Abstract
Proper timing of gene expression is essential for many biological events, but the molecular mechanisms that control timing remain largely unclear. It has been suggested that introns contribute to the timing mechanisms of gene expression, but this hypothesis has not been tested with natural genes. One of the best systems for examining the significance of introns is the oscillator network in the somite segmentation clock, because mathematical modeling predicted that oscillating expression depends on negative feedback with a delayed timing. The basic helix–loop–helix repressor gene Hes7 is cyclically expressed in the presomitic mesoderm (PSM) and regulates the somite segmentation. Here, we found that introns lead to an ∼19-min delay in the Hes7 gene expression, and mathematical modeling suggested that without such a delay, Hes7 oscillations would be abolished. To test this prediction, we generated mice carrying the Hes7 locus whose introns were removed. In these mice, Hes7 expression did not oscillate but occurred steadily, leading to severe segmentation defects. These results indicate that introns are indeed required for Hes7 oscillations and point to the significance of intronic delays in dynamic gene expression.
Keywords: differential equation, time-lapse imaging
Proper timing of activation and repression of gene expression is essential for many biological events, but the molecular mechanisms that control timing remain largely unclear. It has been suggested that introns contribute to the timing mechanisms of gene expression (1–3), but this hypothesis has not been tested with natural genes. One of the best systems for examining the significance of introns is the oscillator network in the somite segmentation clock (4–9).
During somite segmentation of mouse embryos, the basic helix–loop–helix repressor gene Hes7 is cyclically expressed with a period of about 2 h in the presomitic mesoderm (PSM) (10). The Hes7 expression domain is propagated from the posterior to the anterior PSM, and each cycle leads to formation of a bilateral pair of somites (10). Both loss of expression and persistent expression of Hes7 lead to somite fusion, suggesting that oscillatory expression is required for periodic somite segmentation (10, 11). This oscillatory expression is regulated by negative feedback similar to the regulation of Hes1 in fibroblasts (12): Hes7 protein represses transcription from the Hes7 promoter, and this repression down-regulates both Hes7 mRNA and Hes7 protein levels. This down-regulation results in relief from the negative feedback and allows the next round of expression (13). Transcription of the Hes7 gene and accumulation of Hes7 protein occur in a mutually exclusive manner, and therefore these two events proceed alternately every 2 h, indicating that Hes7 protein accumulation is substantially delayed relative to Hes7 gene transcription (Fig. 1A) (13). This negative feedback-mediated oscillatory expression has been mathematically simulated by differential equations (14–17). These models predict that the delay from transcription to protein expression and then to negative autoregulation must be sufficiently long for sustained oscillations and that short delays would abolish oscillations. However, this prediction has not been experimentally tested yet.
Fig. 1.
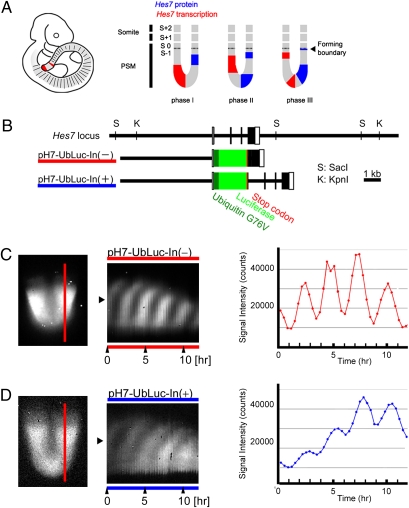
Dynamic expression of the intron (+) and intron (−) Hes7 reporters. (A) Schematic view of Hes7 transcription and Hes7 protein expression in the presomitic mesoderm. Hes7 transcription and Hes7 protein expression occur in a mutually exclusive manner. (B) Hes7 reporter gene constructs. The Hes7 promoter-driven firefly luciferase gene fused with human ubiquitin variant (G76V) was used as a reporter. pH7-UbLuc-In(−) has no intron, whereas pH7-UbLuc-In(+) has all introns. The total size of three introns is 1,843 bp. (C and D) (Left) Reporter expression from pH7-UbLuc-In(−) (C) and pH7-UbLuc-In(+) (D). (Middle) The spatiotemporal profiles of the reporter expression measured along the vertical red line in the Left. (Right) Quantification of the reporter expression. The signal intensity was measured at the middle region of the PSM (indicated by arrowheads in the Center). Similar expression patterns were observed in four independent experiments for each reporter.
To determine the significance of introns in the timing of gene expression, we focused on the Hes7 gene in the mouse segmentation clock. We first found that the Hes7 promoter-driven reporter without any introns led to expression ∼19 min earlier than the reporter with full introns, indicating that introns lead to about a 19-min delay in Hes7 expression. According to our previous mathematical model (17), if the delay is 19 min shorter, the oscillatory expression of Hes7 would be abolished, although a different model predicted sustained oscillations without delays (18). To determine whether intronic delays are required for sustained oscillations of Hes7 expression, we generated mice carrying the Hes7 locus whose introns were removed. We found that Hes7 oscillations were abolished in these mutant mice and that somites were not properly segmented. These data indicate that intronic delays are essential for sustained Hes7 oscillations and periodic somite segmentation.
Results and Discussion
Both Intron-Plus and Intron-Minus Hes7 Reporters Display Oscillatory Expression.
To assess the significance of introns in the timing of gene expression, we first generated two Hes7 promoter-driven reporters: One carried no introns [pH7-UbLuc-In(−), 3,070-bp transcript, Fig. 1B], whereas the other carried all Hes7 gene introns [pH7-UbLuc-In(+), 4,913-bp transcript, Fig. 1B]. This promoter fragment was previously shown to contain regulatory elements required for normal Hes7 expression (11). Because the half-life of Hes7 protein is ∼20 min (17), that of a reporter should be ≤20 min. Otherwise, a reporter protein would be accumulated after several cycles. We previously showed that a ubiquitinated luciferase (Ub-luc) can be successfully used as a reporter for Hes1, which also displays oscillatory expression with a period of ∼2 h (19, 20). Therefore, we decided to use Ub-luc to assess the significance of introns (Fig. 1B).
Transgenic mice carrying either the pH7-UbLuc-In(−) or the pH7-UbLuc-In(+) reporter were generated, and explants of caudal parts of embryonic day (E)10.5 embryos were cultured. Time-lapse imaging of the reporter expression was done by monitoring bioluminescence with a highly sensitive CCD camera. Bioluminescence from both pH7-UbLuc-In(−) and pH7-UbLuc-In(+) explant cultures oscillated dynamically (Movies S1 and S2), and each cycle corresponded to formation of a bilateral pair of somites (Fig. 1 C and D). Reporter expression from both cultures was propagated from the posterior to the anterior PSM, indicating that expression of both reporters accurately reflected endogenous Hes7 expression. However, we noticed that the timing of the reporter expression was different between the two constructs.
Introns Delay the Timing of Gene Expression.
We next compared the timing of reporter expression with that of endogenous Hes7 transcription and of Hes7 protein expression. Hes7 transcription was examined by in situ hybridization with the first intron probe, as previously described (13). Because expression domains move periodically from the posterior to the anterior PSM, the difference in timing was reflected by the difference in the position of expression domains. The length difference between the anterior ends of expression domains was transformed into a time difference by calculating the propagation speed of the time-lapse imaging of Hes7 reporter expression (Fig. S1).
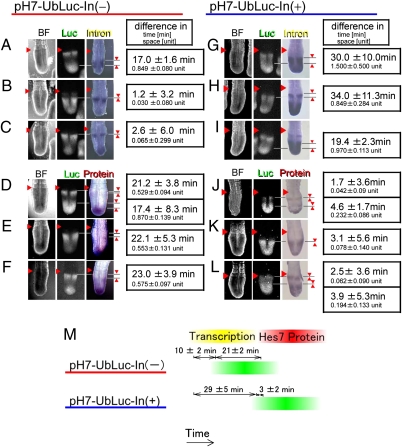
Reporter expression from pH7-UbLuc-In(−) mice occurred 1.2–17.0 min (on average 10 ± 2 min) more slowly than the endogenous Hes7 intron expression (Fig. 2 A–C and M) but 17.4–23.0 min (on average 21 ± 2 min) earlier than the endogenous Hes7 protein expression (Fig. 2 D–F and M). Thus, reporter expression from pH7-UbLuc-In(−) occurred temporally between the endogenous Hes7 transcription and Hes7 protein expression (Fig. 2M). In contrast, reporter expression from pH7-UbLuc-In(+) mice occurred 19.4–34.0 min (on average 29 ± 5 min) more slowly than the endogenous Hes7 intron expression (Fig. 2 G–I and M) but 1.7–4.6 min (on average 3 ± 2 min) earlier than the endogenous Hes7 protein expression (Fig. 2 J–M). Thus, reporter expression from pH7-UbLuc-In(+) was very similar to the endogenous Hes7 protein expression in timing (Fig. 2M); indeed, it occurred on average ∼19 min more slowly than did reporter expression from pH7-UbLuc-In(−). These results suggest that the introns caused an ∼19-min delay in Hes7 expression, which was within the expected range of in vivo splicing kinetics (21, 22). The ratio of this delay to the segmentation period is comparable to that of zebrafish (23).
Fig. 2.
Introns delay the timing of Hes7 reporter expression. (A–L) Comparison of reporter expression (Luc) with either Hes7 intron expression or Hes7 protein expression in pH7-UbLuc-In(−) mice (A–F) or pH7-UbLuc-In(+) mice (G–L). Reporter expression was classified into three phases on the basis of luciferase activity: phase A, relatively narrower expression in the anterior PSM and broader expression in the posterior PSM (A, D, G, and J); phase B, strong expression in the middle part of the PSM (B, E, H, and K); and phase C, relatively broader expression in the anterior PSM and narrower expression in the posterior PSM (C, F, I, and L). After acquisition of luciferase activity images, posterior parts of the embryos were fixed immediately and analyzed for either Hes7 intron expression (n = 16 in A–C, n = 7 in G–I) or Hes7 protein expression (n = 22 in D–F, n = 22 in J–L). The first column shows bright field (BF) images. The difference in space (1 unit is defined as a one-somite length) was converted into the difference in time on the basis of the movies. The averages with SEs are shown. The arrowhead on the left side of each section indicates a boundary between a newly formed somite and the PSM. (M) Comparison of Hes7 reporter expression with Hes7 gene transcription and Hes7 protein expression.
To measure the intronic delay in a different system, we transfected the same reporters but under the control of Hes1 promoter instead of Hes7 promoter into cultured fibroblasts, because Hes1 promoter, but not Hes7 promoter, was active in these cells (Fig. S2). We found that the introns led to a similar delay in appearance of the reporter expression (Fig. S2). These results confirmed that introns produce a substantial delay in gene expression.
We also noticed that the amplitude of the Hes7 reporter oscillation was different depending on whether the introns were present (Fig. 1 C and D). The smaller amplitude of pH7-UbLuc-In(+) expression could be due to nonsense-mediated mRNA decay because the stop codon was present in the first exon. To prevent this decay, we performed explant cultures for the pH7-UbLuc-In(+) reporter in the presence of PTC124 (24), but the patterns were not improved (Fig. S3). Alternatively, the smaller amplitude of pH7-UbLuc-In(+) expression could be due to distributed intronic delays (see mathematical modeling in SI Materials and Methods and Fig. S4D).
Hes7 Gene Introns Are Required for Periodic Somite Segmentation.
We previously proposed a mathematical model consisting of differential equations (SI Materials and Methods), which accurately described the dynamics of Hes7 oscillations (Fig. S4A) (17). On the basis of this modeling, we found that a 19-min shorter delay would abolish Hes7 oscillations (Fig. S4C), although different models predict sustained oscillations without delays (12, 18).
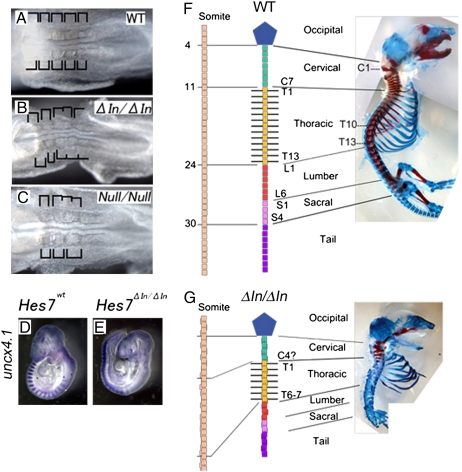
To determine whether intronic delays are required for sustained oscillations of Hes7 expression, we decided to remove all introns from the Hes7 locus. A targeting vector with the intronless Hes7 gene was introduced into mouse embryonic stem (ES) cells, and homologous recombinants were obtained (Fig. S5). After the neo gene was deleted by the Cre-LoxP system, the recombinant ES cells were used to make chimeric mice. From these mice, we obtained homozygous mutant mice carrying the Hes7 locus whose introns were removed (ΔIn/ΔIn; Fig. 3 and Fig. S6). In these intronless mice, somites were irregular in size and partially fused (Fig. 3B), compared with the wild type (Fig. 3A), indicating that somites were not properly segmented without Hes7 gene introns. These defects were indistinguishable from Hes7-null mice (Fig. 3C). Furthermore, expression of Tbx18 and Uncx4.1, markers for the anterior- and posterior-half somites, respectively, was not properly segmented (Fig. 3E; compare with wild type in Fig. 3D and Fig. S7 A–D), and the vertebrae and ribs, which are derived from somites, were also severely fused in the intronless mice (Fig. 3G, compare with wild type in Fig. 3F). These results indicate that introns in the Hes7 locus play an essential role in periodic somite segmentation.
Fig. 3.
Segmentation defects in mice carrying the intronless Hes7 alleles. (A–C) Bright field images of E8.5 embryos. Somites were regular in size and symmetric between the left and right in wild-type (A) but not in intronless (ΔIn/ΔIn, B) and Hes7-null mice (Null/Null, C). (D and E) Uncx4.1 expression in E9.5 embryos. Somites were severely fused in Hes7 intronless mice (ΔIn/ΔIn, E). (F and G) Bones and cartilages of neonates were stained in red and blue, respectively. The vertebrae and ribs were severely fused in the intronless mice (G).
Hes7 Gene Introns Are Required for Oscillatory Expression.
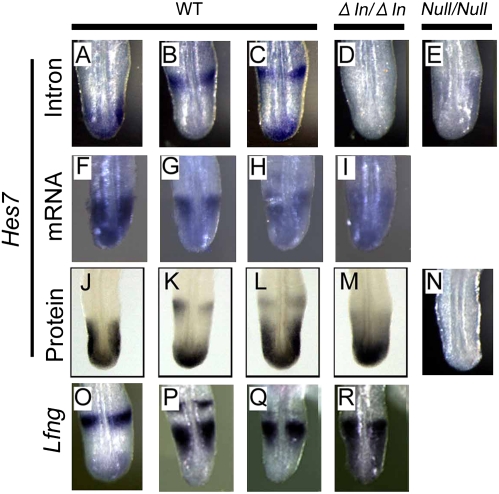
To determine the role of intronic delays in oscillatory expression, we next examined the dynamics of cyclic genes in the intronless mice. Wild-type embryos displayed different patterns of Hes7 transcription, Hes7 mRNA expression, and Hes7 protein expression, suggesting that Hes7 expression oscillates in the PSM (Fig. 4 A–C, F–H, and J–L). Similarly, Lunatic fringe (Lfng), a gene for a glycosyltransferase of Notch, and Dusp4/MKP2, a phophatase gene downstream of Fgf signaling, whose expression is regulated by Hes7 oscillations, displayed different expression patterns in the PSM of wild-type embryos (Fig. 4 O–Q and Fig. S7E). It has been shown that oscillatory expression of Lfng is essential for somite segmentation (25–28). In Hes7-null mice, however, intron regions of Hes7 were constitutively expressed throughout the PSM (Fig. 4E) but no Hes7 protein was expressed (Fig. 4N), as previously described (13). In contrast, in the intronless mice, both Hes7 mRNA and Hes7 protein were steadily expressed in the PSM (Fig. 4 I and M). Furthermore, Lfng and Dusp4/MKP2 expression was also steady (Fig. 4R and Fig. S7E). Axin2 expression was still variable but less dynamic in Hes7 intronless mice (Fig. S7F). In these mice, Hes7 intron signals were not detectable, confirming that introns were deleted from the Hes7 locus (Fig. 4D). We also introduced pH7-UbLuc-In(+) reporter into Hes7 intronless mice, but oscillatory reporter expression was not detectable (Fig. S8). These results indicate that Hes7 expression does not oscillate; rather, it occurs steadily in the PSM when introns are deleted from the Hes7 locus.
Fig. 4.
Oscillatory expression was abolished in mice carrying the intronless Hes7 alleles. There were different patterns of Hes7 gene transcription (n = 16, A–C), which were detected by intron in situ hybridization, Hes7 mRNA expression (n = 3, F–H), Hes7 protein expression (n = 13, J–L), and Lfng mRNA expression (n = 5, O–Q) in E10.5 wild-type embryos (WT). This result suggested that Hes7 and Lfng expression oscillates in the WT. In contrast, Hes7 intron signals were undetectable in the intronless mice (n = 6, ΔIn/ΔIn, D) but were observed steadily in the PSM of Hes7-null mice (n = 13, Null/Null, E) at E10.5. Furthermore, Hes7 mRNA was expressed steadily in the PSM of the intronless mice (n = 4, ΔIn/ΔIn, I). In addition, Hes7 protein was steadily expressed in the PSM of intronless mice (n = 10, ΔIn/ΔIn, M) but not detectable in the PSM of Hes7-null mice (n = 8, Null/Null, N). Lfng mRNA was also expressed steadily in the PSM of the intronless mice (n = 4, ΔIn/ΔIn, R).
Introns are known to play an important role in regulating gene expression levels (29). We thus examined Hes7 protein expression levels in the PSM. Extracts of posterior parts of embryos were subjected to Western blot analysis. The intronless mice (ΔIn/ΔIn) expressed ∼34% of wild-type levels (Fig. S9). Heterozygous mutant mice (+/ΔIn) had normal vertebrae and ribs but displayed a kinked tail, indicating that segmentation defects occurred only at later stages. This late defect is probably because the wild-type allele, which expresses a higher level of Hes7 protein, is dominant over the intronless allele.
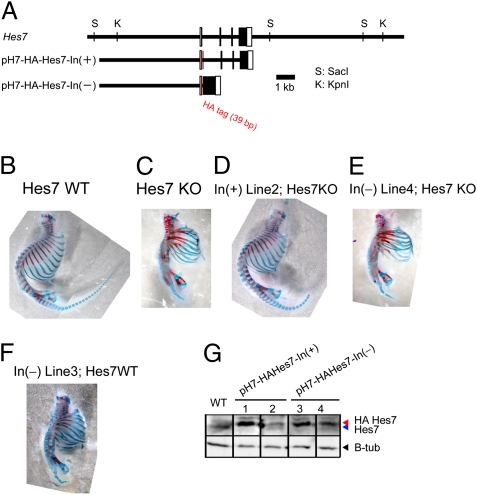
It was previously shown that oscillatory expression was maintained in the chick PSM even though >80% of protein synthesis was blocked by cycloheximide (30). This phenomenon was also mathematically simulated (14). These data suggest that loss of Hes7 oscillations is not due to a reduced expression level but to a lack of intronic delays. To overcome the expression level, multiple copies of the intron(+) and intronless Hes7 transgenes (Fig. 5A) were introduced into Hes7-null mice. We generated four independent lines carrying the intron(+) Hes7 transgene and four independent lines carrying the intronless Hes7 transgene that expressed Hes7 protein at levels comparable to or higher than the wild type in the PSM (two lines for each transgene are shown in Fig. 5G). All of the lines carrying the intron(+) Hes7 transgene successfully rescued the defect of Hes7-null mice (one line is shown in Fig. 5D), whereas all of the lines carrying the intronless Hes7 transgene failed to rescue the segmentation defects of Hes7-null mice (one line is shown in Fig. 5E). Furthermore, the intronless Hes7 transgene caused segmentation defects even in the wild-type background (one line is shown in Fig. 5F). These results support the idea that the intronic delay but not the expression level is important for Hes7 oscillations.
Fig. 5.
Segmentation defects of Hes7-null mice were rescued by the intron(+) Hes7 transgene but not by the intron(−) Hes7 transgene. (A) Structures of intron(+) and intron(−) Hes7 transgenes. The HA tag was added to the amino terminus to differentiate between the endogenous and transgene expression. This tag did not affect the segmentation (D). (B) Wild-type mouse. (C) Hes7-null mouse. (D) Hes7-null mouse containing the intron(+) Hes7 transgene. Four independent lines containing the intron(+) Hes7 transgene including the one shown here (line 2) rescued the segmentation defects. (E) Hes7-null mouse containing the intron(−) Hes7 transgene. Four independent lines containing the intron(−) Hes7 transgene including the one shown here (line 4) did not rescue the segmentation defects. (F) Segmentation defects were caused even in the Hes7(+/+) backgroud by the intron(−) Hes7 transgene (line 3). Bones and cartilages of neonates were stained in red and blue, respectively (B–F). (G) Western blot analysis of the PSM in the Hes7(+/+) background. Hes7 protein levels in two independent lines of the intron(+) transgene (lines 1 and 2) and two independent lines of the intron(−) transgene (lines 3 and 4) are shown. Hes7 protein made from the transgene is larger in size than the endogenous one because of the HA tag. β-Tubulin was used as a control. Note that the endogenous Hes7 protein expression was down-regulated by the exogenous Hes7 protein expressed from the transgenes. Segmentation defects of Hes7-null mice were rescued by the intron(+) transgene lines 1 and 2 but not by the intron(−) transgene lines 3 and 4.
Significance of Intronic Delays in Oscillatory Expression.
The Hes7 reporter without any introns directed reporter expression ∼19 min earlier than the reporter with introns, indicating that introns cause a 19-min delay in the timing of gene expression. Our mathematical modeling predicted that a delay 19 min shorter than the wild type would abolish Hes7 oscillations. We demonstrated that Hes7 oscillations were indeed abolished in embryos carrying the Hes7 locus whose introns were removed, indicating that introns are essential for Hes7 oscillations. It has been shown that introns and splicing are important for gene expression levels, and we found that only 34% of the wild-type levels of Hes7 protein was expressed in the intronless mice. However, reduction to the 34% level did not seem to be responsible for loss of oscillations because oscillations were not damped even though >80% of protein synthesis was blocked (31). Moreover, we showed that the segmentation defects of Hes7-null mice were rescued by introduction of multiple copies of the intron(+) Hes7 transgene but not by introduction of the intronless Hes7 transgene, although the Hes7 expression levels were rescued. All these data suggest that intronic delays, but not intron/splicing-dependent gene expression levels, are important for sustained Hes7 oscillations.
Our current mathematical modeling also predicted that the instability of Hes7 gene products is essential for sustained oscillations and that if the products are stabilized, oscillations would be damped (17). We previously demonstrated that Hes7 oscillations are indeed damped in embryos with a point mutation that makes the Hes7 protein half-life ∼8 min longer than the wild type (17). Thus, we have to date evaluated two important predictions of our mathematical modeling, that the instability of gene products and sufficient delays in the timing of gene expression are required for sustained oscillations (14–16), and verified both predictions by introducing specific mutations into mouse embryos. Taken together, our current mathematical modeling, which is based on delayed negative feedback, accurately describes the dynamics of Hes7 oscillations. However, recent studies have revealed that multiple signaling molecules are involved in oscillatory expression in the PSM (11, 31), and incorporation of such oscillators will be required to understand the whole structure of the segmentation clock.
Materials and Methods
Hes7 Reporter Mice.
The reporters consisted of the genomic fragment of the Hes7 promoter region (5,393-bp upstream fragment from the first codon), the firefly luciferase gene fused with human ubiquitin variant (G76V) with a termination codon (TAA) at the 3′ end (19), and a genomic sequence from the second codon to 76 bp downstream of the putative polyadenylation signal. For pH7-UbLuc-In(−), all introns were removed. Transgenic mice were generated by injecting the linearized constructs without any vector sequence into the pronucleus of fertilized eggs. All animals used for this study were maintained and handled according to protocols approved by Kyoto University.
Bioluminescence Imaging of the PSM.
All images were recorded in 16 bit with IMAGE-PRO PLUS (Media Cybernetics), and other equipment was described previously (19) with the following modifications. To increase the number of samples at a single capturing, a 10× UPlan FLN (NA 0.30) objective lens was used with 4 × 4 binning and exposure for 10 min. Tyrode's solution was used when dissecting embryos and capturing images. For movies, recording was performed, as previously described (19). Briefly, a 20× UPlan Apo Objective (NA 0.80) was used with 4 × 4 binning and exposure of 19 min, 24 s. For processing of bioluminescence images, cosmic ray-induced signals were first removed, and then the images were converted to 8 bit with 1,024 × 1,024 pixels in size by setting the maximum intensity to 255 and the minimum to 0. The 8-bit images of the PSM were aligned in the same anterior–posterior axis by Photoshop (Adobe Systems) to correct the moving of the tissue during recording. The images were trimmed into 15 pixels in width.
Generation of the Intronless Hes7 Mutant Mice.
The targeting vector was constructed by replacing the Hes7 coding and intron region with Hes7 cDNA to remove all introns. The Floxed Neor cassette was inserted into the SacI site in the 3′-downstream region of Hes7, and the diphtheria toxin A gene was ligated to the 3′-homologous region of the vector (Fig. S4A). The vector was electroporated into TT2 ES cells, and G418-resistant clones were analyzed by Southern blotting to isolate homologous recombinant ES cells. The Neor cassette was next removed by transient expression of Cre recombinase. Chimeras were generated by injecting recombinant ES cells into eight-cell stage mouse embryos according to standard procedures.
Genotyping and Analysis of Mice.
Genomic DNA was prepared from either tails of adult mice or amnion of embryos. Genotypes were determined by PCR. Primer sequences are described in SI Materials and Methods. Whole-mount in situ hybridization and whole-mount immunochemistry were performed, as described previously (13). Cartilage and bone of neonates were stained with alcian blue and alizarin red, respectively, as described previously (10).
Western Blotting.
The posterior parts of embryos were mixed with 30 μL of lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 0.5% Nonidet P-40, 1× Proteinase inhibitor mixture, 1 mM PMSF, 250 units/mL Benzonase) and incubated on ice for 30 min. After addition of 3 μL of 10% SDS, the samples were boiled, and the protein concentrations were measured. The protein solution after boiling in sample buffer was run on 12.5% SDS/PAGE. After transferring protein from the gel to the PVDF membrane (Millipore; IPVH15150), the membrane was immersed in buffer containing 5% skim milk, anti-Hes7 antibody (1/500) (13), and peroxidase-conjugated anti-guinea pig IgG (1/5,000; Chemicon) in sequence. Immunoreactive bands were visualized with ECL-plus (GE Healthcare) and LAS 3,000mini (Fujifilm). Intensity of each band was calculated with Image Gauge (Fujifilm). After stripping of antibodies, the membrane was immersed in buffer containing 5% skim milk, anti–β-tubulin IgG (Santa Cruz), and peroxidase-conjugated anti-rabbit IgG (GE Healthcare) in sequence. Immunoreactive bands were visualized with ECL (GE Healthcare).
Supplementary Material
Acknowledgments
We thank A. Isomura for discussion. This work was supported by the Genome Network Project and Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Uehara Memorial Foundation. Y.T. was supported by the 21st Century Center of Excellence Program of the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.I.-H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014418108/-/DCSupplemental.
References
- 1.Thummel CS. Mechanisms of transcriptional timing in Drosophila. Science. 1992;255:39–40. doi: 10.1126/science.1553530. [DOI] [PubMed] [Google Scholar]
- 2.Swinburne IA, Silver PA. Intron delays and transcriptional timing during development. Dev Cell. 2008;14:324–330. doi: 10.1016/j.devcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swinburne IA, Miguez DG, Landgraf D, Silver PA. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 2008;22:2342–2346. doi: 10.1101/gad.1696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saga Y, Takeda H. The making of the somite: Molecular events in vertebrate segmentation. Nat Rev Genet. 2001;2:835–845. doi: 10.1038/35098552. [DOI] [PubMed] [Google Scholar]
- 5.Aulehla A, Herrmann BG. Segmentation in vertebrates: Clock and gradient finally joined. Genes Dev. 2004;18:2060–2067. doi: 10.1101/gad.1217404. [DOI] [PubMed] [Google Scholar]
- 6.Giudicelli F, Lewis J. The vertebrate segmentation clock. Curr Opin Genet Dev. 2004;14:407–414. doi: 10.1016/j.gde.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Kageyama R, Ohtsuka T, Kobayashi T. The Hes gene family: Repressors and oscillators that orchestrate embryogenesis. Development. 2007;134:1243–1251. doi: 10.1242/dev.000786. [DOI] [PubMed] [Google Scholar]
- 8.Mara A, Holley SA. Oscillators and the emergence of tissue organization during zebrafish somitogenesis. Trends Cell Biol. 2007;17:593–599. doi: 10.1016/j.tcb.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Dequéant M-L, Pourquié O. Segmental patterning of the vertebrate embryonic axis. Nat Rev Genet. 2008;9:370–382. doi: 10.1038/nrg2320. [DOI] [PubMed] [Google Scholar]
- 10.Bessho Y, et al. Dynamic expression and essential functions of Hes7 in somite segmentation. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niwa Y, et al. The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell. 2007;13:298–304. doi: 10.1016/j.devcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 12.Hirata H, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 13.Bessho Y, Hirata H, Masamizu Y, Kageyama R. Periodic repression by the bHLH factor Hes7 is an essential mechanism for the somite segmentation clock. Genes Dev. 2003;17:1451–1456. doi: 10.1101/gad.1092303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- 15.Monk NAM. Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Curr Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- 16.Jensen MH, Sneppen K, Tiana G. Sustained oscillations and time delays in gene expression of protein Hes1. FEBS Lett. 2003;541:176–177. doi: 10.1016/s0014-5793(03)00279-5. [DOI] [PubMed] [Google Scholar]
- 17.Hirata H, et al. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- 18.Tiedemann HB, et al. Cell-based simulation of dynamic expression patterns in the presomitic mesoderm. J Theor Biol. 2007;248:120–129. doi: 10.1016/j.jtbi.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Masamizu Y, et al. Real-time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Audibert A, Weil D, Dautry F. In vivo kinetics of mRNA splicing and transport in mammalian cells. Mol Cell Biol. 2002;22:6706–6718. doi: 10.1128/MCB.22.19.6706-6718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giudicelli F, Özbudak EM, Wright GJ, Lewis J. Setting the tempo in development: An investigation of the zebrafish somite clock mechanism. PLoS Biol. 2007;5:e150. doi: 10.1371/journal.pbio.0050150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welch EM, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 25.Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. Lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 26.Zhang N, Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- 27.Serth K, Schuster-Gossler K, Cordes R, Gossler A. Transcriptional oscillation of lunatic fringe is essential for somitogenesis. Genes Dev. 2003;17:912–925. doi: 10.1101/gad.250603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shifley ET, et al. Oscillatory lunatic fringe activity is crucial for segmentation of the anterior but not posterior skeleton. Development. 2008;135:899–908. doi: 10.1242/dev.006742. [DOI] [PubMed] [Google Scholar]
- 29.Tange TØ, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 30.Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 31.Dequéant M-L, et al. A complex oscillating network of signaling genes underlies the mouse segmentation clock. Science. 2006;314:1595–1598. doi: 10.1126/science.1133141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.