in JW2553 was reintroduced into a single colony-purified BW25113 strain by P1kc-mediated transduction (27) to construct isogenic strains, because a slight but obvious difference in the growth rate was observed among the population of BW25113 we obtained. Replacement of the lepA gene in BW25113 by a kanamycin-resistant cassette was verified by PCR. M9 media were prepared according to ref. 28, and the pH of medium was adjusted using tables for the Sörensen buffer (29), buffer 19, and http://www.aeisner.de/rezepte/puffer2.html. KCl (200 mM) or MgCl2 (100 mM) were added for growth under stress conditions.
Growth competition.
The same amount of cells from an LB overnight culture of wild-type and mutant strain were mixed, yielding a final OD600 of 0.01 in a volume of 5 mL, and incubated with mild shaking at the indicated conditions of pH, Mg2+ concentration, and temperature. Aliquots were withdrawn every 4 h for a total of 24 h and the OD600 was measured. Simultaneously, 10-5, 10-6, and 10-7 dilutions were made and 250 μL of each was plated in duplicates on LB plates with and without 50 μg/mL kanamycin and the number of colonies were counted after incubation at 37 °C for overnight.
Growth of cells, cell disruption, fractionation of cytosol, and membranes.
E. coli wild-type strain BW25113 and the mutant derivative were grown overnight in LB medium at pH 7 or 6 with or without 100 mM MgCl2 at 37 °C. The overnight cultures were diluted 1∶100 in LB medium and grown at the indicated conditions of pH, Mg2+ concentration, and temperature to an OD600 of ∼0.6. Portions (10 mL) were pelleted (3,000 × g for 15 min). Aliquots were quick-frozen and stored at -80 °C. The fractionation procedure followed ref. 30. Briefly, a pellet was dissolved in 400 μL standard buffer (20 mM Hepes pH 7.5, 6 mM MgCl2, 150 mM  , 2 mM spermidine, and 0.05 mM spermine). Fractions of 100 μL were removed (total cell extract control). The remainder was centrifuged at 4 °C for 15 min at 3,000 × g, and the pellet was dissolved in 250 μL of lysis buffer (20 mM Hepes pH 7.8, 100 mM NaCl, 0.5 M sucrose, 2 mM EDTA, 200 μg/mL lysozyme) and incubated on ice for 2 min. Mg2+ was then added to a final concentration of 10 mM. After three cycles of freezing and thawing the samples were centrifuged (51,000 × g for 60 min), leaving the membrane fraction in the pellet and the cytoplasm fraction in the supernatant. The membrane fraction was resuspended in 250 μL of urea SDS buffer (0.5 M Tris•HCl pH 6.8, 140 mM β-mercaptoethanol, 0.6 % SDS, 3 M urea) and 2X Laemmli loading buffer (Bio-Rad) was added to both cytosol and membrane fractions. The total cell extract fraction was resuspended in 330 μL cracking buffer [0.5 M Tris•HCl pH 6.8, 140 mM β-mercaptoethanol, 0.6% SDS, 3 M urea, 0.2 (wt/vol) bromophenol blue]. The samples were denatured for 5 min at 95 °C and equal volumes containing material from roughly the same amount of cells were loaded on a 10% SDS–polyacrylamide gel (PAAG), and subjected to either Coomassie blue staining or Western blot analysis.
, 2 mM spermidine, and 0.05 mM spermine). Fractions of 100 μL were removed (total cell extract control). The remainder was centrifuged at 4 °C for 15 min at 3,000 × g, and the pellet was dissolved in 250 μL of lysis buffer (20 mM Hepes pH 7.8, 100 mM NaCl, 0.5 M sucrose, 2 mM EDTA, 200 μg/mL lysozyme) and incubated on ice for 2 min. Mg2+ was then added to a final concentration of 10 mM. After three cycles of freezing and thawing the samples were centrifuged (51,000 × g for 60 min), leaving the membrane fraction in the pellet and the cytoplasm fraction in the supernatant. The membrane fraction was resuspended in 250 μL of urea SDS buffer (0.5 M Tris•HCl pH 6.8, 140 mM β-mercaptoethanol, 0.6 % SDS, 3 M urea) and 2X Laemmli loading buffer (Bio-Rad) was added to both cytosol and membrane fractions. The total cell extract fraction was resuspended in 330 μL cracking buffer [0.5 M Tris•HCl pH 6.8, 140 mM β-mercaptoethanol, 0.6% SDS, 3 M urea, 0.2 (wt/vol) bromophenol blue]. The samples were denatured for 5 min at 95 °C and equal volumes containing material from roughly the same amount of cells were loaded on a 10% SDS–polyacrylamide gel (PAAG), and subjected to either Coomassie blue staining or Western blot analysis.
Other Procedures.
Western blotting.
After transferring the proteins from the PAAG to PVDF membranes they were blocked with 5% fat-free dry milk in PBS. The primary antibody (polyclonal anti-EF4 serum E. coli, Pineda Antikörper Service; anti-S7 was our own preparation) of 1∶10,000 dilution was prepared in PBS with 0.05% Tween 20 containing 5% fat-free dry milk (precentrifuged for 20 min at 12,000 × g) and used for hybridization for 90 min at room temperature. The secondary antibody of 1∶20,000 dilution (goat anti-rabbit HRP conjugate, Santa Cruz Biotechnology) was prepared in the same way as the primary antibody and was used for incubation for 60 min at room temperature. An ECL Western blotting kit (Amersham Biosciences) was used for protein detection. The distribution of the relative EF4 amounts between membrane and cytosol (Fig. 2) was calculated by taking the sum EF4 in (membrane plus cytosol) as 100%. The molar ratio of EF4∶ribosomes (Fig. 2) was determined by adding defined amounts (picomole) of both purified EF4 and proteins from 70S ribosomes as reference to the SDS–PAAG with the cytosol samples. The ribosomal protein S7 was taken as ribosomal reference. After Western blotting the molar ratios EF4∶70S were calculated by means of the pixel numbers of the reference bands corresponding to S7 and EF4.
Poly(U)-dependent poly(Phe) synthesis and misincorporation assay.
The procedure was performed under optimized ion conditions [binding buffer: 20 mM Hepes–KOH (pH 7.6 at 0 °C), 4.5 mM Mg(acetate)2, 150 mM NH4(acetate), 4 mM β-mercaptoethanol, 2 mM spermidine, and 0.05 mM spermine] as described in detail in ref. 15 with the following modifications: Where indicated, 14 mM Mg2+ was present instead of 4.5 mM. The incubation mixture contained 0.3 μM 70S ribosomes, 2.3 μg/μL poly(U) mRNA, 0.4 mM L-[U-14C]phenylalanine with specific activity of 15 dpm/pmol, 6 μM [3H]leucine (6,000 dpm/pmol), 11.5 μM of E. coli tRNAbulk, 2.5 μM tRNAPhe, 3 mM ATP, 1.5 mM GTP, 5 mM acetylphosphate and S-100 fraction (125 μL/mL assay). Purified E. coli EF4 was added when indicated (final concentration 0.9 μM). The reaction was performed at 37 °C for up to 60 min. At indicated time points samples of 17.5 μL were withdrawn and mixed with 2 mL of cold 5% trichloroacetic acid (TCA) containing one drop 1% bovine serum albumin as a precipitation carrier to stop the translation. Samples were incubated for 15 min at 95 °C to hydrolyze RNAs. After cooling down, the samples were filtered through glass filters, which were washed 12 times with 2 mL 10% TCA and twice with 3 mL of ether/ethanol (1∶1 vol/vol) to remove the TCA and dry the filters. Radioactivity resulting from incorporated [14C]phenylalanine and [3H]leucine could be accurately determined by the properly programmed Wallac 1409 Liquid Scintillation Counter after incubation of the filters in 5 mL of Filter-CountTM (complete liquid scintillation cocktail, Perkin Elmer) overnight at 4 °C.
Poly(U)-dependent oligo(Phe) synthesis with precharged Phe-tRNA and purified factors.
The posttranslocational (POST) complex reactions were prepared in binding buffer as described (11), except that 70S ribosomes were programmed with poly(U) (30 μg per 3 pmol 70S) and the P site blocked with tRNAPhe before the A site was loaded with Ac[3H]Phe-tRNAPhe followed by EF-G-dependent translocation (10 min at 37 °C). Six picomole of the resulting POST complex with or without 18 pmol of EF4 in 70 μL were added to 30 μL of the ternary complex mix (Mg2+; usually 4.5, when indicated 14 or 30 mM) containing 60 pmol [14C]Phe-tRNAPhe, 90 pmol EF-Tu, 90 pmol EF-Ts, 1.5 mM GTP in binding buffer. The ternary complex mix was preincubated 5 min at 37 °C. Kinetics (0, 2.5, 5, 10, 20, and 40 min) were performed at 30 °C withdrawing at the times indicated 10 μL for hot TCA precipitation, filtered through glass filters, and counted.
ACKNOWLEDGMENTS.
We thank Dr. Daniel N. Wilson (Gene Center, University of Munich) for support and discussions. This work was supported by the Deutsche Forschungsgemeinschaft Grant NI 174/15-1 (to K.H.N.), a Bundesministerium für Bildung und Forschung/Deutscher Akademischer Austauschdienst Grant 0315181 (to M.P.), and an Alexander-von-Humboldt Grant GAN 1127366 STP-2 (to H.Y.).
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Nierhaus KH. The elongation cycle. In: Nierhaus KH, Wilson DN, editors. Protein Synthesis and Ribosome Structure. Weinheim, Germany: Wiley-VCH; 2004. pp. 323–366. [Google Scholar]
- 2.Qin Y, et al. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell. 2006;127:721–733. doi: 10.1016/j.cell.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, et al. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- 4.March PE, Inouye M. Characterization of the Lep operon of Escherichia coli. Identification of the promoter and the gene upstream of the signal peptidase I gene. J Biol Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]
- 5.March PE, Inouye M. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc Natl Acad Sci USA. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dibb NJ, Wolfe PB. Lep operon proximal gene is not required for growth or secretion by Escherichia coli. J Bacteriol. 1986;166:83–87. doi: 10.1128/jb.166.1.83-87.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijlsma JJ, Lie ALM, Nootenboom IC, Vandenbroucke-Grauls CM, Kusters JG. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J Infect Dis. 2000;182:1566–1569. doi: 10.1086/315855. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Pan D, Pech M, Cooperman BS. Interrupted catalysis: The EF4 (LepA) effect on back-translocation. J Mol Biol. 2010;396:1043–1052. doi: 10.1016/j.jmb.2009.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans RN, Blaha G, Bailey S, Steitz TA. The structure of LepA, the ribosomal back translocase. Proc Natl Acad Sci USA. 2008;105:4673–4678. doi: 10.1073/pnas.0801308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connell SR, et al. A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation. Nat Struct Mol Biol. 2008;15:910–915. doi: 10.1038/nsmb.1469. [DOI] [PubMed] [Google Scholar]
- 11.Di Giacco V, et al. Shine–Dalgarno interaction prevents incorporation of noncognate amino acids at the codon following the AUG. Proc Natl Acad Sci USA. 2008;105:10715–10720. doi: 10.1073/pnas.0801974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nierhaus KH. Decoding errors and the involvement of the E-site. Biochimie. 2006;88:1013–1019. doi: 10.1016/j.biochi.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Shoji S, Janssen BD, Hayes CS, Fredrick K. Translation factor LepA contributes to tellurite resistance in Escherichia coli but plays no apparent role in the fidelity of protein synthesis. Biochimie. 2010;92:157–163. doi: 10.1016/j.biochi.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karim Z. Berlin: Freie Universität Berlin; 2010. The importance of the ribosomal elongation factor 4 (LepA) PhD dissertation. [Google Scholar]
- 15.Szaflarski W, et al. New features of the ribosome and ribosomal inhibitors: Non-enzymatic recycling, misreading and back-translocation. J Mol Biol. 2008;380:193–205. doi: 10.1016/j.jmb.2008.04.060. [DOI] [PubMed] [Google Scholar]
- 16.Wilson DN. Antibiotics and the inhibition of ribosome function. In: Nierhaus K, Wilson D, editors. Protein Synthesis and Ribosome Structure. Weinheim, Germany: Wiley-VCH; 2004. pp. 449–527. [Google Scholar]
- 17.Lusk JE, Williams RJP, Kennedy EP. Magnesium and the growth of Escherichia coli. J Biol Chem. 1968;243:2618–2624. [PubMed] [Google Scholar]
- 18.Froschauer EM, Kolisek M, Dieterich F, Schweigel M, Schweyen RJ. Fluorescence measurements of free [Mg2+] by use of mag-fura 2 in Salmonella enterica. FEMS Microbiol Lett. 2004;237:49–55. doi: 10.1016/j.femsle.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 19.Schindl R, Weghuber J, Romanin C, Schweyen RJ. Mrs2p forms a high conductance Mg2+ selective channel in mitochondria. Biophys J. 2007;93:3872–3883. doi: 10.1529/biophysj.107.112318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richey B, et al. Variability of intracellular ionic environment of Escherichia coli. J Biol Chem. 1987;262:7157–7164. [PubMed] [Google Scholar]
- 21.Csonka LN, Hanson AD. Prokaryotic osmoregulation: Genetics and physiology. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 22.Kunte H. Osmoregulation in bacteria: Compatible solute accumulation and osmosensing. Environ Chem. 2006;3:94–99. [Google Scholar]
- 23.Morbach S, Krämer R. Structure and function of the betaine uptake system BetP of Corynebacterium glutamicum: Strategies to sense osmotic and chill stress. J Mol Microbiol Biotechnol. 2005;10:143–153. doi: 10.1159/000091561. [DOI] [PubMed] [Google Scholar]
- 24.Wood JM, et al. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp Biochem Physiol Part A: Mol Integr Physiol. 2001;130:437–460. doi: 10.1016/s1095-6433(01)00442-1. [DOI] [PubMed] [Google Scholar]
- 25.Shoji S, Walker SE, Fredrick K. Reverse translocation of tRNA in the ribosome. Mol Cell. 2006;24:931–942. doi: 10.1016/j.molcel.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller JH. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1972. Experiment 28. Generalized transduction : Use of P1 in strain construction; pp. 201–205. [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Anonymous. Wissenschaftliche Tabellen. 6th Ed. Basel: Geigy; 1960. [Google Scholar]
- 30.Remme J, Margus T, Villems R, Nierhaus KH. The third ribosomal tRNA-binding site, the E site, is occupied in native polysomes. Eur J Biochem. 1989;183:281–284. doi: 10.1111/j.1432-1033.1989.tb14925.x. [DOI] [PubMed] [Google Scholar]