Abstract
Despite some appealing similarities of protein synthesis across all phyla of life, the final phase of mRNA translation has yet to be captured. Here, we reveal the ancestral role and mechanistic principles of the newly identified twin-ATPase ABCE1 in ribosome recycling. We demonstrate that the unique iron-sulfur cluster domain and an ATP-dependent conformational switch of ABCE1 are essential both for ribosome binding and recycling. By direct (1∶1) interaction, the peptide release factor aRF1 is shown to synergistically promote ABCE1 function in posttermination ribosome recycling. Upon ATP binding, ABCE1 undergoes a conformational switch from an open to a closed ATP-occluded state, which drives ribosome dissociation as well as the disengagement of aRF1. ATP hydrolysis is not required for a single round of ribosome splitting but for ABCE1 release from the 30S subunit to reenter a new cycle. These results provide a mechanistic understanding of final phases in mRNA translation.
Keywords: ABC proteins, Archaea, ribonucleoprotein particle, X-ray structure
During mRNA translation, the ribosome and associated translation factors orchestrate the rapid and precise conversion of genetic information into proteins (1, 2). Whereas the basic principles of protein synthesis are conserved between Eubacteria, Archaea, and Eukarya, the underlying molecular mechanisms, regulatory principles, and cellular requirements differ markedly between the three domains of life. In addition to a core set of translation factors, noncanonical enzymes facilitate and regulate protein synthesis. Among them, several ATP-binding cassette (ABC) ATPases have been identified (3). ABC enzymes represent a large protein family with members in all phyla of life. Most of them function as energy-dependent transporters responsible for vectorial movement of solutes across cell membranes (4, 5). A distinct subclass includes soluble mechanochemical engines involved in chromosome segregation, DNA double-stranded break repair, ribosome biogenesis, and translation regulation (3, 6, 7). Despite their large functional diversity, ABC proteins appear to follow unifying structural and functional principles, according to which ATP binding and hydrolysis power mechanochemical work (8). Two ATP molecules are bound at the interface of two head-to-tail oriented nucleotide-binding domains (NBD) by the Walker A and B motifs of one NBD and the C-loop (ABC signature) of the opposite NBD (9, 10). Hence, ABC-type ATPases function as either homo- or heterodimers, operating in a tightly controlled cycle of engagement and disengagement (11, 12). During the catalytic cycle, ATP binding, hydrolysis, and subsequent release of Pi and ADP are proposed to coordinate a clamp-like motion of the two NBD motors, transmitting conformational rearrangements to associated domains or interacting cellular clients. Remarkably, all ABC-type ATPases linked to translation regulation are twin-ATPases with two NBDs encoded in one polypeptide chain. The twin-ABC ATPases can be phylogenetically grouped into subfamily E and F, but their molecular function remains largely elusive, so far.
ABCE1, the sole member of subfamily E, is universally found in Eukarya and Archaea, but not in Bacteria. It is essential in all eukaryotes examined to date (13–15) and one of the most conserved proteins in evolution. In addition to two head-to-tail oriented NBDs (16), ABCE1 contains an extra N-terminal region coordinating two nonequivalent, diamagnetic [4Fe-4S]2+ clusters with the Cx4Cx3Cx3CPxnCx2Cx2Cx3P consensus sequence (17). ABCE1 was originally discovered in vertebrates as RNase L inhibitor (18) and as host-factor HP68 required for HIV-1 capsid assembly (19). Apart from these nonessential functions, the evolutionarily conserved distribution of this ATPase points to a more fundamental role in cell homeostasis and indeed ABCE1 turned out to be involved in ribosome biogenesis and translation regulation (14, 20–23). ACBE1 was found to interact with a number of translation factors, eIF3 in Drosophila melanogaster, eIF2, 3, 5, and eRF1 in Saccharomyces cerevisiae, as well as eIF2, 5, and eRF1 in vertebrates (14, 21–25). Furthermore, a colocalization with ribosomal subunits has been demonstrated (14, 24). In yeast, ABCE1 depletion leads to a breakdown in protein synthesis accompanied by an accumulation of unprocessed rRNA and ribosomal subunits in the nucleus (7, 20, 22). The role of ABCE1 in eukaryotic translation has been linked to several stages during protein synthesis, including initiation and most recently termination and ribosome recycling (21, 23). However, the primeval function shared with its archaeal homologs is unknown. So far, the modus operandi of the twin-ATPase ABCE1 is largely unexplored due to the lack of a functional recombinant enzyme.
Here, we reveal the role of ABCE1 in ribosome recycling in Archaea. We further demonstrate that ABCE1 binding to and splitting of ribosomes proceeds in a closed ATP-occluded enzyme conformation and strictly requires the FeS cluster domain. Ribosome dissociation is promoted by a direct and stoichiometric interaction with the release factor aRF1. Altogether, ABCE1 is an evolutionarily conserved molecular machine, ribosome recycling and ribonucleoprotein (RNP) particle (dis)assembly.
Results
Structure and Function of the Twin-ATPase.
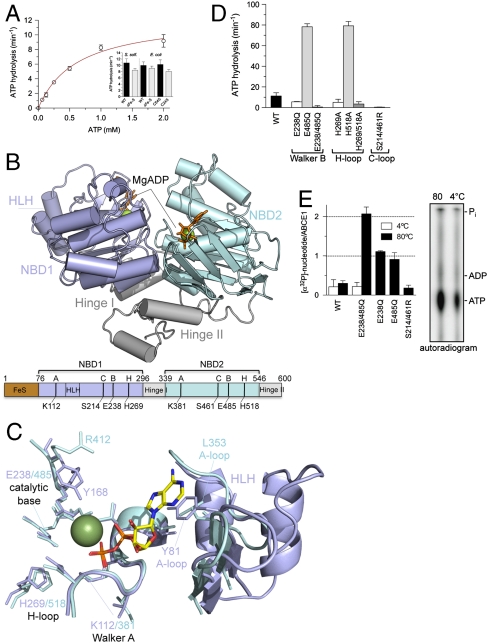
Based on our expression and purification strategy, which preserves the structural and functional integrity of the ATPase and otherwise unstable [4Fe-4S]2+ clusters, we uncovered the enzymatic fingerprint of ABCE1 for the first time. ABCE1 isolated from Sulfolobus solfataricus shows the highest ATP turnover at 80–90 °C, matching the growth conditions of this Crenarchaeon (Fig. S1). The wild-type enzyme (ABCE1WT) has a Michaelis–Menten constant KM(MgATP) of 0.7 mM and a turnover rate kcat of 12 ATP/ min (Fig. 1A and Table S1). N-terminal truncated ABCE1ΔFeS (lacking the entire FeS cluster domain) as well as ABCE1C24S (no assembled FeS cluster) (17) and ABCE1C54S (conversion of one diamagnetic [4Fe-4S]2+ into a paramagnetic [3Fe-4S]+ cluster) (17) showed ATP turnover rates and substrate affinities similar to the wild-type. These results demonstrate that neither the integrity of the prosthetic group nor the lack of the entire FeS cluster domain influence the basal ATPase cycle of ABCE1. In addition, the ATP turnover rate is independent of the protein concentration and no oligomerization was observed upon incubation with ADP, ATP, or adenosine 5′-(β,y-imido)triphosphate (AMPPNP) (Fig. S1). A Hill coefficient of 1.0 attests to a noncooperative process for the rate-limiting step in ATP hydrolysis, indicating that ABCE1 acts as monomer and that the twin NBDs undergo cycles of intramolecular engagement and disengagement to harness the chemical energy of ATP.
Fig. 1.
Structure and function of ABCE1. (A) ATPase activity of ABCE1 (5 μM) at 80 °C. Experiments were performed in triplicates. Data represented as mean ± SEM are fitted to the Michaelis–Menten equation. The ATP turnover rate kcat of various FeS cluster mutants (5 μM) is shown as insert. (B) X-ray structure of ABCE1ΔFeS-E238/485Q from S. solfataricus (side view). In the open ADP-bound state, ABCE1 shows a V-like architecture with NBD1 (pale blue) and NBD2 (cyan) preoriented via the hinge domain (gray). The two bound Mg-ADP molecules are shown as green spheres and orange sticks. Functional domains and critical residues in the Walker A and B motifs, C- and H-loop are illustrated below. (C) Overlay of the conserved motifs and catalytic residues of NBD1 (pale blue) and NBD2 (cyan) with regard to the bound Mg-ADP (ADP as sticks, Mg2+ as green sphere). Notably, the catalytic Glu 238 and 485 are exchanged to Gln in the X-ray structure. (D) ATP turnover rate kcat of ABCE1 mutants (5 μM) with substitutions in conserved motifs (Walker B motifs, H- and C-loop). All experiments were performed in triplicates at 80 °C. (E) Stoichiometry of occluded nucleotides. ABCE1 mutants (5 μM) were incubated with 500 μM ATP (traced with [α-32P]-ATP) at 4 °C (white) or 80 °C (black bars). Free and bound nucleotides were separated by spin-down gel filtration and quantified by Cerenkov counting. Data from two independent experiments performed in triplicates are represented as mean ± SEM. The nucleotides occluded in ABCE1E238/485Q were identified by thin layer chromatography (Right).
To correlate functional and structural data, we crystallized ABCE1ΔFeS-E238/485Q from S. solfataricus and determined its X-ray structure at 2.0 Å resolution [Rwork/Rfree 0.186/0.244, Protein Data Bank (PDB) ID code 3OZX; Table S2]. In the Mg-ADP-bound state, ABCE1 displays an overall V-shape architecture, thereafter referred to as the open conformation (Fig. 1B). The structure resembles orthologs from Pyrococcus furiosus and Pyrococcus abyssi, documented by an overall rms deviation of 1.5 Å (16, 26). Two head-to-tail oriented nucleotide-binding domains (NBD1 and NBD2) are positioned by a hinge region, which is built up from the linker between both NBDs and the C-terminal stretch. This structural framework supports a clamp-like motion of both NBDs, which cycle between the open (crystallized) and a closed state upon ATP binding and hydrolysis. The hinge region determines the preorientation and the magnitude of displacement of the two NBDs during the catalytic cycle. The formation of the ATP-bound closed state may result in a large movement of the FeS domain (26).
Despite the high overall similarities of both NBDs, a superposition illustrates an asymmetry between the two ATP-binding pockets (site I and II), which has not been reported in any ABC protein so far, including the Pyrococcus ABCE1 (Fig. 1C). Strikingly, the phenyl group of Y168 adjacent to the Q-loop in NBD1 points to active site I and binds via solvent molecules to the α-, β- and, potentially, to the γ-phosphate, whereas R412 in the equivalent position of NBD2 is flipped out and does not contact the nucleotide in active site II. Even if its side chain is flipped toward the nucleotide in an ATP-bound closed state, the arginyl and tyrosinyl residue would influence ATP hydrolysis differently. Furthermore, whereas a canonical aromatic (A)-loop is found in NBD1 (Y81), forming π-π-stacking interactions toward the adenine base, the A-loop in NBD2 (L353) is degenerate. In NBD1, the region associated to the A-loop from the backside is enlarged by a prolonged loop between F127 and V134 and a helix-loop-helix insertion compared to NBD2. These differences are conserved among orthologs of Crenarchaea and Eukarya, implicating a more rigid active site I. The weak interaction of the nucleotide base in the ATP-binding sites explains why the enzyme is rather promiscuous for the nucleoside triphosphates (Figs. S1 and S2), which is typical for ABC proteins (5).
To dissect the function of the two active sites, we analyzed a set of mutants including the catalytic glutamate next to the Walker B motif (E238Q, E485Q) and the H-loop (H269A, H518A). As expected, the double mutants are ATPase inactive (Fig. 1D). ABCE1 with mutations in NBD1 (E238Q and H269A) showed only 30–50% activity compared to wild-type ABCE1 as a consequence of the inactivation of site I. Strikingly, mutations in NBD2 (E485Q and H518A) are hyperactive with 10-fold higher ATPase activity as compared to wild-type. Hyperactivity cannot be explained by an altered substrate affinity, cooperativity, or oligomeric state (Table S1 and Fig. S1). This unusual behavior has not been observed in any other ABC system and underlines the functional and structural asymmetry of the two ATP-binding sites.
Importantly, if ATP hydrolysis is blocked by mutation of the two catalytic glutamates (ABCE1E238/485Q), a stoichiometric occlusion of two ATP molecules is observed (Fig. 1E). Correspondingly, only one ATP is bound in each single Walker B mutant. ATP occlusion strictly requires ATPase permissive temperatures at which a conformational switch into a closed state can occur. In the closed state, the conserved serines 214 and 461 in the C-loop (LSGGQ) coordinate the γ-phosphates of ATP in the opposite NBD. As shown in Fig. 1 D and E, the engagement mutation (S214/461R) prevents ATP occlusion and hydrolysis. In conclusion, ABCE1 undergoes a cycle of closure (ATP occlusion) and opening upon ATP binding and hydrolysis.
ABCE1 Binds to 30S Ribosomes in an ATP-Occluded State.
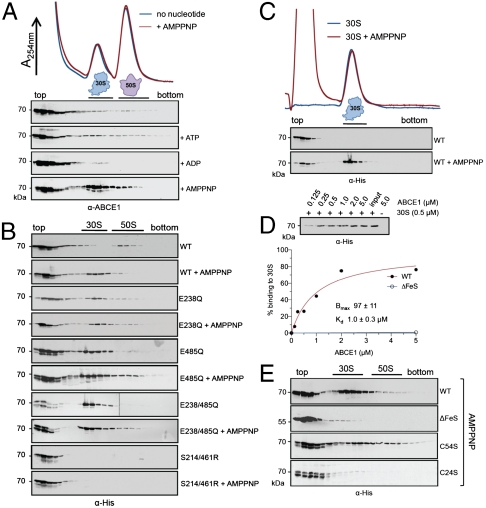
We next examined whether the different conformational states of ABCE1 interact with components of the translation machinery. Whole cell extracts (WCE) from S. solfataricus were fractioned by sucrose density gradients (SDG). Notably, Sulfolobus 70S ribosomes are very labile and detectable only after cross-linking (27). ABCE1 was mainly found on the top of the gradient and only in a minor degree with the 30S and 50S subunits (Fig. 2A). Neither addition of ATP nor ADP could enhance the fraction of ABCE1 bound to ribosomal subunits. However, its accumulation at the 30S particle was strongly promoted when arresting ABCE1 in its ATP-occluded state by addition of nonhydrolyzable AMPPNP, whereas no binding to the 50S subunit was observed above background.
Fig. 2.
The ATP-occluded state and the FeS cluster domain of ABCE1 are essential for stable ribosome association. (A) Ribosome association of ABCE1 was analyzed by SDG in 100 μL of WCE (15 mg/mL) from S. solfataricus incubated in the presence of different nucleotides at 73 °C for 4 min. Fractions (0.5 mL) were analyzed by SDS-PAGE and immunoblotting using an anti-ABCE1 antibody. (B) Isolated ABCE1 mutants (0.4 μM) were incubated with 100 μL of S. solfataricus WCE (15 mg/mL) with and without 5 mM of AMPPNP and analyzed as described above using an anti-His antibody. (C) Binding of ABCE1WT (1 μM) to isolated 30S particles (1 μM) assayed by SDG analysis in the presence and absence of AMPPNP (5 mM) at 73 °C for 4 min. (D) Ribosome pelleting assays of ABCE1WT and isolated 30S subunits (0.5 μM) reveal a stoichiometric (1∶1) binding in the presence of AMPPNP. The amount of ABCE1WT expected for 100% binding (0.5 μM) is given as input. Data were analyzed by quantitative immunoblotting and fitted according to a one-site binding isotherm. (E) SDG analysis of FeS cluster mutants. Purified ABCE1WT, ABCE1ΔFeS, ABCE1C54S, or ABCE1C24S (0.2 μM of each) was added to 100 μL S. solfataricus WCE (15 mg/mL) and incubated with 5 mM of AMPPNP at 73 °C for 4 min. Fractions were probed with an anti-His antibody.
To correlate the conformational changes and ATP-binding/hydrolysis events with ribosome association, ABCE1 mutants were incubated with S. solfataricus lysates. Ribosome profiles were probed with an anti-His antibody to detect selectively the recombinant enzyme. Association of recombinant ABCE1WT with 30S particles was strongly enhanced upon addition of AMPPNP as observed for endogenous ABCE1. Remarkably, nonhydrolytic ABCE1E238/485Q interacts with the 30S subunit already without addition of AMPPNP as it can occlude two ATPs from the lysate (Fig. 2B and Fig. S3). Likewise, both single Walker B mutants, ABCE1E238Q (50% activity) and ABCE1E485Q (hyperactive), are attached to the 30S subunit without AMPPNP. These data suggest that a conformational switch in ABCE1 plays a key role in 30S association. Indeed, the engagement mutant ABCE1S214/461R, which cannot switch into the closed state and occlude ATP, does not associate with the 30S subunit under any condition assayed (Fig. 2B). Thus, the occlusion of one ATP molecule (independent whether positioned in active site I or II) appears to be necessary and sufficient to adopt the closed state and to promote the formation of a stable ABCE1/30S complex.
The FeS Cluster Domain Is Essential for Ribosome Interaction.
We next explored whether 30S binding of ABCE1 is direct or indirect by sedimentation assays using isolated, high-salt washed ribosomal subunits. Similar to the previous experiments in whole cell extracts, ABCE1 binds to isolated 30S particles in an AMPPNP dependent manner (Fig. 2C), whereas no specific interaction with 50S was observed. Importantly, a direct (1∶1) binding of ABCE1 to the 30S subunit with Kd of 1.0 μM was determined (Fig. 2D). Strikingly, ABCE1ΔFeS neither cofractionates with 30S in WCE (Fig. 2E) nor binds to isolated 30S particles in the presence of AMPPNP (Fig. 2D). Under the same conditions ABCE1C54S (conversion of one diamagnetic [4Fe-4S]2+ into a defined paramagnetic [3Fe-4S]+ cluster) (17), can bind to 30S, whereas a FeS cluster defective mutant ABCE1C24S does not interact with 30S (Fig. 2E). Because all FeS cluster mutants exhibit a similar ATPase activity (Fig. 1A and Table S1), the observed binding behavior suggests a pronounced contact area between the FeS cluster domain and the 30S subunit provided by the conformation switch of ABCE1. The electronic state of the FeS clusters is not changed upon 30S binding as revealed by electron spin resonance. These results point to a structural, rather than a redox-catalytic role of the FeS cluster domain.
ABCE1 Acts Downstream of Translation Initiation in Archaea.
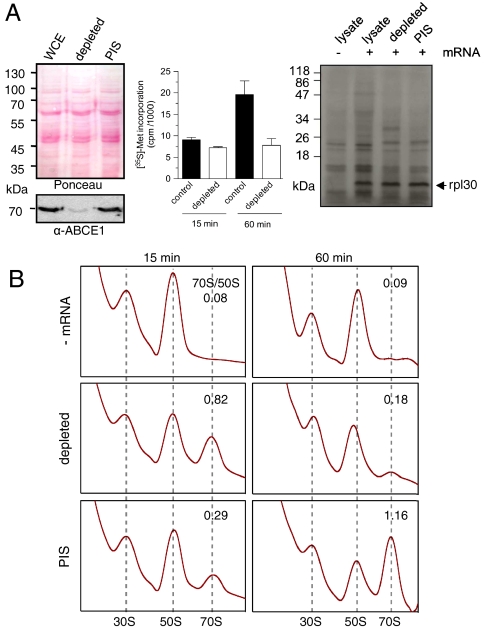
We next addressed whether the ABCE1/30S complex is built up in the initiation phase or in the final stages of translation. Because an archaeal translation system reconstituted from purified components has not been established, we used a cell-free translation system employing in vitro transcribed mRNAs (28). The mRNA-104 contains a canonical Shine–Delgarno (SD) sequence in the 5′ untranslated region (UTR) and translation initiation follows a bacterial-like SD-anti-SD base pairing mechanism. Surprisingly, after immunodepletion of ABCE1 (> 95%, Fig. 3A), lysates are still translation active based on the total translation capacity or defined mRNAs as reporters. Notably, a similar behavior was observed for the leaderless mRNA-aIF1, lacking the entire 5′UTR that initiates on archaeal ribosomes by a still poorly understood mechanism.
Fig. 3.
ABCE1 acts downstream of translation initiation. (A) Immunodepletion of ABCE1 from S. solfataricus WCE (Left). Immunodepletion by preimmune serum serves as control (PIS). The effect of ABCE1 depletion on protein synthesis was assayed by measuring the total capacity of protein synthesis via [35S]-methionine incorporation in TCA precipitates (Center) and by in vitro translation in lysates programmed for mRNA-104 translation coding for rpl30 (Right). (B) Effect of ABCE1 depletion on 70S formation. After in vitro translation of mRNA-104 for 15 or 60 min at 73 °C, reactions were formaldehyde cross-linked and analyzed by SDG centrifugation. The peak areas (A254 nm) corresponding to 70S and 50S were calculated.
After 15-min incubation, the ABCE1-depleted lysate displayed a consistently higher 70S/50S ratio than the preimmune serum depleted controls (PIS), indicating that ribosome recycling was slowed down in dearth of ABCE1 (Fig. 3B). However, after 60 min, only a small amount of 70S ribosomes was detected in the depleted lysates, whereas the controls showed a large peak of 70S ribosomes. These results suggest that an adequate amount of ABCE1 is required both for dissociating 70S and enabling the released subunits to enter a new translation cycle.
A Conformational ATP-Switch of ABCE1 Powers Ribosome Splitting.
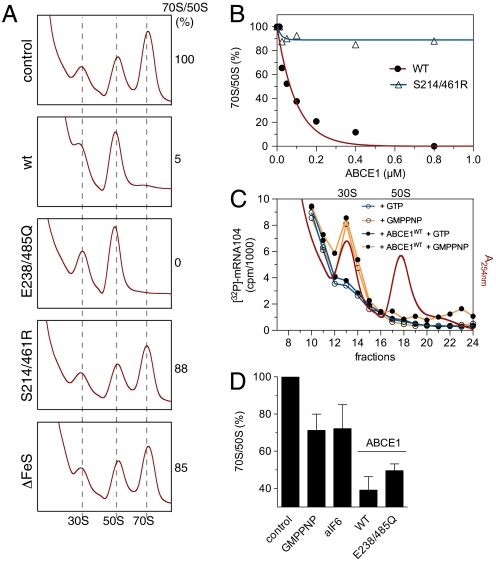
To critically examine the previous hypothesis, we investigated the effects of ABCE1 on 70S formation. When either ABCE1WT or ABCE1E238/485Q was added to lysates programmed for translation, we observed a dose-dependent decrease in the 70S/50S ratio (Fig. 4 A and B). This effect is ABCE1 specific because mutants incompetent in stable 30S interaction (ABCE1ΔFeS or ABCE1S214/461R) do not promote 70S breakdown. Hence, the 70S/50S ratio in the translationally active lysate inversely correlates with the amount of ABCE1, with more 70S at lower ABCE1 concentration at least during the first rounds of translation.
Fig. 4.
A conformational switch of ABCE1 powers 70S breakdown. (A) ABCE1 mutants (0.8 μM of each) were added to lysates programmed for translation at 73 °C for 30 min and the amount of 70S particles formed were examined by SDG fractionation. The peak area (A254 nm) of 50S and 70S ribosomes was calculated. The 70S/50S ratio of the control reaction was set to 100%. (B) 70S breakdown at different concentrations of ABCE1. Reactions were performed and analyzed as described in (A). The initial 70S/50S ratio was set to 100%. (C) Preinitiation complex formation of lysates programmed with [32P]-labeled mRNA-104 was visualized by Cerenkov counting of the corresponding SDG fractions (10–20% sucrose). ABCE1WT (0.8 μM) was added to these lysates in combination with different nucleotides (2 mM of each). (D) Recombinant ABCE1, aIF6 or GMPPNP were added to lysates programmed for in vitro translation. After 2-min incubation, samples were cross-linked on ice and analyzed by SDG fractionation. Addition of ABCE1WT but also the ATPase inactive mutant ABCE1E238/485Q leads to a significantly larger 70S breakdown than GMPPNP (2 mM) or aIF6 (5 μM). The initial 70S/50S ratio without additional factors (control) was set to 100%.
To exclude that the above results were caused by a defect in the formation of preinitiation complexes, lysates programmed for translation were incubated with an excess of ABCE1WT in the presence of GMPPNP, which blocks translation in the initiation phase (29). The formation of preinitiation complexes was analyzed by monitoring mRNA retained by the 30S subunit (Fig. 4C). Under these conditions, ABCE1WT neither arrests the translation machinery nor prevents the formation of the initiation complex. In a second approach, we pulsed the in vitro translation reaction for 2 min with either GMPPNP or aIF6 to prevent 50S subunits to reenter the translation cycle after termination (Fig. 4D). Addition of either ABCE1WT or ABCE1E238/485Q leads to a significantly higher breakdown of 70S ribosomes as the controls (GMPPNP or aIF6). These results suggest that ABCE1 functions in ribosome recycling by active splitting of 70S ribosomes.
ABCE1 Acts as a Ribosome Recycling Factor Cooperatively with aRF1.
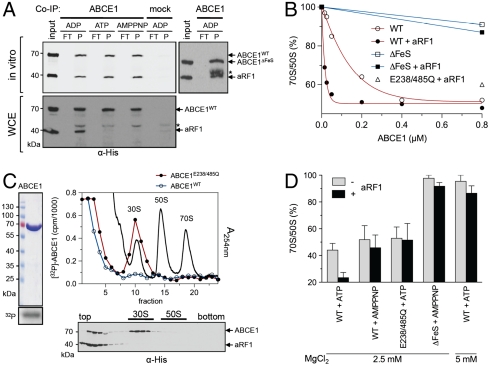
By coimmunoprecipitation using ABCE1 specific antibodies, we show that ABCE1 interacts directly and stoichiometrically (1∶1) with aRF1 (Fig. 5A). Importantly, this interaction is independent of the FeS cluster domain and the catalytic state of ABCE1. In contrast, in WCE, ABCE1 binds to aRF1 only in its ADP-bound open conformation. To address the physiological relevance of this interaction, we followed the 70S dissociation in a pulse assay as introduced above. ABCE1 induces 70S breakdown in a dose-dependent manner (Fig. 5B). However, in the presence of aRF1, 70S dissociation is promoted. This synergistic effect is lost with ABCE1E238/485Q, suggesting that ATP hydrolysis is required for a proper cooperation between ABCE1 and aRF1 and multiple rounds of ribosome recycling. Significantly, aRF1 alone cannot split 70S ribosomes (Fig. S4). We noticed that not all 70S particles are dissociated, likely because ABCE1 encounters translating 70S ribosomes at different phases. ABCE1 mutants lacking the FeS cluster domain are unable to promote ribosome splitting, even in combination with aRF1.
Fig. 5.
ABCE1 functions synergistically with aRF1 in posttermination ribosome disassembly. (A) Purified ABCE1 and aRF1 (2 μM of each) were incubated in the presence of different nucleotides (5 mM) at 73 °C for 10 min. Coimmunoprecipitation by a polyclonal anti-ABCE1 antibody was performed in Co-IP buffer (in vitro) or in 100 μL of WCE (15 mg/mL). Preimmune serum coupled beads served as a control (mock). The asterisk marks the cross-reaction of the eluted antibody heavy chain. (B) Lysates programmed for translation were pulsed for 2 min with increasing concentrations of ABCE1 alone or in combination with aRF1 (1 μM). After the pulse, samples were cross-linked on ice. Ribosome profiles were analyzed as in Fig. 4B. The initial 70S/50S ratio (no additional ABCE1) was set to 100%. (C) Localization of [32P]-labeled ABCE1WT or ABCE1E238/485Q (0.8 μM of each, left panel) in lysates programmed for translation in the presence of aRF1 (1 μM). The ribosome profile at A254 nm in the presence of ABCE1E238/485Q is shown. After ribosome splitting by ABCE1WT and aRF1 (1 μM of each) in the presence of AMPPNP (Lower), the ribosome association of ABCE1 and aRF1 was analyzed by SDG fractionation and immunoblotting by an anti-His antibody. (D) In vitro 70S ribosome breakdown. Isolated 70S ribosomes from T. celer (1 μM) were incubated with ABCE1 (1 μM) in the presence and absence of aRF1 (1 μM) for 4 min at 73 °C and different nucleotides (2 mM of each). Ribosome reassociation was prevented by addition of aIF6 (5 μM). Data were analyzed by SDG fractionation as described in Fig. 4B. The 70S/50S ratio in the absence of added ABCE1 served as control (100%). Experiments were performed in duplicates.
To monitor the localization of ABCE1 after 70S disassembly, [32P]-labeled ABCE1 was added for improved sensitivity and quantification. In agreement with our previous data, ABCE1WT was only found on top of the gradient, whereas ABCE1E238/485Q was stably bound to 30S (∼30%), but not found on 50S or 70S particles (Fig. 5C). Strikingly, either ABCE1WT or ABCE1E238/485Q lead to a fast 70S splitting (Fig. 5B). However, only ABCE1E238/485Q in its ATP-occluded closed state remains stably bound to the 30S subunit after ribosome dissociation. Based on the fact that ABCE1E238/485Q can split ribosomes, we concluded that ATP binding and the conformational switch into the closed state but not ATP hydrolysis powers ribosome dissociation. Notably, whereas ABCE1E238/485Q or ABCE1WT (in the presence of AMPPNP) are arrested at the 30S subunit after 70S splitting, aRF1 has been released (Fig. 5C, Lower). This suggests that the conformational switch of ABCE1 not only powers 70S splitting but also leads to the dissociation of aRF1 from the 30S/ABCE1 complex.
Because Sulfolobus 70S ribosomes are intrinsically unstable even at very high MgCl2 concentrations (27), we searched for an appropriate model to reconstitute ribosome splitting by ABCE1 entirely from purified components. We found that 70S ribosomes from the hyperthermophilic Euryarchaeon Thermococcus celer are stable at temperatures around 70–80 °C and therefore fulfill this crucial requirement. Confirming our data above, we observed a direct (1∶1) and nucleotide-dependent binding of ABCE1 to T. celer 30S particles (Fig. S5). Strikingly, ABCE1 specifically dissociates isolated Thermococcus 70S ribosomes within a short 4-min pulse (Fig. 5D). Importantly, ATP hydrolysis is not required for a single round of ribosome splitting, because ABCE1WT in the presence of AMPPNP or ABCE1E238/485Q is able to dissociate 70S ribosomes. Nevertheless, ATP hydrolysis of ABCE1 significantly enhanced 70S disassembly. The behavior of ABCE1WT and ABCE1E238/485Q is clearly different with respect to the synergy with aRF1. The mutant does not cooperate with aRF1, so ATP hydrolysis is important for 30S release and reentering a new cycle of ribosome splitting. No 70S breakdown was observed for ABCE1ΔFeS or aRF1 alone (Fig. S4). In conclusion, ribosome recycling depends on an allosteric coupling between the FeS cluster domain and a conformational ATP-switch of ABCE1.
Discussion
Although the basic steps of protein synthesis are conserved between all three domains of life, marked differences exist. Archaea combine features from bacterial as well as eukaryotic mRNA translation including very specific properties. In our study, the final phase of archaeal translation and key mechanistic steps of posttermination ribosome recycling powered by ABCE1 have been disclosed. Several lines of evidence argue against a primary role of ABCE1 in translation initiation as shown for its eukaryotic homologs: (i) the previously described interaction partners eIF3 and eIF5 (14, 22, 24, 25) do not exist in Archaea; (ii) ABCE1 has no effect on the formation of the preinitiation complex (Fig. 4C); and (iii) short-term mRNA translation was not affected in ABCE1-depleted lysates (Fig. 3A). In contrast, even more 70S ribosomes are built up within the short-term mRNA translation after ABCE1 depletion (Fig. 3B). Indeed, our in vitro assays reconstituted from isolated components reveal that ABCE1 is essential for ribosome recycling.
ABCE1 function is promoted by the synergistic interaction with the release factor aRF1, thus coupling translation termination to ribosome recycling. Interestingly, the class II release factor eRF3 and homologs of bacterial recycling factors do not exist in Archaea. Furthermore, an energy-independent process of ribosome recycling, as found in Eukarya (30), can be excluded in Archaea, because homologs of the initiation factor eIF3 are absent. Thus, ribosome recycling in Archaea resembles a unique and minimalistic version of the eukaryotic process. However, it is still unknown which regulatory events may exist to suppress futile cycles of ribosome dissociation by ABCE1. In Eukarya, eRF3 seems to have a control function by preventing premature association of ABCE1 with posttermination complexes (21). Additionally, translation termination by eRF1 depends on the cooperative action of eRF3, in which GTP hydrolysis of eRF3 triggers the peptide release by eRF1 (31). It has most recently been shown that archaeal elongation factor 1α (aEF1α) may function as a homolog of eRF3 triggering polypeptide release by aRF1 (32). Hence, it will be interesting to analyze the exact conformational state at which ABCE1 encounters the 70S ribosome and how aEF1α regulates posttermination ribosome recycling driven by ABCE1.
Our data not only shed light on the process of ribosome recycling in Archaea, but also reveal fundamental principles of the twin-ATPase machine. We show that a conformational switch of ABCE1 is required for ribosome splitting. However, the two catalytic sites fulfill distinct roles. Based on our functional and structural data, we propose that NBD2 serves an important regulatory function by sensing different cellular clients; e.g., aRF1 positioned at the postterminated ribosome. In line with this, the interaction with eRF1 has been mapped genetically to the NBD2 of ABCE1 (23). Apart from the conformational ATP-switch, the FeS cluster domain of ABCE1 is essential for ribosome recycling. Interestingly, the FeS cluster domain of ABCE1 displays a ferrodoxin-like fold. This superfamily includes ferrodoxin from Desulfovibrio vulgaris (33) and the poly(A) binding protein (34), which bind RNA. The FeS cluster domain of ABCE1 exposes highly conserved, positively charged residues. Thus, we assume that this domain plays an important role in RNA binding. This provides a mechanistic link between two key cellular processes, FeS cluster biogenesis and ribosome recycling. In addition, the FeS clusters of ABCE1 could have a sensory function to regulate translation, for example under oxidative stress.
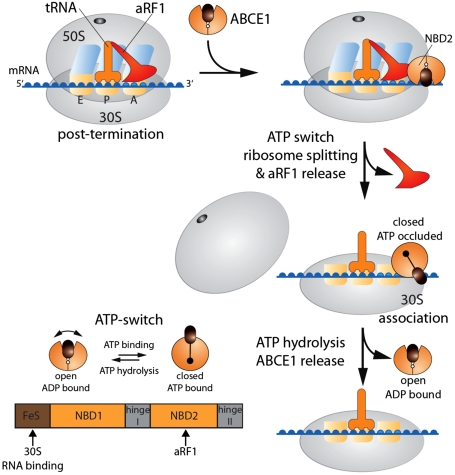
Based on our data, we derive the following model for ribosome recycling (Fig. 6). After termination and release of the polypeptide chain catalyzed by aRF1 and presumably aEF1α (32), ABCE1 docks to stop-codon programmed ribosomes via aRF1 positioned at the A-site. ATP binding to ABCE1 induces a conformational change into its closed state, which reorganizes the FeS cluster domain to drive ribosome splitting and release of aRF1. Because the ATPase inactive mutant ABCE1E238/485Q is able to dissociate ribosomes, we propose that the ATP-switch into the closed conformation and the movement of the FeS domain drives ribosome dissociation as well as the disengagement of aRF1 from the ribosome. After 70S splitting, ABCE1 can be arrested in its ATP-occluded state at the 30S subunit. ATP hydrolysis is finally required to release ABCE1 (and probably tRNA and mRNA by the aid of several other factors still to be identified) from the 30S particle to initiate a new translation cycle. The question remains why ABCE1 has been linked to so many different cellular pathways in Eukaryota. The presented data raise the possibility that ABCE1 is a RNP remodeling factor using the ATP-switch as power stroke for the (dis)assembly of macromolecular RNP complexes, such as in translation control, ribosome biogenesis, and lentiviral RNA/gag capsid assembly.
Fig. 6.
Model of ribosome recycling in Archaea. After termination, ABCE1 is recruited to the 70S ribosome by binding to aRF1. ATP and ribosome binding induces a conformational switch into a closed state of ABCE1, which powers ribosome dissociation and release of aRF1. Finally, ATP hydrolysis triggers the dissociation of ABCE1 from the 30S subunit. In addition, the conformational changes that drive 70S ribosome dissociation (ATP-switch) and putative interaction sites with aRF1, 30S and RNA are illustrated.
Materials and Methods
Details of materials, cloning, expression, protein purification, and activity assays are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Jennifer Hüllein and Katja Rudolph for technical assistance and Ulrike Demmer (MPI for Biophysics, Frankfurt/M.) for help with protein crystallization. We thank Christian Schölz and Drs. Rupert Abele, David Parcej, and Roger Draheim for stimulating discussions and critical reading of the manuscript. We thank Dr. Harald Huber (University of Regensburg) for fermenting T. celer. We kindly acknowledge Dr. Thomas Prisner and Reza Dastvan for the EPR measurements (Physical Chemistry, Goethe University Frankfurt). This work was supported by the Otto Stern Graduate School Frankfurt (D.B.), the Fonds of the Chemical Industry (FCI), and the Cluster of Excellence Frankfurt—Macromolecular Complexes (R.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3OZX).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015953108/-/DCSupplemental.
References
- 1.Dever TE. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- 2.Ramakrishnan V. Ribosome structure and the mechanism of translation. Cell. 2002;108:557–572. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 3.Chakraburtty K. Translational regulation by ABC systems. Res Microbiol. 2001;152:391–399. doi: 10.1016/s0923-2508(01)01210-4. [DOI] [PubMed] [Google Scholar]
- 4.Rees DC, Johnson E, Lewinson O. ABC transporters: The power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitt L, Tampé R. Structure and mechanism of ABC transporters. Curr Opin Struct Biol. 2002;12:754–760. doi: 10.1016/s0959-440x(02)00399-8. [DOI] [PubMed] [Google Scholar]
- 6.Hopfner KP, Putnam CD, Tainer JA. DNA double-strand break repair from head to tail. Curr Opin Struct Biol. 2002;12:115–122. doi: 10.1016/s0959-440x(02)00297-x. [DOI] [PubMed] [Google Scholar]
- 7.Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis. Mol Cell Biol. 2005;25:9859–9873. doi: 10.1128/MCB.25.22.9859-9873.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: Unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 9.Hopfner KP, et al. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 10.Smith PC, et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Janas E, et al. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J Biol Chem. 2003;278:26862–26869. doi: 10.1074/jbc.M301227200. [DOI] [PubMed] [Google Scholar]
- 13.Braz AS, Finnegan J, Waterhouse P, Margis R. A plant orthologue of RNase L inhibitor (RLI) is induced in plants showing RNA interference. J Mol Evol. 2004;59:20–30. doi: 10.1007/s00239-004-2600-4. [DOI] [PubMed] [Google Scholar]
- 14.Dong J, et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Z, Fang LL, Johnsen R, Baillie DL. ATP-binding cassette protein E is involved in gene transcription and translation in Caenorhabditis elegans. Biochem Biophys Res Commun. 2004;323:104–111. doi: 10.1016/j.bbrc.2004.08.068. [DOI] [PubMed] [Google Scholar]
- 16.Karcher A, Buttner K, Martens B, Jansen RP, Hopfner KP. X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure. 2005;13:649–659. doi: 10.1016/j.str.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Barthelme D, et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J Biol Chem. 2007;282:14598–14607. doi: 10.1074/jbc.M700825200. [DOI] [PubMed] [Google Scholar]
- 18.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2–5 Å pathway. J Biol Chem. 1995;270:13308–13317. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 19.Zimmerman C, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 20.Yarunin A, et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–588. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisarev AV, et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kispal G, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 2005;24:589–598. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40S ribosome in an ATP-dependent manner and is required for translation initiation. J Biol Chem. 2007;282:14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- 25.Chen ZQ, et al. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 26.Karcher A, Schele A, Hopfner KP. X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem. 2008;283:7962–7971. doi: 10.1074/jbc.M707347200. [DOI] [PubMed] [Google Scholar]
- 27.Londei P, Altamura S, Cammarano P, Petrucci L. Differential features of ribosomes and of poly(U)-programmed cell-free systems derived from sulphur-dependent archaebacterial species. Eur J Biochem. 1986;157:455–462. doi: 10.1111/j.1432-1033.1986.tb09689.x. [DOI] [PubMed] [Google Scholar]
- 28.Condo I, Ciammaruconi A, Benelli D, Ruggero D, Londei P. Cis-acting signals controlling translational initiation in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 1999;34:377–384. doi: 10.1046/j.1365-2958.1999.01615.x. [DOI] [PubMed] [Google Scholar]
- 29.Benelli D, Maone E, Londei P. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol Microbiol. 2003;50:635–643. doi: 10.1046/j.1365-2958.2003.03721.x. [DOI] [PubMed] [Google Scholar]
- 30.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- 32.Saito K, et al. Omnipotent role of archaeal elongation factor 1 alpha (EF1alpha in translational elongation and termination, and quality control of protein synthesis. Proc Natl Acad Sci USA. 2010;107:19242–19247. doi: 10.1073/pnas.1009599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arendsen AF, Schalk JS, van Dongen WM, Hagen WR. Characterization of a ferredoxin from Desulfovibrio vulgaris (Hildenborough) that interacts with RNA. Eur J Biochem. 1995;231:352–357. doi: 10.1111/j.1432-1033.1995.tb20707.x. [DOI] [PubMed] [Google Scholar]
- 34.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.