Abstract
HET-s is a prion protein of the fungus Podospora anserina which, in the prion state, is active in a self/nonself recognition process called heterokaryon incompatibility. Its prionogenic properties reside in the C-terminal “prion domain.” The HET-s prion domain polymerizes in vitro into amyloid fibrils whose properties depend on the pH of assembly; above pH 3, infectious singlet fibrils are produced, and below pH 3, noninfectious triplet fibrils. To investigate the correlation between structure and infectivity, we performed cryo-EM analyses. Singlet fibrils have a helical pitch of approximately 410 Å and a left-handed twist. Triplet fibrils have three protofibrils whose lateral dimensions (36 × 25 Å) and axial packing (one subunit per 9.4 Å) match those of singlets but differ in their supercoiling. At 8.5-Å resolution, the cross-section of the singlet fibril reconstruction is largely consistent with that of a β-solenoid model previously determined by solid-state NMR. Reconstructions of the triplet fibrils show three protofibrils coiling around a common axis and packed less tightly at pH 3 than at pH 2, eventually peeling off. Taken together with the earlier observation that fragmentation of triplet fibrils by sonication does not increase infectivity, these observations suggest a novel mechanism for self-propagation, whereby daughter fibrils nucleate on the lateral surface of singlet fibrils. In triplets, this surface is occluded, blocking nucleation and thereby explaining their lack of infectivity.
Keywords: assembly nucleation, cryoelectron microscopy, protein template, polymorphism, three-dimensional image reconstruction
A wide range of diseases including Alzheimer’s disease, type II diabetes, Creutzfeldt-Jakob disease, and other spongiform encephalopathies are associated with the accumulation of cross-β-structured protein fibrils called amyloids. (we use the term “fibril” for a protein polymer that is all or almost all amyloid [like a HET-s prion domain (PD) fibril] and “filament” for a structure that is part amyloid and part nonamyloid. A “protofibril” is a substituent of a fibril and a “protofilament” of a filament). Proteinaceous infectious agents called prions constitute a subclass of amyloids. Prion infection involves conversion of proteins from their normal functional conformations—usually, via an unfolded intermediate state—into amyloids. Once seeded, this process is self-propagating, i.e., existing amyloid fibrils provide templates for further assembly (1, 2).
HET-s is a prion protein of the filamentous fungus Podospora anserina, which in its prion form participates in a genetic proofing process called heterokaryon incompatibility (3, 4). When two cells respectively containing het-s (prion) and the allele het-S(nonprion) fuse, programmed cell death is induced. Thus [HET-s] is a gain-of-function prion and contrasts in this respect with other fungal prions such as [PSI+] and [URE3] which are loss-of-function prions (5).
HET-s has two domains, an N-terminal domain (residues 1–217) of bundled α-helices (HeLo domain) (6) and the PD (residues 218–289), which is unfolded in the nonprion protein and amyloid in the prion (7, 8). The PD is necessary and sufficient for prion formation and fibrillizes readily in vitro (9–11). The HeLo domain interacts with the PD to affect its fibrillation but as yet has no other assigned function (6). Like other amyloids (12, 13), HET-s PD fibrils have cross-β structures with β-strands running perpendicular to the fibril axis (10, 14). A structure has been determined by solid-state NMR spectroscopy (ssNMR) for fibrils assembled at pH 7 (14, 15), whose well-resolved resonances were taken to indicate a high degree of structural homogeneity. This structure is a stack of β-solenoids to which each subunit, consisting of two sequence pseudorepeats, contributes two coils. Each coil has four β-strands, and the coils are connected by an extended, flexible loop.
HET-s PD fibrillation has been investigated under a variety of conditions: In particular, fibril morphology was found to vary with the pH of assembly (9). Between pH values 5 and 7, bundles of thin (approximately 5 nm) fibrils are obtained. On decreasing the pH to 3-4, dispersed fibrils are observed; and below pH3, they are replaced by multistranded fibrils of which a majority are triplets. Fibril infectivity, measured after electroporating in vitro assembled preparations into spheroplasts, also correlates with pH of assembly. Fibrils formed at pH 7 (loosely bundled singlets) or pH 4 (singlets) are infectious, and those formed at pH 2 (multistranded fibrils) are essentially noninfectious (9). These properties establish HET-s as a model system to study the correlation between structure and infectivity in amyloids. Here we have used cryoelectron microscopy (cryo-EM) to investigate the structures of HET-s PD fibrils formed at various pH values. Our goals were to test the current model (14), as well as to investigate the relationship between fibril structure and self-replication ability.
Results
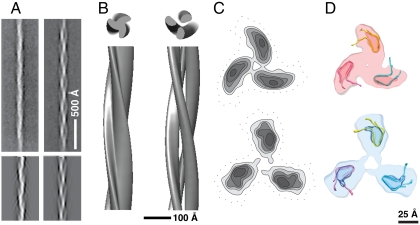
First, we extended the range of conditions under which fibrillation has been studied (Table S1). In particular, at pH 3, we observed mixed populations of singlets and triplets (Fig. 1A). Occasionally, we observed triplet fibrils separating into protofibrils whose width and appearance (Fig. S1) closely resemble those of singlet fibrils.
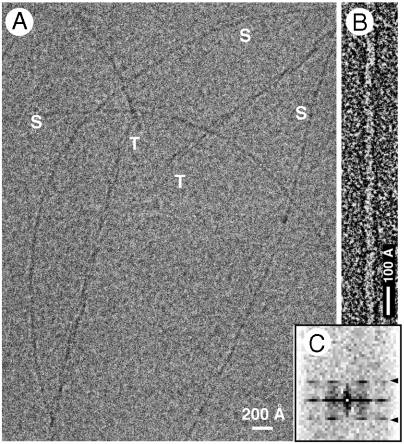
Fig. 1.
(A) Cryoelectron micrograph of pH 3 fibrils. S, singlets; T, triplets. (B) Negatively stained pH 3 singlet fibril. (C) Averaged diffraction pattern from 43 negatively strained fibrils, showing the layer line at approximately (400 Å)-1 (arrowheads). The scaling in the vertical (meridional) and horizontal (equatorial) dimensions differs by a factor of approximately 6 (the vertical scaling is finer).
Singlet Fibrils Have a 410-Å Axial Repeat with a Left-Handed Twist.
Many amyloid fibrils exhibit a small, usually left-handed, helical twist, giving a characteristic axial repeat. In some cases, fibrils of the same amyloidogenic protein exhibit different repeats (5, 16). These parameters have not been determined experimentally for HET-s PD fibrils. We measured them, as described below. At pH 4, the assembly products included regular meshworks (“angled-layer aggregates”) of singlet fibrils (Fig. 2). The meshes have a rhomboid unit cell. We measured its edge length in terms of internode distances, obtaining a unimodal distribution with a mean of 393 ± 42 Å (SD) (Fig. 2B). The edge length corresponds to the pitch of the interlocked helical fibrils. Their pitch is also expressed in terms of a strong reflection with an axial spacing of approximately 400 Å in averaged power spectra of singlet fibril images (Fig. 1 B and C). Image analysis further refined the mean helical pitch to 410 Å.
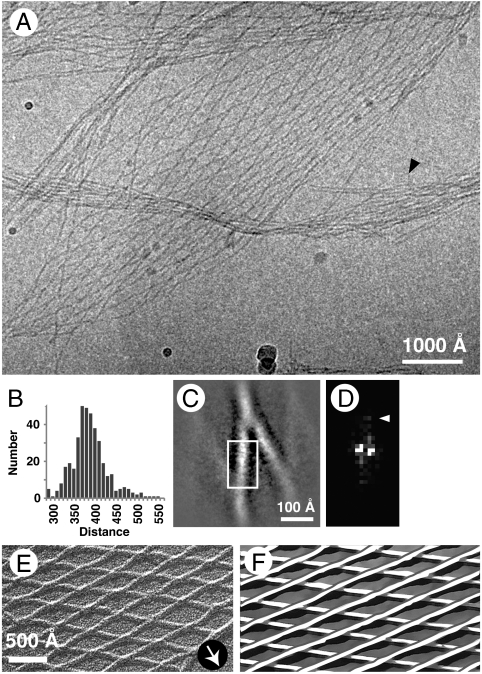
Fig. 2.
(A) Cryoelectron micrograph of an angled-layer aggregate formed at pH 4. Arrowhead points to a loose bundle of fibrils resembling a pH 7 bundle. (B) Histogram of internodal distances. N = 394, average = 393 ± 42 Å (SD). (C) Averaged image of a rhombic segment. The crossing angle is approximately 30°. (D) Diffraction pattern of the boxed area. Arrowhead marks the reflection at approximately (20 Å)-1. (E) Unidirectional shadowing of a pH 4 angled-layer aggregate. Arrow shows the direction of shadowing. (F) Schematic interpretation of E. The upper layer consists of fibrils running from bottom left to top right, whereas in the lower layer they run from top left to bottom right.
To determine the hand of this twist, we performed unidirectional metal shadowing (Fig. 3A). The images show striations with an axial spacing of about 400 Å, which are accentuated by axial compression (Fig. 3B, arrows) and confirmed by diffraction (Fig. 3C). Referring to a standard of known handedness (T4 polyheads, ref. 17), we determined that the fibrils’ twist is left-handed. Triplet fibrils were also found to be left-handed.
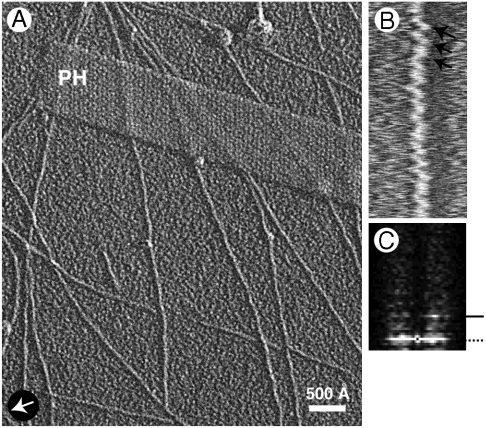
Fig. 3.
(A) EM of pH 3 fibrils contrasted by unidirectional shadowing. Arrow marks shadowing direction. PH denotes a T4 polyhead, included as a handedness reference (its low-pitch helices are right-handed). (B) A singlet fibril compressed 10-fold axially. Arrows marks slanting striations that indicate left-handedness. (C) Power spectrum of a singlet fibril image. Arrowhead marks the (400 Å)-1 layer line with strong intensity only on one side, confirming the fibril’s left-handed twist.
Three-Dimensional Reconstruction of Singlet Fibrils.
Being very thin, singlet fibrils are subject to bending, which hampers structural analysis by Fourier methods (18). Accordingly, we employed a “single particle” approach by the iterative helical real space reconstruction (IHRSR) method (19). Digitized fibril images were segmented to squares, 471 Å on a side (i.e., slightly longer than one axial repeat), the helical symmetry refined, and a 3D reconstruction calculated. During classification to obtain a uniform dataset, 93% of the original segments were discarded (see Methods). On averaging the individual power spectra of 94,200 aligned segments, we observed the expected layer line at approximately 400 Å, but also one at approximately 20 Å (Fig. S2). The 20-Å periodicity is also discernible in averaged images of fibril segments from the rhombic meshes (Fig. 2 C and D, arrowhead).
IHRSR converges to a unique helical symmetry provided that the starting values assigned to the helical parameters are reasonably close to the final solution (19). We used the observed repeats to set a 20-Å axial translation with left-handed twist of 18° as starting values for the asymmetric unit, which corresponds to 20 steps along a 400-Å-pitch helix. After 50 cycles, these values shifted to a 19.5-Å translation with 17.1° twist. We chose to impose the very close value of 18.8 Å for the axial translation, corresponding to a pair of HET-s molecules and four times the 4.7-Å cross-β repeat. To maintain the 410-Å pitch indicated by the helical refinement, the twist angle was changed to 16.5°. Thus there are 21.8 (= 360/16.5) of these 18.8-Å-long units in one 410-Å helical turn. As the differences are so small, we could equally well have chosen a value of 19.2 Å for the unit length (4 × 4.8 Å, the axial repeat of most crystalline cross-β peptides) and 17.1° for the twist (in better accord with the refined helical parameters), which would give a pitch of 404 Å for the final helical reconstruction.
The reconstruction (Fig. 4A) depicts the singlet fibril as having an asymmetric cross-section of ∼36 × 25 Å, whose center is positioned approximately 7 Å off-axis, accounting for the slightly wavy appearance of fibrils (Fig. 1B). At this resolution (Fig. S3), single HET-s subunits (9.4 Å apart) are partially resolved (Fig. 4A, arrows). As expected from the approximately 20-Å reflection, there appears to be a pairing of successive subunits (Fig. 4 A, bracket, and D).
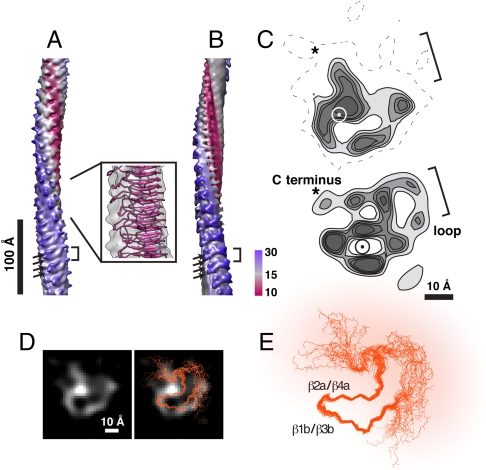
Fig. 4.
(A) Three-dimensional reconstruction of the pH 3 singlet fibril. Box, ssNMR atomic model (PDB ID 2 RNM) fitted to the reconstruction. (B) Fibril incorporating the ssNMR structure, resolution limited to 8 Å (see Methods). Brackets in A and B denote densities protruding at axial intervals of 18.8 Å. Arrows mark densities from subunits spaced 9.4 Å apart. Densities are colored according to radius from the fibril axis. (C) Transverse sections from the reconstruction (Upper) and the model (chain C, 20 models averaged) with loops (Lower), contoured at discrete levels and resolution limited to 10 Å. The circle marks the position of the fibril axis. Brackets show the location of flexible loops. Asterisks show the location of the C terminus. (D) Densities of transverse sections of the reconstruction overlaid by the Cα superposition of the HET-s backbone from 20 energy-minimized models (orange, chain C). (E) Cα superposition of the HET-s backbone used in D. The flexible region is overlaid with diffuse orange color. The reconstruction and the model in panel C agree best in the regions occupied by the β-sheets (marked in panel E) where the density is relatively well defined.
Segmentation of the Loop and Backbone Densities.
To evaluate the outer surface of the reconstruction, the densities corresponding to fibril backbone and the intercoil loops were segmented and extracted, and the 2D (Fig. S4C, backbone in middle, in pink, and loop in purple) and 1D (Fig. S4D, backbone in pink and loop in purple) projections along the fibril axis were calculated. The backbone density has a rather smooth profile in the projections with a faint 18.8-Å periodicity. The projections of the loop/backbone junction show peaks repeating every 9.4 Å (Fig. S4D, blue). On the other hand, the projections of the loop densities show a definite 18.8 Å periodicity (Fig. S4C, arrow and Fig. S4D, purple), suggesting a pairing of the loops between adjacent subunits.
Comparing the Singlet Fibril Reconstruction and the ssNMR-Derived Structure.
To make this comparison, we used the coordinates for one and two subunit(s) from the model [Protein Data Bank (PDB) ID 2RNM] (19) to computationally synthesize fibrils with the helical symmetry determined in our analysis (see Methods). Helical parameters were calculated from the 20 energy-minimized solutions provided. Of these, model 1 has helical parameters that are very close to those we have determined, i.e., a rotation angle of 8.45° and an axial step of 9.5 Å (405 Å pitch) but others are quite different, e.g., model 17 gives 1.3° for rotation, an axial step of 9.7 Å, and a pitch of 2,600 Å. An averaged fibril structure (Fig. 4B) was constructed from the ssNMR-based models (19) by superposing the central pair of chains (C and D in PDB ID 2RNM) from each of the 20 energy-minimized models and filtering to 8-Å resolution. This structure shows diffuse loop-associated density due to the variability of this part of the molecule. On the other hand, the sites occupied by the first and last few residues that graft the loop onto the β-solenoid have rather solid densities (Fig. 4B, bracket) as observed in the reconstruction (Fig. 4A, bracket). These grafting densities also contribute to the surface with 9.4-Å periodicity (Fig. 4 A and B, arrows). The fitting of the model 1 from PDB ID 2RNM is shown in a box in Fig. 4A. Our reconstruction and the modeled fibril agree in terms of dimensions, overall shape, and in some major features, although the match is not perfect, as considered further in Discussion.
Three-Dimensional Reconstruction of Triplet Fibrils.
Triplet fibrils assembled at pH 2 and pH 3 were also reconstructed using IHRSR (Fig. 5 A and B). Both have been found to have features characteristic of amyloid (11). The helical symmetries initially assigned were obtained by measuring the distance between cross-over points, which corresponds to a 120° rotation around the central axis for a protofibril. Although similar amounts of data were processed as for the singlet, the resolutions of these reconstructions were lower (Fig. S3), possibly reflecting some variability in inter-cross-over spacing. The reconstructions show three protofibrils of approximately 40 × 25 Å in cross-section, giving outer diameters of 88 and 107 Å, for the pH 2 and pH 3 triplets, respectively (Fig. 5 B and C). In both cases, the protofibrils are dimensionally consistent with the singlet fibril; in the pH 3 triplet, they are less closely associated than in the pH 2 triplet (Fig. 5C).
Fig. 5.
Three-dimensional reconstructions of triplet fibrils. (A, Upper) Averaged pH 2 triplet fibril (Left) and pH 3 triplet fibril (Right). (Lower) Reprojections of the reconstructions shown in B. (B) Surface renderings of the pH 2 (Left) and pH 3 (Right) triplet fibrils. (C) Contour plots of transverse sections of the reconstructions for pH 2 (Upper) and pH 3 triplet (Lower). (D) Modeling of triplet fibrils by fitting the ssNMR-derived subunit structure into cryo-EM densities for the corresponding protofibrils (see C).
Comparison of Subunit Stackings in Singlet and Triplet Fibrils.
The pH 2 triplet fibrils were found to have a 361-Å axial repeat (i.e., 1,083 Å for a protofibril to rotate 360° about the central axis) and the center of each protofibril to be positioned approximately 22 Å off-axis. The corresponding numbers for the pH 3 triplet are 335 Å (1,005 Å for 360°) and 31 Å. These parameters differ considerably from those of the singlet fibril (410 and 9.4 Å) (Fig. S5A and Table S2).
To investigate these interactions further, we compared the helical stacking of HET-s PD subunits (Fig. S5 and Table S3) in singlet fibrils and triplet protofibrils. Radial projections of the corresponding helical lattices show that the arrangements in pH 3 singlets and pH 2 triplets are quite similar (1.06 and 1.20 Å of circumferential displacement between adjacent subunits on the surface lattice, respectively; Figs. S5 B and C and Table S2). In the pH 3 triplet, this spacing increases to 1.81 Å. We take the singlet fibril to represent an energetically minimized state. Relative to this state, its supercoiling to form the protofibrils of the pH 2 triplet encurs an energy deficit that is compensated by attractive interactions between the protofibrils. In the pH 3 triplet, these interactions are weakened and the protofibril twist reverts close to its value in the singlet, and marginal perturbations may cause the protofibrils to separate into singlet fibrils.
A minor assembly product at pH 2 is three-stranded ribbons (width 140 Å, 1,300 Å axially per 180° cross-over) (9). Analyzing their outer protofibrils in the same way yielded a value of 1.02 Å per circumferential step between adjacent subunits, essentially the same as in the pH 2 triplet and the pH 3 singlet (Fig. S5). However, the central protofibril in the ribbon does not follow the same packing.
Modeling the protofibrils in triplet fibril reconstructions.
To explore the interaction between protofibrils further, we fitted the ssNMR model into the cryo-EM envelopes of the triplet fibrils (Fig. 5D). To do so, a “core” subunit lacking the intercoil loop and the N and C termini was used to generate protofibrils (Fig. 5D). Thanks to the asymmetric cross-section of the protofibril and the known handedness, the docking was readily accomplished, although there was insufficient information to allow specification of the axial register of the protofibrils. Charge–charge or polar interactions may contribute to the stability of triplet fibrils, according to experiments that we performed to test for morphological changes at different ionic strengths (0–300 mM NaCl) and pH 2. In the absence of NaCl, there are no singlet fibrils. As the ionic strength increases, so does the fraction of singlet fibrils.
Discussion
HET-s prion fibrils are of major interest from both functional and structural perspectives. Although the mechanism whereby they play their role in heterokaryon incompatibility remains obscure, they represent a prototypic gain-of-function prion. A recent study of a translation regulator expressed in sensory neurons suggests the existence of other such prions (20). As for structure, HET-s PD fibrils have been amenable to structure determination by ssNMR (14). Our cryo-EM analysis confirms certain key features of that structure and, in addition, gives insight into how singlet fibrils interact to form higher-order structures: triplet fibrils and angled-layer aggregates. HET-s was the first prion system in which fibrils assembled in vitro were demonstrated to be infectious (4). Subsequently, a striking difference was observed between fibrils formed at pH 4 and above (singlets, infectious) and at pH 3 and below (triplets, noninfectious). Prion infectivity entails both the ability to penetrate a host cell and the ability to self-replicate. In the infectivity assay used, cell entry was accomplished by electroporation so that infectivity, as measured, essentially came down to self-replication.
Structural Analysis of Singlet Fibrils.
Our analysis shows that the fibril has a left-handed twist and an average pitch of 410 Å. The reconstruction partially resolves the β-sheets in the fibril cross-section (Fig. 4 C and D) and represents the second such determination by cryo-EM for an amyloid fibril, following work on β-amyloid (21) and showing a very different structure. It is noteworthy that HET-s PD singlets are exceptionally narrow fibrils, containing only about 17 amino acids per β-strand in cross-section. Furthermore, our reconstruction shows density extending from the core towards the C terminus (Fig. 4C, marked with asterisks), consistent with a recently published NMR analysis in which the C terminus is defined (22). Finally, we observed an apparent pairing of subunits along the fibril. The significance of this pairing is not evident, although we note that similar effects have been observed in some other protein filaments: tobacco mosaic virus (23), SH3 amyloid (24), the mammalian prion protein PrP (25), and myosin (26).
Experimental Uncertainties.
Our analysis has assumed that the singlet fibril pitch is a uniquely defined quantity. The strongest evidence in support of this proposition is the sharp (400 Å)-1 reflections, which are not split or doubled, in the averaged diffraction pattern of 43 randomly chosen, negatively stained singlets (Fig. 1C and Fig. S6). Further support comes from the reproducibility (± 2.7 Å SD) of the value obtained for the fibril pitch when different subsets of data were analyzed by IHRSR. This inference is also consistent with the well-resolved resonances in the NMR spectra of singlet (pH 7) fibrils (21).
However, these considerations do not rule out variations around the mean pitch value on a given fibril. The low contrast and density of the fibrils make it difficult to measure the scale of such variations directly on filament images. An empirical estimate is given by the spread of internodal distances in the angled-layer aggregates (± 42 Å SD; i.e., approximately 10%). This value is likely to be an overestimate because these apparently flimsy structures are likely to undergo distortions when prepared for EM. Nevertheless local variations in pitch may be significant and, in fact, could account for the low survival rate (7%) from the original set of singlet fibril segments. We note that a stringent restriction on cross-over length was needed to achieve subnanometer resolution in a cryo-EM study of β-amyloid fibrils (21).
As noted above, the cross-sections of the singlet reconstruction and the NMR-derived model, as limited to the same resolution, are similar but not identical (Fig. 4). The discrepancies may arise from residual noise in the EM analysis. However, they may also reflect conformational differences between fibrils assembled at pH 7 (as in the NMR study) and those assembled at pH 2 or pH 3 (as studied here). In this context, Sabate et al. detected pH-dependent spectral differences by FTIR (9).
Interactions Between Protofibrils in Triplet Fibrils.
Triplet protofibrils and singlet fibrils differ only slightly in the twists implicit in their long-range coiling. Thus their core of stacked β-solenoids is largely preserved: The intercoil loops are on the outside of the triplet (Fig. 5D) where they can have little effect on interactions between protofibrils. However, as the packing of the three protofibrils slackens in the pH 3 triplet compared to the pH 2 triplet, their interactions should undergo some adjustment between these two conditions.
A Lateral Seeding Model Can Explain the Noninfectivity of Triplet Fibrils.
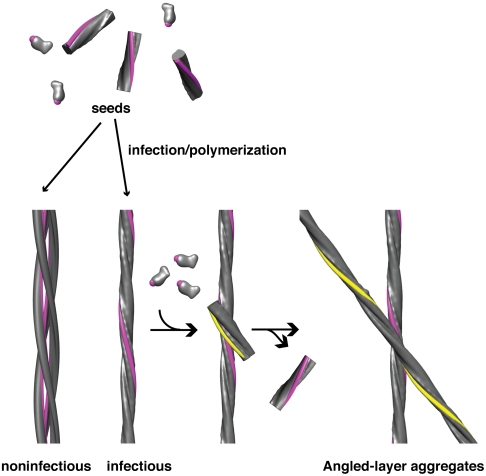
Self-replication of prions is dependent on the availability of oligomeric seeds that promote fibril growth (27, 28). In the simplest and most commonly considered seeding model, one or both ends of a fibril serves as a template for the deposition of additional subunits. For a given mass of fibrillar protein, the more free ends (shorter fibrils), the greater the potential for seeding—hence the greater the infectivity. Consistent with this picture, propagation of the yeast prions [PSI+] and [URE3] in vivo depends on the ability of the chaperone Hsp104 (29–32) to fragment existing fibrils (33). Similarly, the infectivity of in vitro assembled fibrils of the corresponding proteins, Sup35p and Ure2p, is enhanced by sonication (34–36). The situation is somewhat different with [Het-s]. First, its requirement for Hsp104 is less critical than for the yeast prions (37). Second, sonic fragmentation of triplet fibrils does not increase their infectivity (9). To explain this observation, we surmise that the lateral surface of the singlet fibril may serve as a nucleation site for daughter fibrils (Fig. 6), whose subsequent elongation is end-templated, as in the conventional model. These surfaces are sequestered in triplet fibrils and therefore unable to nucleate fibril assembly. As a daughter fibril elongates in assembly at pH 3 to 4, a point is reached at which its twist becomes incompatible with further in-phase growth along the surface of the mother fibril (triplets are marginally stable at pH 3 and unstable at pH > 3; see above). At this point, the daughter detaches from the surface that supported its formation, in a separation comparable to the parting of protofibrils from triplets at pH 3 (Fig. S1).
Fig. 6.
Lateral surface seeding model for assembly of HET-s PD fibrils. The exposed lateral surface of singlet fibrils (colored in pink) nucleates the polymerization of HET-s monomers. This surface is occluded in triplet fibrils and retains this configuration when shifted to higher pH. Accordingly, the triplets are noninfectious. Singlet fibrils may associate to form angled-layer aggregates whose crossing-points may mimic the proposed nucleation complexes (see Fig. S7).
The lateral seeding model can also explain the formation of angled-layer aggregates at pH 4. A growing daughter fibril offers nucleation sites for granddaughters which gradually build up a network of interfibril connections that counter the fibrils’ tendency to detach if secured by only a single interaction and thereby stabilize the aggregate. The lateral interaction involves a specific crossing angle which we assume to be the same as the crossing angle of angled-layer aggregates, i.e., 30° (Fig. S7).
Methods
Purification of HET-s (218-289) and Fibril Assembly.
HET-s PD protein was prepared as described (10) and stored at -80 °C in guanidine-HCl-containing buffer. For fibril assembly, the buffer was changed appropriately. Buffers containing 20 mM citrate titrated with either NaOH or HCl were used for assembly at pH 2—4. Buffers with 300 mM acetic acid at pH 2.8 and with 40 mM boric acid, 10 mM citric acid, and 6 mM NaCl at pH 3 (11) were used for the polymorphism tests. Protein samples in appropriate buffers were divided into two aliquots, one incubated at 4 °C, and the other agitated mildly at 4 °C for the first 3 d. Fibril formation starts from between 3 d to a few weeks and is slower at lower pH values.
Electron microscopy.
For cryo-EM, 4 μL drops of suspensions of fibrils assembled at pH 2, 3, or 4 were blotted onto glow-discharged holey carbon grids R2/2 (Quantifoil) and vitrified in a Vitrobot cryostation (FEI). Micrographs were recorded with a CM200-FEG microscope (FEI) at 120 kV on SO-163 film (Kodak) with defocus values between 1.0 and 2.5 μm, at a nominal magnification of 38,000 and digitized with a SCAI scanner (Carl Zeiss), giving 1.84 Å/pixel at the specimen. For shadowing, specimens were adsorbed to a carbon film, washed, and freeze-dried in a Baltec BAF060 freeze-fracture machine (Technotrade). A 0.7-nm layer of tantalum/tungsten was deposited from an elevation angle of 30°, and stabilized by adding a carbon layer. For negative staining, 2% uranyl acetate was used.
Image analysis.
EMAN (38), SPIDER (39), and Bsoft (40) software were used. The contrast transfer function (CTF) was corrected by phase flipping. Selected fibrils were segmented with a box size of 256 pixels and 90% overlap of adjacent segments. 17,515 boxes were collected for pH 3 singlet fibrils (corresponding to 87,800 subunits), 6,102 boxes (91,500 subunits) for pH 3 triplets, and 4,829 boxes (72,400 subunits) for pH 2 triplets. For averaging of fibril segments in rhomboid meshes, a box size of 300 pixels (552 Å) was used. A total of 442 segments were aligned and averaged.
Reconstruction.
Singlet fibrils.
Reference-free classification of fibril segments was performed using EMAN and SPIDER. Combinations of K-means clustering and Principal Component Analysis were employed. Data assigned to classes whose averages showed unusual features or curvature were discarded as they would otherwise exaggerate the thickness of the reconstructed fibril. Images belonging to classes that failed to exhibit diffraction intensity at (400 Å)-1 were also discarded. Several reconstructions were calculated with datasets further refined according to several strategies, and we ultimately selected the one that gave the highest resolution. Reconstructions were performed using a cylinder as initial reference. Helical symmetry was searched locally and imposed at every cycle of IHRSR. Because there are approximately 20 20-Å units in the approximately 400-Å helix pitch, we used the 20-Å axial translation with left-handed twist of 18° (360/20°). During the iterations, these values moved to 19.5 Å and 17.1°. A value of 18.8 Å and the corresponding angle 16.5° were chosen for the final reconstruction. The final reconstruction was performed using 1,145 segments. Because the images in this set were classified independently of each other, they are no longer overlapping. The original locations of the segmented boxes in the micrographs were traced and we calculated the number of independent subunits included in the final set to be 26,391, corresponding to 24.8 μm. The reconstruction was low-pass filtered to 8 Å for further evaluation. For cross-section visualization, CTF baseline correction and B-factor correction (B factor = -300 Å2) were introduced and the reconstruction was further limited to 10-Å resolution. Segmentation of the fibril into the backbone and the loops was done by evaluating the corresponding areas of transverse sections of 20 pixels (36.8 Å) and creating binary masks by averaging the selected areas of the 20 pixels and extending helically along the z direction. The segmented densities were projected toward the y axis and then the x axis, using bproject in BSOFT (40). The z axis defined the fibril axis.
Triplet fibrils.
Classification and averaging was performed using EMAN, as above. Inter-cross-over spacings were initially estimated to be approximately 390 Å for pH 2 and approximately 350 Å for pH 3 fibrils. Three-dimensional reconstructions were performed by IHRSR with the axial translation step fixed at 9.4 Å, and the corresponding rotations to be searched resulting in 3.12° and 3.37° for pH 2 and pH 3 triplets, corresponding to helical pitches of 361 and 335 Å. Threefold symmetry was applied. Reconstructions were also performed assuming a C31 screw symmetry but the density profiles did not change significantly reflecting the limited resolution. Differences from these alternative symmetrizations would become relevant if the resolution were to reach 9.4 Å or so, the spacing of HET-s monomers along the axis.
Fibril Modeling.
Helical parameters of adjacent subunits in the 20 energy-minimized ssNMR models (PDB ID 2RNM) were calculated using LSQMAN (41). Chain A of model 1 was centered and used as a reference. Cα coordinates for residues 227–246 and 261–281 were used to calculate the rigid body superposition. For computational fibril synthesis, coordinates for the central two subunits (chains C and D) in a stack of five HET-s subunits were extracted from the models and fitted manually to the EM density maps. Using the rotation and translation values obtained from IHRSR, fibrils were modeled with MOLEMAN2 (42). To create density maps for comparison with the EM reconstructions, the electron densities of these 20 modeled fibrils were averaged and then subjected to a low-pass (8 Å) filter. To convey the β-solenoid backbone, structure factors were calculated from the PDB coordinates, corrected for bulk solvent contribution, and density maps were calculated for 20 models and averaged. X-ray scattering factors were used instead of electron scattering.
Supplementary Material
Acknowledgments.
We thank Dr. D.L.D. Caspar for insightful discussions and many suggestions; Dr. S. Saupe for advice on sample preparation, Drs. C. Biertümpfel and G. Cardone for helpful discussions; and Drs. B. Heymann and D. Winkler for support with computational and microscopy resources, respectively. This work was supported by the Intramural Research Program of the National Institute of Arthritis, Musculoskeletal, and Skin Diseases.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011342108/-/DCSupplemental.
References
- 1.Serio TR, et al. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 2.Collins SR, Douglass A, Vale RD, Weissman JS. Mechanism of prion propagation: Amyloid growth occurs by monomer addition. PLoS Biol. 2004;2:e321. doi: 10.1371/journal.pbio.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saupe SJ. A short history of small s: A prion of the fungus Podospora anserina. Prion. 2007;1:110–115. doi: 10.4161/pri.1.2.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baxa U, Cassese T, Kajava AV, Steven AC. Structure, function, and amyloidogenesis of fungal prions: Filament polymorphism and prion variants. Adv Protein Chem. 2006;73:125–180. doi: 10.1016/S0065-3233(06)73005-4. [DOI] [PubMed] [Google Scholar]
- 6.Greenwald J, et al. The mechanism of prion inhibition by HET-S. Mol Cell. 2010;38:889–899. doi: 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coustou V, Deleu C, Saupe SJ, Begueret J. Mutational analysis of the [Het-s] prion analog of Podospora anserina. A short N-terminal peptide allows prion propagation. Genetics. 1999;153:1629–1640. doi: 10.1093/genetics/153.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balguerie A, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabate R, et al. Prion and non-prion amyloids of the HET-s prion forming domain. J Mol Biol. 2007;370:768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Sen A, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 11.Wasmer C, et al. Infectious and noninfectious amyloids of the HET-s(218-289) prion have different NMR spectra. Angew Chem Int Ed Engl. 2008;47:5839–5841. doi: 10.1002/anie.200704896. [DOI] [PubMed] [Google Scholar]
- 12.Kajava AV, Steven AC. Beta-rolls, beta-helices and other beta-solenoid proteins. Adv Protein Chem. 2006;73:55–96. doi: 10.1016/S0065-3233(06)73003-0. [DOI] [PubMed] [Google Scholar]
- 13.Jahn TR, et al. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–727. doi: 10.1016/j.jmb.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 14.Wasmer C, et al. Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 15.Ritter C, et al. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxa U, Speransky V, Steven AC, Wickner RB. Mechanism of inactivation on prion conversion of the Saccharomyces cerevisiae Ure2 protein. Proc Natl Acad Sci USA. 2002;99:5253–5260. doi: 10.1073/pnas.082097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kistler J, Aebi U, Onorato L, ten Heggeler B, Showe MK. Structural changes during the transformation of bacteriophage T4 polyheads: Characterization of the initial and final states by freeze-drying and shadowing Fab-fragment-labelled preparations. J Mol Biol. 1978;126:571–590. doi: 10.1016/0022-2836(78)90059-1. [DOI] [PubMed] [Google Scholar]
- 18.DeRosier DJ, Klug A. Reconstruction of three-dimensional structures from electron micrographs. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 19.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 20.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Sachse C, Fandrich M, Grigorieff N. Paired beta-sheet structure of an Abeta(1-40) amyloid fibril revealed by electron microscopy. Proc Natl Acad Sci USA. 2008;105:7462–7466. doi: 10.1073/pnas.0712290105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Melckebeke H, et al. Atomic-resolution three-dimensional structure of HET-s(218-289) amyloid fibrils by solid-state NMR spectroscopy. J Am Chem Soc. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 23.Caspar DL, Holmes KC. Structure of dahlemense strain of tobacco mosaic virus: a periodically deformed helix. J Mol Biol. 1969;46:99–133. doi: 10.1016/0022-2836(69)90060-6. [DOI] [PubMed] [Google Scholar]
- 24.Jimenez JL, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tattum MH, et al. Elongated oligomers assemble into mammalian PrP amyloid fibrils. J Mol Biol. 2006;357:975–985. doi: 10.1016/j.jmb.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 26.Oshima K, et al. Axial dispositions and conformations of myosin crossbridges along thick filaments in relaxed and contracting states of vertebrate striated muscles by X-ray fiber diffraction. J Mol Biol. 2007;367:275–301. doi: 10.1016/j.jmb.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 27.Ness F, Ferreira P, Cox BS, Tuite MF. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox BS, Byrne LJ, Tuite MF. Prion stability. Prion. 2007;1:170–178. doi: 10.4161/pri.1.3.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 30.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 33.Wegrzyn RD, Bapat K, Newnam GP, Zink AD, Chernoff YO. Mechanism of prion loss after Hsp104 inactivation in yeast. Mol Cell Biol. 2001;21:4656–4669. doi: 10.1128/MCB.21.14.4656-4669.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 35.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 36.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malato L, Dos Reis S, Benkemoun L, Sabate R, Saupe SJ. Role of Hsp104 in the propagation and inheritance of the [Het-s] prion. Mol Biol Cell. 2007;18:4803–4812. doi: 10.1091/mbc.E07-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 39.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 41.Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr, Sect D: Biol Crystallogr. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 42.Kleywegt GJ, Jones TA. Model building and refinement practice. Methods Enzymol. 1997;277:208–230. doi: 10.1016/s0076-6879(97)77013-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.