Abstract
Point mutations of the NADP+-dependent isocitrate dehydrogenases 1 and 2 (IDH1 and IDH2) occur early in the pathogenesis of gliomas. When mutated, IDH1 and IDH2 gain the ability to produce the metabolite (R)-2-hydroxyglutarate (2HG), but the downstream effects of mutant IDH1 and IDH2 proteins or of 2HG on cellular metabolism are unknown. We profiled >200 metabolites in human oligodendroglioma (HOG) cells to determine the effects of expression of IDH1 and IDH2 mutants. Levels of amino acids, glutathione metabolites, choline derivatives, and tricarboxylic acid (TCA) cycle intermediates were altered in mutant IDH1- and IDH2-expressing cells. These changes were similar to those identified after treatment of the cells with 2HG. Remarkably, N-acetyl-aspartyl-glutamate (NAAG), a common dipeptide in brain, was 50-fold reduced in cells expressing IDH1 mutants and 8.3-fold reduced in cells expressing IDH2 mutants. NAAG also was significantly lower in human glioma tissues containing IDH mutations than in gliomas without such mutations. These metabolic changes provide clues to the pathogenesis of tumors associated with IDH gene mutations.
Keywords: cancer, liquid chromatography-MS/MS, GC-MS, N-acetylated amino acids
Differences in cellular metabolism between cancer and normal cells have long been noted by cancer researchers (1). Genetic alterations that occur in cancer, such as mutations and copy number changes that alter K-Ras and c-Myc, are thought to be responsible for at least some of these metabolic differences (2, 3). The genetic alterations that drive cancer pathogenesis may do so in part by deregulating cellular metabolism. Such deregulation could aberrantly signal cells to proliferate and provide molecular building blocks for cellular replication (4). This possibility has generated enthusiasm for the idea that that drug targets for the specific killing of cancer cells can be identified by studying the metabolic differences between normal and cancer cells.
Gliomas are tumors of the central nervous system that respond poorly to therapy and are associated with a heterogeneous collection of genetic alterations (5, 6), including mutations in IDH1 and IDH2 (7, 8). IDH1 and IDH2 are the cytoplasmic and mitochondrial NADP+-dependent isocitrate dehydrogenases, respectively, and are homologs. Isocitrate dehydrogenase 3 (IDH3), which is unrelated to IDH1 and IDH2, is a NAD+-dependent isocitrate dehydrogenase and has not been found to be mutated in cancer (Fig. S1A). These enzymes convert isocitrate to α-ketoglutarate (Fig. S1B). IDH1 catalyzes this reaction in the cytosol and peroxisome to mediate a variety of cellular housekeeping functions, whereas IDH2 and IDH3 catalyze a step in the tricarboxylic acid (TCA) cycle (reviewed in ref. 9). IDH1-R132 mutations occur frequently (50–93%) in astrocytomas and oligodendrogliomas, as well as in secondary glioblastomas, and may be the initiating lesion in these glioma subtypes (7, 8). Mutations in the analogous IDH2-R172 codon also occur at a lower rate (3–5%) in these cancers (8). Interestingly, mutations in IDH1 and IDH2 were observed subsequently in 22% of acute myelogenous leukemias (10). In gliomas, R132H is the most common IDH1 mutation, and R172K is the most common IDH2 mutation (8). IDH1 and IDH2 mutations are mutually exclusive and alter only one allele, apparently in a dominant fashion (8, 11). These observations suggest that IDH1 and IDH2 are proto-oncogenes that are activated by mutation of R132 and R172, respectively. Mutation of these codons abolishes the normal ability of IDH1 and IDH2 to convert isocitrate to α-ketoglutarate (8). Moreover, it has been reported that the mutated IDH1-R132H enzyme can dominant-negatively inhibit IDH1-WT isocitrate dehydrogenase activity in vitro (12). A separate line of research revealed that IDH1-R132 and IDH2-R172 mutants gain the neomorphic ability to convert α-ketoglutarate to (R)-2-hydroxyglutarate (2HG) (Fig. S1B) and that 2HG is highly elevated in IDH-mutated cancer tissues (10, 13, 14).
These data suggest that IDH mutations might cause changes in global cellular metabolism that favor cancer pathogenesis, either through downstream effects of increased 2HG or through inhibition of isocitrate metabolism. Techniques that profile metabolism by determining the level of metabolites, by tracing the fate of metabolites, or by monitoring dynamic changes in metabolite levels are called “metabolomics.” Studies using metabolomics have illuminated important aspects of metabolism at the cellular, tissue, and organism levels (15–19). In this study, we investigate the effects of IDH mutant expression on cellular metabolism using a glioma cell line, a homologous expression system, and a comprehensive metabolomics platform. We identify reproducible alterations and show that a subset of these changes is recapitulated in human glioma tissues.
Results
Glioma Cells Expressing IDH1-R132H and IDH2-R172K Have Similar Metabolomes.
To test whether IDH mutants alter the metabolic profile of glioma cells, we performed unbiased metabolic profiling on sister clones of the human oligodendroglioma (HOG) cell line that stably express IDH1-R132H or IDH2-R172K. As controls, we expressed IDH1-WT, IDH2-WT, or vector alone in sister clones (Fig. S1 C and D). We analyzed lysates prepared from cells in logarithmic growth phase using three mass spectrometry platforms, liquid chromatography (LC)-MS/MS with or without electrospray ionization (ESI) and gas chromatography (GC)-MS [with electron ionization (EI)], in six replicates per sample. This analysis yielded MS ion counts corresponding to 315 biochemicals, of which 215 were known metabolites and 100 were unique biochemicals with unknown identity. We normalized these data to protein concentration and mapped the mean level of each biochemical to pathways (Dataset S1) based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) (20). To determine which clones shared global metabolic profile features, we used unsupervised hierarchical clustering, univariate comparisons, and correlation analysis. We also used principal components analysis (PCA), a dimension-reduction strategy that transforms a large number of variables, in this case metabolites, into a small number of variables describing the variation among groups (21). Our procedures for technical and statistical analysis are summarized in Fig. S2.
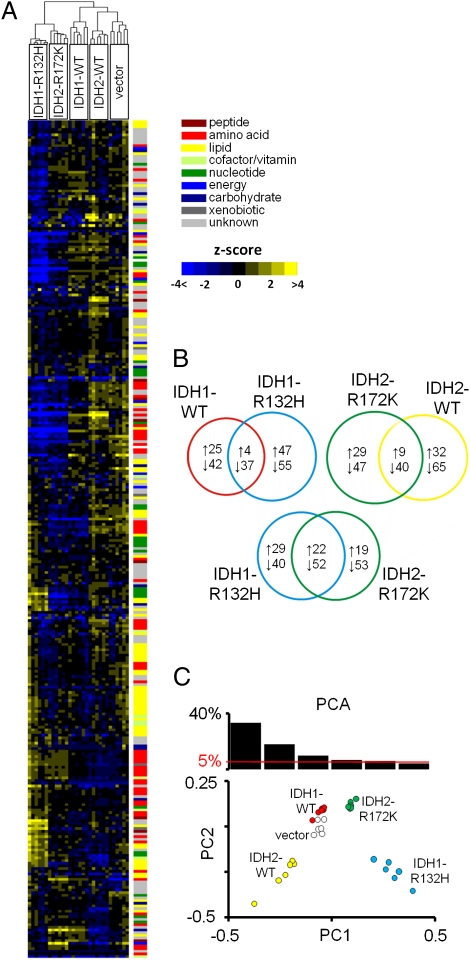
Hierarchical clustering revealed that IDH1-R132H and IDH2-R172K cells cluster together, separately from controls (Fig. 1A). IDH1-R132H cells had 143 biochemicals and IDH2-R172K cells had 146 biochemicals with significantly changed (cutoff value for significance: P < 0.05, Welch's t test) levels compared with vector control cells. Seventy-four of these biochemicals were altered in the same direction in both IDH-mutant groups, more than for any comparison of an IDH mutant with its respective WT control (Fig. 1B). The levels of biochemicals in the IDH-mutant groups had a weak but significant correlation (r = 0.15, P = 0.008) and did not correlate well with controls (Table S1). PCA showed that cells expressing IDH1-R132H and IDH2-R172K were distinguished from controls by their principal component 1 (PC1) value (33.3% of variance; Fig. 1C and Fig. S1 E and F). These analyses demonstrate that IDH1-R132H and IDH2-R172K expression is associated with a specific set of shared metabolic alterations.
Fig 1.
Metabolite profile of a glioma cell line expressing IDH1-R132H or IDH2-R172K. (A) Heat map showing 314 biochemicals in lysates from six replicates each of HOG cells expressing IDH1-WT, IDH1-R132H, IDH2-WT, IDH2-R172K, or vector alone, arranged by unsupervised hierarchical clustering. The level of each biochemical in each sample is represented as the number of SDs above or below the mean level of that biochemical (z score). (B) Venn diagrams indicating the number of biochemicals with mean levels that are significantly (P < 0.05) higher or lower in cells expressing each transgene compared with the vector. (C) PCA of metabolite profile dataset. The percentage of variance in the dataset reflected by the first six PCs is shown in the histogram, and PC1 and PC2 for each sample are plotted.
We next investigated whether differences in metabolism in cells expressing IDH1-R132H might cause those cells to have altered uptake or excretion of specific metabolites. To test this notion, we analyzed spent medium incubated for 48 h with HOG clones that express IDH1-R132H, IDH1-WT, or vector and also fresh medium (Dataset S2). Hierarchical clustering, correlation analysis, and PCA of 111 biochemicals in these samples demonstrated that medium incubated with cells expressing IDH1-R132H has a distinct metabolic profile compared with medium incubated with controls (Fig. S3 A–C). In the IDH1-R132H group, 2HG, kynurenine, and glycerophosphocholine (GPC), were increased, whereas the branched-chain amino acid (BCAA) catabolites 4-methyl-2-oxopentanoate, 3-methyl-2-oxovalerate, and 3-methyl-2-oxobutyrate were decreased compared with controls (Fig. S3D). These six metabolites are a subset of those that were altered in lysates of cells expressing either IDH1-R132H or IDH2-R172K (Dataset S1).
Glioma Cells Expressing IDH1-R132H Share Metabolomic Features with Cells Treated with 2HG but Not with Cells in Which IDH1-WT Expression Is Inhibited.
We next sought to obtain information on whether any of the known functions of IDH1-R132H could be responsible for the metabolomic changes that we observed. Currently, the two suspected functions of IDH1 mutants are (i) gain of a neomorphic enzymatic activity required to convert α-ketoglutarate to 2HG (13) and (ii) inhibition of normal IDH1 activity by binding to the WT enzyme (12). To test whether 2HG alone could produce metabolic changes similar to those resulting from IDH mutant expression, we analyzed cells treated with medium containing 7.5 mM or 30 mM 2HG, representing the range of concentrations of 2HG found in IDH1-mutated human glioma tissues (13). To test whether a loss of IDH1-WT function can produce the same metabolite changes as IDH1-R132H expression, we analyzed sister HOG clones that expressed shRNA targeted to IDH1 (22). The IDH1-targeted shRNA reduced IDH1 protein levels by more than 90% and lowered IDH activity accordingly (22). We obtained data on the levels of 204 known biochemicals in cells treated with 2HG, cells stably expressing IDH1-targetted shRNA, and analogous control cells (Fig. S4A and Dataset S3).
Hierarchical clustering revealed that 2HG-treated cells clustered together with IDH1-R132H–expressing cells, whereas IDH1-knockdown and control cells clustered separately (Fig. 2A). The levels of 107, 117, and 130 biochemicals were altered in the IDH1-R132H expression, 7.5 mM 2HG-treated, and 30 mM 2HG-treated groups, respectively, and 43 of these alterations occurred in all three groups (Fig. 2B). Additionally, the biochemical levels were correlated for the IDH1-R132H and 30 mM 2HG groups (r = 0.22; P = 0.001; Table S2). Fewer alterations were shared by the IDH1-knockdown and IDH1-R132H expression groups, and these groups were inversely correlated (r = −0.15; P = 0.03). PCA revealed that 2HG-treated and IDH1-R132H expression groups shared large PC1 values (37.7% of variance) compared with the other groups, but IDH1-knockdown and IDH1-R132H expression groups did not share any PCs that distinguished these groups from controls (Fig. 2C, and Fig. S4 B and C).
Fig 2.
Metabolite profile of a glioma cell line expressing IDH1-R132H, treated with 2HG, or with knocked-down IDH1. (A) Heat map showing z scores for 202 biochemicals in HOG cell lysates arranged by unsupervised hierarchical clustering. Six replicates each of cells stably expressing IDH1-R132H, IDH1 shRNA, or scrambled shRNA and cells treated with medium containing 0 (vector), 7.5 mM, or 30 mM 2HG for 72 h before analysis are shown. (B) Venn diagrams indicating the number of biochemicals with mean levels that are significantly (P < 0.05) higher or lower in each group of cells compared with vector and the number of these changes shared by cells in the indicated groups. (C) PCA of this metabolite profile dataset. The percentage of variance in the dataset reflected by the first six PCs is shown in a histogram, and PC1 and PC2 for each sample are plotted.
To integrate our findings and identify biochemicals that were most altered in cells expressing IDH1-R132H, we analyzed the 28 biochemicals that were reproducibly and significantly altered by at least twofold by IDH1-R132H expression (Fig. 3). We found that many of these same biochemicals were altered in cells expressing IDH2-R172K and, to a lesser extent, in cells treated with 2HG. However, IDH1-WT, IDH2-WT, and IDH1 shRNA-treated cells shared only a few (0–2) of these alterations.
Fig 3.
Metabolites altered twofold or more by IDH1-R132H expression. Biochemicals that were on average more than twofold higher or lower in HOG IDH1-R132H cells relative to vector cells are displayed. The fold-change of these biochemicals in cells expressing IDH1-WT, IDH2-R172K, IDH2-WT, treated with 2HG, or expressing IDH1 shRNA is shown also. All changes shown here that were greater than twofold were significant (P < 0.05). Note that this scale colors only findings with changes greater than twofold. Detailed information on these changes can be found in Dataset S1 and S3.
Amino Acid, Choline Lipid, and TCA Cycle Metabolite Levels Are Altered in Cells Expressing IDH-Mutants or Treated with 2HG.
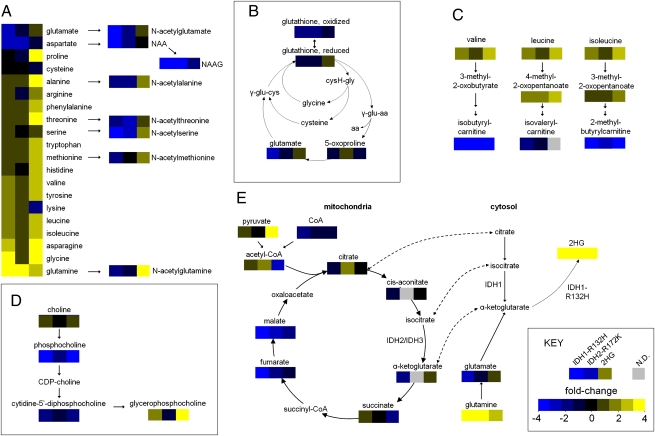
Next, we used information from the above analyses of cell lysates (Datasets S1 and S3) to identify metabolic pathways that were affected by IDH1-R132H expression, IDH2-R172K expression, or 2HG treatment. Because treatment with 30 mM 2HG, as opposed to 7.5 mM 2HG, achieved intracellular 2HG levels and global changes more similar to those observed for IDH mutant expression, we chose to focus on this level of 2HG treatment. We selected KEGG subpathways (as delineated in Datasets S1 and S3, Heatmap tabs) that had significant and reproducible alterations in >50% of biochemicals from that subpathway in cells expressing IDH1-R132H. After selecting subpathways that were altered in cells expressing IDH1-R132H in this manner, we determined the level of metabolites in these subpathways in the IDH1-R132H, IDH2-R172K, and 2HG-treated cells. We then mapped these data to simplified versions of these pathways (Fig. 4).
Fig 4.
Alterations in metabolic pathways observed in cells expressing IDH1-R132H, expressing IDH2-R172K, or treated with 2HG. The fold difference in metabolite in each experiment relative to vector is indicated by the color of each box. (A) Amino acids and N-acetylated amino acids (IDH1-R132H, left boxes; IDH2-R172K, center boxes; 30 mM 2HG, right boxes). (B) Glutathione and metabolites involved in its regeneration. (C) BCAAs and catabolites. (D) Choline, GPC, and intermediates. (E) TCA and shuttling of citrate, isocitrate, and α-ketoglutarate to the cytosol. Dashed lines indicate exchange of a metabolite between the mitochondria and cytosol. cysH-gly, cysteinylglycine; γ-glu-aa, γ-glutamyl amino acids; γ-glu-cys, γ-glutamyl-cysteine.
This analysis revealed that amino acids and their derivatives were altered in the IDH1-R132H, IDH2-R172K, and 2HG groups (Fig. 4A). Many amino acids, including glycine, serine, threonine, asparagine, phenylalanine, tyrosine, tryptophan, and methionine, were increased (range: 1.2- to 5.6-fold; P < 0.05) in all three groups. Aspartate, on the other hand, was decreased in all three groups (range: 1.8- to 2.5-fold; P < 0.001 for each). Interestingly, glutamate was decreased in the IDH1-R132H cells (2.6-fold; P < 0.001) and IDH2-R172K cells (1.4-fold; P = 0.003) but was increased in 2HG-treated cells (1.4-fold; P = 0.002). Glutamine was one of only three biochemicals that were significantly altered in opposite directions in two independent analyses of IDH1-R132H cells (P < 0.001 for both). We also observed alterations of N-acetylated amino acids, which are amino acid derivatives synthesized by N-acetyltransferases from free l-amino acids and acetyl-CoA, yielding free CoA as a product. All eight N-acetylated amino acids analyzed were lower in IDH1-R132H–expressing cells (range: 1.7- to 50-fold; P < 0.05 for each), and seven also were lower in IDH2-R172K–expressing cells (range: 1.4- to 8.3-fold; P < 0.05 for each). In contrast, six N-acetylated amino acids were increased by 2HG treatment (range: 1.2- to 3.0-fold; P < 0.05 for each). Although N-acetyl-aspartyl-glutamate (NAAG) was somewhat lower in 2HG-treated cells (1.8-fold; P < 0.001), it was remarkably (50-fold) lower in IDH1-R132H–expressing cells (P < 0.001) and was reduced 8.3-fold IDH2-R172K–expressing cells (P < 0.001). N-acetyl-aspartate (NAA) also was greatly reduced in IDH1-R132H–expressing cells (3.4-fold; P < 0.001) and IDH2-R172K–expressing cells (1.4-fold; P < 0.001) and was not significantly changed in 2HG-treated cells (1.1-fold lower; P = 0.38). Both reduced and oxidized glutathione (an amino acid-derived antioxidant that scavenges reactive oxygen species) were lower in IDH1-R132H– and IDH2-R172K–expressing cells (>1.6-fold; P < 0.001 for all four comparisons), but these compounds were not significantly affected by 2HG treatment (Fig. 4B). We also observed numerous changes in BCAAs and their catabolites (Fig. 4C) and choline lipid derivatives (Fig. 4D), which are detailed in SI Results.
TCA intermediates were markedly affected by IDH mutant expression (Fig. 4E). Most striking were reduced levels of late TCA intermediates fumarate (3.0- and 1.8-fold; P < 0.001 and P = 0.002) and malate (5.6- and 2.2-fold; P < 0.001 for each) in IDH1-R132H and IDH2-R172K cells, respectively. α-Ketoglutarate, which is the substrate for production of 2HG by IDH mutants, tended to be lower in IDH1-R132H–expressing cells (1.8-fold; P = 0.11) and higher in 30 mM 2HG-treated cells (1.3-fold; P = 0.10). As expected, 2HG was highly elevated in all IDH mutant groups, with a 216-fold elevation for IDH1-R132H cells, a 112-fold elevation for IDH2-R172K cells, and a 54-fold elevation in the 30 mM 2HG group (P < 0.001 for each).
N-Acetylated Amino Acids Are Depleted in IDH1-Mutated Gliomas.
One of the most striking findings of our analysis was the association of lowered N-acetylated amino acids with IDH mutant expression. Using targeted-mass LC-MS/MS, we verified that NAA and NAAG were lower in HOG cells expressing IDH1-R132H (mock-treatment group, Fig. 5A). We also noted that NAA and NAAG normally are secreted into culture medium by HOG cells, but cells expressing IDH1-R132H do not secrete detectable levels of NAAG (Fig. 5B). We sought to obtain information about a mechanism that could account for the very low NAAG levels that we observed in cells expressing IDH1-R132H. NAAG normally is synthesized from NAA and glutamate by NAAG synthase (23, 24). HOG cells incubated in 100 μM NAA had higher intracellular NAA levels than controls (NAA levels in NAA-treated cells, Fig. 5A), suggesting that NAA can enter the cell from extracellular media. This treatment increased the level of NAA in IDH1-R132H–expressing cells to the normal level of NAA in the vector control. However, this restoration of NAA levels did not increase the NAAG in HOG cells expressing IDH1-R132H, indicating that these cells cannot synthesize NAAG from NAA even in the presence of normal NAA levels. Finally, we treated cells with 10 μM NAAG and found that intracellular NAAG levels were increased compared with the no-treatment group, as expected (NAAG levels in NAAG-treated cells, Fig. 5A). Additionally, NAA levels were increased modestly in the NAAG-treated cells in all groups, indicating that IDH1-R132H expression does not interfere with NAAG breakdown into NAA and glutamate (NAA levels in NAAG-treated cells, Fig. 5A). Finally, we determined whether NAA or NAAG depletion occurs in IDH1-mutated cells in vivo by analyzing tissue from 26 intermediate-grade gliomas. We found that IDH1-mutated tumors had lower mean levels of NAA (2.1-fold; P = 0.049) and NAAG (2.4-fold; P = 0.019) than tumors without IDH1 mutations (Dataset S3, Tumor NAA_NAAG tab).
Fig 5.
NAA and NAAG in cell lines containing IDH1-R132H determined by targeted LC-MS/MS. (A) NAA and NAAG levels in HOG cells expressing vector, IDH1-WT, or IDH1-R132H mock-treated or incubated in 100 μM NAA or 10 μM NAAG medium for 48 h. (B) NAA and NAAG in media incubated for 48 h with HOG cells expressing IDH1-R132H, IDH1-WT, or a vector control (v).
Discussion
2HG Can Mediate Metabolic Changes Associated with IDH Mutants.
Here, we show that IDH1-R132H expression and IDH2-R172K expression induce multiple changes in the cellular metabolome. Knockdown of IDH1-WT produced few changes that also were caused by IDH1-R132H expression, indicating that dominant negative inhibition of the functional IDH1 allele by IDH1-R132H is not likely to be responsible for the metabolic changes associated with IDH1 mutations. In contrast, 2HG treatment resulted in global metabolic changes more similar to those in IDH1-R132H–expressing cells than to controls. In particular, both 2HG treatment and IDH expression were associated with changes in free amino acids, BCAAs, and choline phospholipid synthesis.
Although 2HG treatment and IDH mutant expression induced many similar changes, 56 of the 107 significant alterations that we observed in IDH1-R132H–expressing cells were not observed in 2HG-treated cells. 2HG-independent changes included depletion of glutamate and several metabolites that are directly or indirectly derived from glutamate, including glutathiones, N-acetylglutamate, NAAG, α-ketoglutarate, malate, and fumarate. IDH1-R132H expression results in elevated flux from glutamine to 2HG through glutamate and α-ketoglutarate (13) (see pathway in Fig. 4E). Thus, glutamate may become depleted as it is converted first to α-ketoglutarate and then to 2HG. Interestingly, glioma cells expressing IDH1-R132H are susceptible to knockdown of glutaminase, the enzyme which converts glutamine to glutamate (25). This observation suggests that glutamine-to-glutamate conversion could be a metabolic bottleneck for IDH-mutated cells. Because treatment with exogenous 2HG did not deplete glutamate, some differences between 2HG-treated and IDH mutant-expressing cells could reflect the different levels of glutamate in these cells. Alternative explanations for the differences between cells expressing IDH mutants and cells treated with 2HG include the possibility that the metabolite profiles reflect different levels of 2HG in the cells expressing mutant IDH1 vs. those treated with 2HG or that IDH mutants exert effects on cellular metabolism that are independent of their generation of 2HG.
NAA and NAAG Are Depleted in IDH1-Mutated Human Gliomas.
We found that N-acetylated amino acids are lowered in glioma cells expressing IDH1 or IDH2 mutants. Low levels of acetyl-CoA or free amino acids, the substrates for N-acetyltransferases, cannot explain this phenomenon, because these compounds were not decreased consistently in cells expressing IDH1-R132H and IDH2-R172K (Datasets S1 and S3). More likely explanations are that N-acetyltransferase enzymes are down-regulated or that breakdown of N-acetylated amino acids is up-regulated. NAAG differs from the other N-acetylated amino acids analyzed here in that it is synthesized from another N-acetylated amino acid, NAA, by NAAG synthetase. In cells expressing mutant IDH1, NAAG synthetase cannot synthesize NAAG even when NAA is restored to normal levels (Fig. 5A), suggesting that down-regulation of this enzyme underlies the depletion of NAAG. Because glutamate also is a substrate for NAAG synthetase, it is reasonable to expect that this enzyme has a low reaction rate in cells expressing IDH mutants, which we proved to have low glutamate levels. Future experiments using cell-permeable glutamate mimetics could determine whether restoration of normal cellular glutamate levels can rescue NAAG synthetase function in cells expressing IDH mutants. Also, analysis of RNA levels, protein expression, and enzymatic activity for NAAG synthetase and N-acetyltransferases in IDH-mutated gliomas models may further pinpoint the mechanism(s) underlying N-acetylated amino acid depletion.
NAA is the second-most abundant compound in brain, and NAAG is the most abundant dipeptide in brain, but their normal physiological function is poorly understood. Both metabolites take part in a brain metabolic cycle that includes synthesis of NAAG in neurons, breakdown of NAAG to NAA and glutamate in association with astrocytes, and breakdown of NAA into aspartate and acetate in oligodendrocytes (24, 26). The finding that NAA and NAAG are lowered in cell lines homologously expressing IDH mutants and also in human glioma tissues with IDH1 mutations suggests that glioma cell lines homologously expressing IDH mutants recapitulate features of human gliomas with somatic IDH mutations in situ. The difference in NAA and NAAG levels in IDH1-mutated compared to wild-type tumors (2.4-fold for NAAG) was not as large as the difference we observed between cells expressing IDH1-R132H and vector control cells (50-fold for NAAG). However, because cancer cells are “contaminated” with normal vascular and inflammatory cells in glioma tissue (5), normal cells containing higher amounts of NAA and NAAG could mask more striking differences in the cancer cells themselves. Whether NAA or NAAG depletion contributes to glioma pathogenesis is unclear. However, if NAA or NAAG is found to exert a tumor-suppressing function that is relieved in IDH-mutated gliomas, therapeutics that replenish these compounds in tumors could have clinical utility.
Other Metabolic Pathways and Future Studies.
Some of the most conspicuous features of cells expressing IDH mutants included elevation of numerous free amino acids, elevation of lipid precursors such as glycerol-phosphates and GPC, and depletion of the TCA cycle intermediates citrate, cis-aconitate, α-ketoglutarate, fumarate, and malate. We did not observe changes in intermediates related to glycolysis such as 1,6-glucose phosphate, pyruvate, or lactate that were reproducible and shared by IDH1 and IDH2 mutant-expressing cells. Thus, the levels of biosynthetic molecules were increased and TCA intermediates were decreased in cells expressing the either IDH mutant, without consistent accumulation or depletion of glycolytic intermediates. These changes could result from shunting of carbons from glycolysis into de novo synthesis of amino acids and lipids rather than into the TCA. Alternatively, they could reflect a lower rate of amino acid and lipid catalysis into carbon backbones that ultimately enter the TCA. A possible explanation for the increase in free amino acids in cells expressing IDH mutants or treated with 2HG could be that 2HG inhibits α-keto acid transaminases, which are enzymes that normally transfer amine groups from free amino acids to α-ketoglutarate as a first step in amino acid breakdown for oxidation in the TCA. This possibility is consistent with the hypothesis that 2HG can competitively inhibit α-ketoglutarate–dependent enzymes (27, 28). It has been proposed that TCA down-regulation is a major effect of some genetic alterations in cancer. Furthermore, it has been suggested that this down-regulation is associated with a selective advantage for cancer cells because nutrients then are converted to building blocks such as amino acids and lipids to be used for proliferation rather than being oxidized in the TCA (4). Treatment of chick neurons with 2HG has been observed to impair complex V (ATP synthase) of the mitochondrial electron transport chain (29). Thus, a possible mechanism through which IDH mutants deregulate the TCA could be by producing 2HG that disrupts the normal transfer of electrons from TCA intermediates into the electron transport chain.
Future research will focus on determining whether the metabolic alterations that we observed in glioma cell lines expressing IDH mutants are recapitulated in other models of IDH-mutated cancer and in primary tumors with IDH mutations. Also, although this analysis provides a snapshot of the level of metabolites in cells, it does not provide information on their compartmentalization or the rate of their synthesis, breakdown, and exchange with extracellular media. Metabolomic analysis of specific cellular compartments, such as the mitochondria, will be needed to determine whether the changes in metabolite levels reported here have a compartmental bias. Furthermore, isotope-labeling experiments will be necessary to explore the metabolic networks and kinetics of metabolite conversion underlying these observations. These studies may help elucidate a mechanism responsible for the changes that we observed. Finally, future work will test whether any of the metabolic changes reported here serve as a mechanistic link between IDH mutations and cancer-related phenotypes. This information could inform the design of therapies that target the altered metabolism of cancer cells while sparing noncancer tissue.
Materials and Methods
Stable HOG clones were created by expansion of single cells transduced with lentiviruses or retroviruses for gene or shRNA expression, respectively. Metabolomic profiling was carried out in collaboration with Metabolon. Hierarchical clustering, Welch's t tests, Pearson correlation, and PCA were performed in R. Human tissue was obtained with consent and analyzed at the Preston Robert Tisch Brain Tumor Center at Duke Biorepository. LC-MS/MS for NAA/NAAG analysis was performed using an Agilent 1200 series HPLC and Sciex/Applied Biosystems API 3200 QTrap in +ESI mode. An extended description of the materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Jennifer Marcello, Dr. James E. Herndon II, and Dr. Paul M. Magwene for comments on the manuscript; Dr. Roger McLendon, Melissa J. Ehinger, and Diane L. Satterfield for assistance with glioma tissue samples; and Ping Fan for assistance with targeted mass LC-MS/MS. This project was supported by American Cancer Society Research Scholar Award RSG-10-126-01-CCE, by Grant 5P30-CA-014236-36 from the National Institutes of Health, and Grants R01-CA-140316 and CA-43460 from the National Cancer Institute, by the Virginia and D.K. Ludwig Fund for Cancer Research, and by grants from the Pediatric Brain Tumor Foundation Institute, American Association for Cancer Research Stand-Up-to-Cancer, and the Duke Comprehensive Cancer Center Core.
Footnotes
Conflict of interest statement: Under agreements between the Johns Hopkins University, Agios Pharmaceuticals, and Personal Genome Diagnostics, B.V. is entitled to a share of the royalties received by the University on sales of products related to IDH genes. B.V. is a cofounder of Personal Genome Diagnostics and is a member of its Scientific Advisory Board. B.V. also owns stock in Personal Genome Diagnostics, which is subject to certain restrictions under University policy. The terms of these arrangements are managed by the Johns Hopkins University in accordance with its conflict-of-interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019393108/-/DCSupplemental.
References
- 1.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yun J, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–1559. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: Diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9:717–726. doi: 10.1016/S1474-4422(10)70105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: Alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartmann C, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: A study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhao S, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross S, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin JL, et al. Assignment of 1H nuclear magnetic resonance visible polyunsaturated fatty acids in BT4C gliomas undergoing ganciclovir-thymidine kinase gene therapy-induced programmed cell death. Cancer Res. 2003;63:3195–3201. [PubMed] [Google Scholar]
- 16.Chen C, Gonzalez FJ, Idle JR. LC-MS-based metabolomics in drug metabolism. Drug Metab Rev. 2007;39:581–597. doi: 10.1080/03602530701497804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazurek S. Pyruvate kinase type M2: A key regulator within the tumour metabolome and a tool for metabolic profiling of tumours. Ernst Schering Found Symp Proc. 2007;4:99–124. doi: 10.1007/2789_2008_091. [DOI] [PubMed] [Google Scholar]
- 18.Fan TW, et al. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009;8:41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane AN, Fan TW, Higashi RM. Stable isotope-assisted metabolomics in cancer research. IUBMB Life. 2008;60:124–129. doi: 10.1002/iub.17. [DOI] [PubMed] [Google Scholar]
- 20.Kanehisa M. Representation and analysis of molecular networks involving diseases and drugs. Genome Inform. 2009;23:212–213. [PubMed] [Google Scholar]
- 21.Jolliffee IT. Principal Component Analysis. 2nd Ed. New York: Springer; 2002. [Google Scholar]
- 22.Jin G, et al. 2-Hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP+-dependent isocitrate dehydrogenase mutations. PLoS One. 2011 doi: 10.1371/journal.pone.0016812. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collard F, et al. Molecular identification of N-acetylaspartylglutamate synthase and beta-citrylglutamate synthase. J Biol Chem. 2010;285:29826–29833. doi: 10.1074/jbc.M110.152629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker I, Lodder J, Gieselmann V, Eckhardt M. Molecular characterization of N-acetylaspartylglutamate synthetase. J Biol Chem. 2010;285:29156–29164. doi: 10.1074/jbc.M110.111765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baslow MH. Evidence that the tri-cellular metabolism of N-acetylaspartate functions as the brain's “operating system”: How NAA metabolism supports meaningful intercellular frequency-encoded communications. Amino Acids. 2010;39:1139–1145. doi: 10.1007/s00726-010-0656-6. [DOI] [PubMed] [Google Scholar]
- 27.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kölker S, et al. NMDA receptor activation and respiratory chain complex V inhibition contribute to neurodegeneration in d-2-hydroxyglutaric aciduria. Eur J Neurosci. 2002;16:21–28. doi: 10.1046/j.1460-9568.2002.02055.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.