Abstract
Poly(ADP-ribose) polymerase (PARP) inhibitors are strikingly toxic to cells with defects in homologous recombination (HR). The mechanistic basis for these findings is incompletely understood. Here, we show that PARP inhibitor treatment induces phosphorylation of DNA-dependent protein kinase substrates and stimulates error-prone nonhomologous end joining (NHEJ) selectively in HR-deficient cells. Notably, inhibiting DNA-dependent protein kinase activity reverses the genomic instability previously reported in these cells after PARP inhibition. Moreover, disabling NHEJ by using genetic or pharmacologic approaches rescues the lethality of PARP inhibition or down-regulation in cell lines lacking BRCA2, BRCA1, or ATM. Collectively, our results not only implicate PARP1 catalytic activity in the regulation of NHEJ in HR-deficient cells, but also indicate that deregulated NHEJ plays a major role in generating the genomic instability and cytotoxicity in HR-deficient cells treated with PARP inhibitors.
Keywords: chemotherapy, DNA repair, synthetic lethality, double-strand break repair
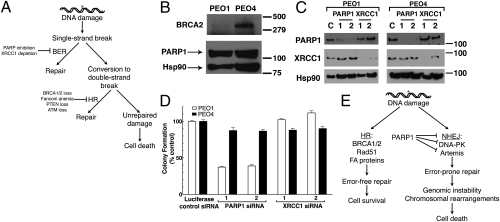
Poly(ADP-ribose) polymerase 1 (PARP1) is an abundant nuclear enzyme that synthesizes poly(ADP-ribose) polymer when activated by DNA nicks or breaks. Activation of PARP1 has important effects on a variety of cellular processes, including base excision repair (BER), transcription, and cellular bioenergetics (1). The role of PARP1 in the DNA damage response sparked interest in the development of PARP inhibitors as potential chemosensitizers for the treatment of cancer (1, 2). The more recent observation that PARP inhibition is particularly lethal to cells deficient in homologous recombination (HR) proteins (3–8) has generated additional excitement in the cancer chemotherapy community. The current explanation for this hypersensitivity focuses on a mechanism (Fig. 1A) in which loss of PARP1 activity is thought to result in accumulation of DNA single-strand breaks (SSBs), which are subsequently converted to DNA double-strand breaks (DSBs) by the cellular replication and/or transcription machinery. These DSBs, which are repaired by HR in BRCA-positive cells, are presumed to accumulate in BRCA1- or BRCA2-deficient cells, leading to subsequent cell death. Heightened sensitivity to PARP inhibition has also been observed in cells with other genetic lesions that affect HR, including phosphatase and tensin homolog (PTEN) deficiency (5), ataxia telangiectasia mutated (ATM) deficiency (7, 8), and Aurora A overexpression (6).
Fig. 1.
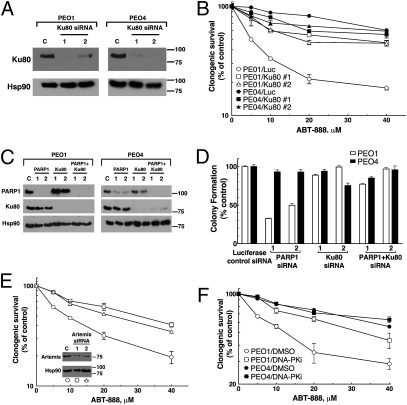
PARP inhibitor synthetic lethality is independent of XRCC1 and BER. (A) Current model explaining synthetic lethality of PARP inhibition and HR deficiency. PARP inhibition is thought to induce accumulation of SSBs, which are converted to DSBs by collisions with replication machinery. The inability of HR-deficient cells to adequately repair DSBs results in genomic instability and eventual cell death. (B) Western blotting analysis of cell lysates from PEO1 and PEO4 cells. Blots were probed for BRCA2, PARP1, and Hsp90 (loading control). (C) Western blots demonstrating siRNA-mediated knockdown with luciferase (control), PARP1, or XRCC1 siRNA in PEO1 or PEO4 cells. (D) Clonogenic viability of cells from C after siRNA knockdown. After knockdown, cells were plated onto triplicate plates and allowed to form colonies. All results are reported as means of triplicate plates ± SEM and are representative of three independent experiments. (E) An alternate model of PARP inhibitor synthetic lethality centering on error-prone NHEJ. In this model, PARP1 catalytic activity regulates NHEJ activity, preventing NHEJ components from binding to sites of DNA damage or DNA ends. In the absence of HR and PARP activity, deregulated NHEJ aberrantly processes DNA and introduces chromosomal instability, leading to cell death.
Although the preceding studies underscore the importance of PARP1 and HR in maintaining genomic stability, they do not address the role of nonhomologous end joining (NHEJ), an alternate DSB repair modality that directly joins broken ends of DNA with little or no regard for sequence homology (9). NHEJ is initiated when free DNA ends are bound by Ku70 and Ku80, which recruit the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs). The resulting complex, known as the DNA-dependent protein kinase (DNA-PK) complex, phosphorylates downstream targets leading to activation of the DNA damage response and initiation of NHEJ. Recent work by two groups has demonstrated that abortive/error-prone NHEJ damages DNA in the absence of HR (10, 11), establishing a model in which NHEJ and HR components compete for DNA ends after DNA damage.
Previous studies have also provided evidence for interplay between NHEJ components and PARP1. In particular, PARP1 interacts with the Ku proteins in vitro and in vivo (12). Moreover, Ku70, Ku80, and DNA-PKcs are capable of binding poly(ADP-ribose) polymer (13–15). In addition, PARP1 and Ku80 compete for DNA ends in vitro (16). Finally, the genetic ablation of KU70 or LIGIV restores the survival of PARP1-deficient cells exposed to agents inducing DSBs (17, 18). These observations raise the question of whether NHEJ is involved in the genomic instability and cytotoxicity observed in HR-deficient cells treated with PARP inhibitors.
Here we demonstrate the critical role of NHEJ in the hypersensitivity of HR-deficient cells to PARP inhibitors. In particular, we show that PARP inhibition preferentially enhances error-prone NHEJ activity in HR-deficient cells, as measured by phosphorylation of DNA-PK substrates and an in vivo reporter assay. Disabling NHEJ reverses the genomic instability induced by PARP inhibitors and rescues HR-deficient cells from the lethality of PARP inhibition or PARP1 knockdown. These results not only highlight the crucial balance between HR and NHEJ, but also implicate NHEJ as a major contributor to the cytotoxicity observed in HR-deficient cells treated with PARP inhibitors.
Results
PARP Inhibitor Synthetic Lethality Is Independent of XRCC1 and BER.
The current model of PARP inhibitor lethality in HR-deficient cells (Fig. 1A) postulates that PARP inhibition induces persistent SSBs through inactivation of BER, and that these breaks are converted to DSBs by collision with replication machinery. This model predicts that disabling BER should recapitulate the effect of PARP inhibition in these cells. To test this model, we induced siRNA-mediated knockdown of XRCC1, an essential protein in BER (19). These experiments used PEO1 and PEO4 cells, a pair of ovarian cancer lines that are derived from the same patient but differ in BRCA2 expression (20) (Fig. 1B). PARP1 depletion significantly and reproducibly decreased the clonogenic survival of BRCA2-deficient PEO1 cells but not BRCA2-expressing PEO4 cells (Fig. 1 C and D), confirming previously published results (3, 4). Depletion of XRCC1 did not alter the viability of either cell line (Fig. 1 C and D), even though the same XRCC1 knockdown sensitized both lines to the alkylating agent methyl methanesulfonate (Fig. S1). This result, coupled with the recent report that PARP inhibitors fail to increase SSBs in BRCA2-deficient cells (21), prompted us to consider the possibility that PARP1 maintains the genomic stability of HR-deficient cells through a mechanism distinct from BER.
PARP Inhibition Induces Phosphorylation of DNA-PK Targets and Enhances NHEJ.
In addition to its role in BER, PARP1 has been implicated in the modulation of a variety of nuclear processes, including classical NHEJ (1, 16, 17). Accordingly, we hypothesized that the simultaneous loss of HR and PARP1 might result in deregulation of NHEJ (Fig. 1E). If this model were correct, one would predict that PARP inhibition in HR-deficient cells would result in increased activation of DNA-PK, increased NHEJ activity, and increased genomic instability resulting from this error-prone pathway. Importantly, this alternative model suggests that inhibition of NHEJ via genetic or pharmacological approaches should diminish the effects of PARP inhibitors on all of these processes.
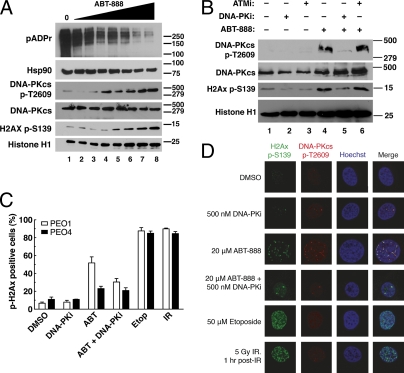
To test these predictions, we incubated PEO1 cells with the PARP inhibitor ABT-888 (22) (Fig. 2A) and examined the phosphorylation of DNA-PK substrates. The epitopes examined included the phosphorylation site of DNA-PKcs at Thr2609, which must be phosphorylated for efficient NHEJ (23), and Ser139 of H2AX, which undergoes DNA damage-induced phosphorylation by several kinases, including activated DNA-PKcs (24). Both of these sites were phosphorylated in a dose-dependent manner as poly(ADP-ribosyl)ation decreased in ABT-888–treated PEO1 cells (Fig. 2A). Addition of the DNA-PK inhibitor AZ12594248 (25) prevented the ABT-888–induced phosphorylation of DNA-PKcs and H2AX, whereas the ATM inhibitor KU55933 (26) did not (Fig. 2B). Likewise, DNA-PKcs autophosphorylation at Ser2056 (27) increased when PEO1 cells were treated with ABT-888 (Fig S2A), and this phosphorylation was reversed by DNA-PK inhibition (Fig. S2 B and C).
Fig. 2.
DNA-PK is activated after PARP inhibitor exposure in PEO1 cells. (A) Western blots for poly(ADP-ribose) polymer (pADPr) and phosphorylation of DNA-PK substrates (DNA-PKcs autophosphorylation at Thr2609 and histone H2AX at Ser139) in PEO1 cells after 72 h of exposure to increasing concentrations of ABT-888 (0, 0.625, 1.25, 2.5, 5, 10, 20, and 40 μM). Hsp90, total DNA-PKcs, and histone H1 are used as loading controls. (B) Phosphorylation of DNA-PK substrates after treatment for 72 h with diluent (0.2% DMSO, lanes 1 and 4), 500 nM DNA-PK inhibitor AZ12594248 (DNA-PKi, lanes 2 and 5), or 5 μM ATM inhibitor KU55933 (ATMi, lanes 3 and 6) alone (lanes 1–3) or in combination with 20 μM ABT-888 (lanes 4–6). (C) Quantitation of cells positive for phospho-H2AX foci in PEO1 and PEO4 cells, after treatment with DMSO, 500 nM DNA-PK inhibitor, 20 μM ABT-888 (ABT), ABT-888 and DNA-PK inhibitor, 50 μM etoposide (Etop), or 5 Gy of ionizing radiation (IR). Cells were exposed to ABT-888 and/or DNA-PK inhibitor for 72 h, etoposide for 1 h, or allowed to recover for 1 h after IR. Results are reported as mean ± SEM of three independent experiments. (D) Confocal images of PEO1 cells treated as in C. Phospho-Ser139-H2AX is shown in green, phospho-Thr2609-DNA-PKcs is shown in red, and Hoechst 33258 is shown in blue.
Additional experiments in PEO1 cells demonstrated that ABT-888 induced phospho-H2AX foci, which could be diminished by inhibiting DNA-PK (Fig. 2 C and D). These phospho-H2AX foci colocalized with phosphorylated DNA-PKcs after PARP inhibition (Fig. 2D, third row). Moreover, formation of foci and phosphorylation of DNA-PKcs were both reduced by the addition of a DNA-PK inhibitor (Fig. 2D, fourth row). Similarly, down-regulation of Ku80 or Artemis, a nuclease responsible for processing DNA ends in NHEJ (28, 29), reduced ABT-888–induced phospho-H2AX foci in PEO1 cells (Fig. S3). In contrast, PARP inhibition failed to induce phosphorylation of both DNA-PKcs and H2AX in PEO4 cells (Fig. S2D). Thus, PARP inhibitors induce DNA-PK activation, as manifested by phosphorylation of DNA-PK substrates and formation of foci containing phosphorylated DNA-PKcs, only in BRCA2-deficient PEO1 cells and not BRCA2-positive PEO4 cells.
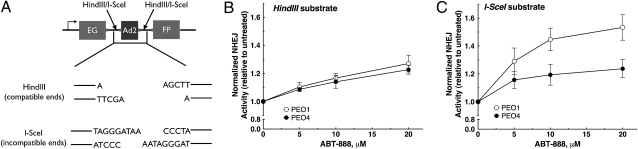
To directly measure the effect of PARP inhibition on NHEJ activity in vivo, we used a validated reporter assay (16, 30, 31) (Fig. 3A). After transfection with linearized Pem1-EGFP-Ad2, PEO1 and PEO4 cells were incubated with diluent or ABT-888. Successful end joining recircularizes the plasmid, restoring EGFP expression that can be detected by flow cytometry. Substrate linearization with HindIII produces cohesive 4-bp overhangs, whereas digestion with I-SceI produces an inverted overhang that requires nucleolytic end processing before successful recircularization. By using this assay, a small increase in end joining was detected after ABT-888 treatment in both PEO1 and PEO4 cells transfected with HindIII-linearized plasmid (Fig. 3B). Strikingly, ABT-888 induced a pronounced increase in end joining of the I-SceI–linearized substrate in PEO1 cells compared with PEO4 cells (Fig. 3C and Fig. S4). Because the I-SceI substrate has ends that require nucleolytic processing before end joining, the disproportionate increase in recircularization of this substrate, but not the HindIII substrate, implies that PARP inhibition increases error-prone repair selectively in BRCA2-deficient PEO1 cells.
Fig. 3.
Error-prone NHEJ activity is enhanced by PARP inhibitors in PEO1 cells. (A) Schematic of the in vivo NHEJ assay. Pem1-Ad2-EGFP is an EGFP-containing vector with a 2.4-kb intron (Pem1) and one exon (Ad2) inserted into the EGFP cassette. Pem1-Ad2-EGFP was cut with either HindIII or I-SceI to produce linearized substrate with compatible overhangs or incompatible inverted overhangs, respectively. Successfully recircularized plasmid will produce intact EGFP, which can be assayed via flow cytometry. Any residual uncut plasmid, caused by the insertion of the Ad2 exon within the EGFP ORF, will be EGFP-negative. A pCherry plasmid was cotransfected with substrate to correct for transfection efficiency. (B and C) Quantitation of NHEJ activity in PEO1 and PEO4 cells transfected with HindIII substrate (B) or I-SceI substrate (C) and exposed to ABT-888 for 72 h. Each data point represents the mean ± SEM from three independent experiments. Representative flow cytometry profiles are shown in Fig. S4.
An alternate form of end joining, microhomology-mediated end joining (MMEJ), has been described in the absence of DNA-PKcs (32). Using an assay for MMEJ (31, 33) (Fig. S5A) that readily detected MMEJ in DNA-PKcs–deficient M059J cells (Fig. S5B, lanes 11 and 12), we failed to detect induction of MMEJ in PEO1 or PEO4 cells exposed to ABT-888 (Fig. S5 B and C), ruling out the induction of MMEJ by PARP inhibition. These results collectively demonstrate that PARP inhibition selectively enhances DNA-PK activity and error-prone NHEJ activity in PEO1 but not PEO4 cells.
PARP Inhibitor-Induced Genomic Instability Is Driven by NHEJ.
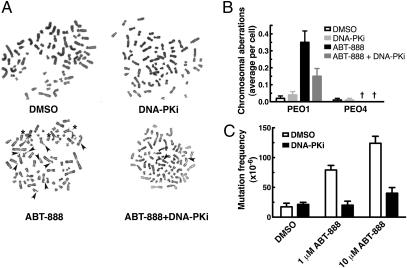
In BRCA-deficient cells, PARP inhibitors induce chromosomal instability typified by the accumulation of chromosomal breaks and radial structures (4, 34). Consistent with these reports, ABT-888 induced the formation of chromosome breaks and aberrant radial structures in PEO1 cells (Fig. 4 A and B) but not in PEO4 cells (Fig. 4B and Fig. S6). Importantly, addition of the DNA-PK inhibitor substantially diminished this effect, indicating that NHEJ plays a role in the development of aberrant chromosomal structures after PARP inhibition in PEO1 cells.
Fig. 4.
PARP inhibitor-induced chromosomal derangement and genomic instability depend on DNA-PK activity. (A) Representative images of metaphase spreads from cells treated with diluent (0.2% DMSO), 500 nM DNA-PK inhibitor (DNA-PKi), 2.5 μM ABT-888, or both ABT-888 and DNA-PK inhibitor for 72 h. Chromosomal breaks are marked with arrowheads, and radial structures are marked with asterisks. (B) Quantitation of data from A showing average radial chromosomes per cell (n = 100 for each data point pooled from two separate experiments, error bars represent SEM). †Zero values. (C) Calculated mutagenesis frequency in BRCA2-mutant CAPAN1 cells after control treatment or exposure to ABT-888 with or without 250 nM DNA-PK inhibitor. Each bar represents the mean ± SEM of five to eight plates. This result is representative of three independent experiments.
To extend these studies to the single-gene level, we performed forward mutagenesis assays to measure the mutation rate of the hypoxanthine-guanine phosphoribosyl transferase (HPRT) locus in BRCA2-mutant cells exposed to a PARP inhibitor. The toxicity of 6-thioguanine (6-TG) depends on the expression of active HPRT; as a consequence, only cells with mutations at the X-linked HPRT locus are able to survive in 6-TG supplemented medium. To perform these experiments, we used CAPAN1 cells, a BRCA2-mutant cell line derived from a male pancreatic cancer patient (35), to ensure that our model system had only one copy of the HPRT gene. CAPAN1 cells treated with PARP inhibitor formed more colonies in the presence of 6-TG, indicating increased mutation frequency compared with diluent controls (Fig. 4C). As was the case with chromosomal aberrations, coadministration of the DNA-PK inhibitor markedly reduced the mutation frequency. Overall, these experiments demonstrate that NHEJ increases genomic damage, at both the chromosomal level and the individual gene level, when PARP is inhibited.
Disabling NHEJ Diminishes PARP Inhibitor Hypersensitivity in BRCA2-Deficient Cells.
To determine whether the previous results extend to cell survival, we performed clonogenic assays in paired cell lines treated with ABT-888 after various alterations in the NHEJ pathway. Knockdown of Ku80, an essential component of NHEJ (9), had little effect by itself but markedly enhanced the survival of BRCA2-deficient PEO1 cells treated with ABT-888 (Fig. 5 A and B). In contrast, BRCA2-positive PEO4 cells were resistant to the effects of ABT-888, which was unaffected by Ku80 siRNA (Fig. 5 A and B). To ensure that the sensitivity of PEO1 cells was not an off-target effect of ABT-888, we performed the same experiment by knocking down PARP1 and/or Ku80 using siRNA (Fig. 5 C and D). Like ABT-888, PARP1 depletion decreased the clonogenic survival of PEO1 cells but not PEO4 cells, and Ku80 knockdown reversed the effect of the PARP1 siRNA. Similar to Ku80 knockdown, siRNA depletion of Artemis also reversed the lethality of ABT-888 in PEO1 cells (Fig. 5E). Likewise, coadministration of the DNA-PK inhibitor AZ12594248 diminished the effects of ABT-888 (Fig. 5F and Fig. S7 A and B) and another PARP inhibitor, AZD2281 (Fig. S7C). Similar results were observed in BRCA2-mutant CAPAN1 cells, where DNA-PK inhibition again mitigated the toxicity of PARP inhibition (Fig. S8). In short, inhibition or down-regulation of multiple components of the NHEJ pathway diminished the toxicity of PARP inhibition in BRCA2-deficient cells, indicating that the toxicity of PARP inhibition depends on NHEJ in this context.
Fig. 5.
NHEJ is a major contributor to PARP inhibitor effects in BRCA2-deficient cells. (A) Western blots showing knockdown of Ku80 in PEO1 and PEO4 cells. (B) Clonogenic survival of PEO1 and PEO4 cells from A, which were treated with the indicated ABT-888 concentration for 72 h, washed, and allowed to form colonies. (C) Western blots after treatment with siRNA targeting luciferase (control), Ku80, PARP1, or both Ku80 and PARP1. (D) Clonogenic viability of PEO1 and PEO4 cells from C. After knockdown, cells were plated onto triplicate plates and allowed to form colonies. (E) Clonogenic survival of PEO1 cells after Artemis knockdown. After treatment with the indicated siRNA, plates were treated with indicated concentration of ABT-888 for 72 h, washed, and allowed to form colonies. (Inset) Western blots showing knockdown with luciferase (control) or Artemis siRNAs in PEO1 cells. (F) Clonogenic survival of PEO1 and PEO4 cells treated for 72 h with ABT-888 in combination with diluent or 500 nM DNA-PK inhibitor (DNA-PKi). All results are reported as means of triplicate plates ± SEM, and are representative of three independent experiments.
NHEJ Is also Responsible for PARP Inhibitor Lethality in Other HR-Deficient Contexts.
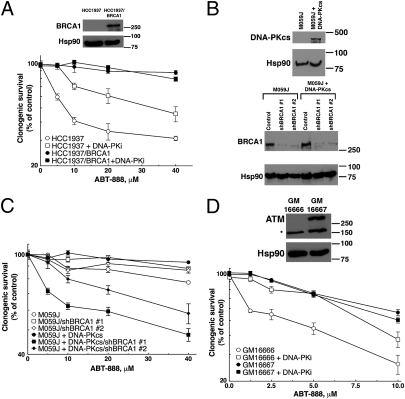
In addition to BRCA2, previous studies have documented synthetic lethality between PARP inhibition and loss of other HR components, such as BRCA1 (4) and ATM (7, 8). In HCC1937 cells, which lack BRCA1 (36) (Fig. 6A Inset), addition of the DNA-PK inhibitor diminished ABT-888 sensitivity (Fig. 6A), just as it did in PEO1 cells. Moreover, in HCC1937 cells, inhibition of DNA-PK also diminished formation of H2AX foci (Fig. S9A) and inhibited ABT-888–induced colocalization of phospho-Thr2609-DNA-PK and phospho-Ser139-H2AX in foci (Fig. S9B). Likewise, BRCA1 knockdown sensitized DNA-PKcs–reconstituted M059J cells to ABT-888 (Fig. 6 B and C). Importantly, parental M059J cells lacking DNA-PKcs were not sensitized by BRCA1 knockdown (Fig. 6 B and C), providing independent genetic evidence for the important role of DNA-PKcs in the synthetic lethality of HR deficiency and PARP inhibition.
Fig. 6.
NHEJ contributes to PARP inhibitor-induced effects in other HR-deficient contexts. (A) BRCA1-deficient HCC1937 and BRCA1-reconstituted HCC1937/BRCA1 cells were continuously exposed to ABT-888 in the presence or absence of 125 nM DNA-PK inhibitor (DNA-PKi) and assayed for clonogenic survival. (Inset) Western blots of cell lysates from HCC1937 and HCC1937/BRCA1. (B) Western blots of M059J and reconstituted M059J?PKcs lines showing the restoration of DNA-PKcs expression and the shRNA-mediated knockdown of BRCA1. (C) Clonogenic survival of shRNA–transfected M059J/M059J?PKcs lines treated with ABT-888 for 72 h. (D) Clonogenic survival of ATM-deficient GM16666 or ATM-reconstituted GM16667 fibroblasts. Cells were exposed to ABT-888 for 48 h in the presence or absence of 250 nM DNA-PK inhibitor (DNA-PKi), washed, and allowed to form colonies. (Inset) Western blots of lysates from GM16666 and GM16667 fibroblasts. Data are displayed as mean ± SEM of triplicate plates. Results are representative of three independent experiments.
To extend these results to ATM deficiency, we examined GM16666 and GM16667 cells, an ATM-deficient line and its ATM-reconstituted counterpart (37) (Fig. 6D Inset). Similar to BRCA1- and BRCA2-deficient cells, GM16666 cells exhibited heightened sensitivity to ABT-888, and inhibition of DNA-PK reversed this effect (Fig. 6D). Collectively, results presented in Fig. 6 not only demonstrate that the effect of DNA-PK inhibition on cellular sensitivity to PARP inhibition extends to other HR-deficient backgrounds but also provide genetic evidence that NHEJ plays a vital role in hypersensitivity of HR-deficient cells to PARP inhibitors.
Discussion
The concept of synthetic lethality centers on the combination of two genetic lesions, each of which is nonlethal, that nevertheless induce lethality together. This approach has been extended to pharmacologic agents that target specific pathways to exploit existing genetic alterations in cancer cells. Most notably, two groups demonstrated the striking sensitivity of BRCA-deficient cells to PARP inhibitors (3, 4), which has since been extended to other HR-deficient backgrounds (5–8). In addition to the clinical potential of these findings, they provide an opportunity to more fully understand the biology of HR as well as the interplay between HR and other modalities of repair. In this study, we evaluated the contribution of NHEJ to the effects of PARP inhibition in HR-deficient cells. Our results strongly support a different model (Fig. 1E) for the mechanism of PARP inhibitor synthetic lethality in these cells.
The original explanation for the antitumor effects of PARP inhibitors in HR-deficient cells invoked the well-defined role of PARP1 in BER. This model postulated that catalytic inhibition of PARP1 disabled the ability of the cell to respond to endogenous DNA damage through BER, resulting in accumulated SSBs (Fig. 1A). However, the inability to demonstrate increased SSBs after PARP inhibition (21) raised questions about this model, and our failure to find synthetic lethality when XRCC1 is down-regulated in BRCA2-deficient cells raised the possibility that the effects of PARP inhibitors may be mediated through a mechanism distinct from BER.
As a corollary to the original model, if accumulated DNA damage were responsible for the toxicity of PARP inhibitors, one would expect HR-deficient cells to depend on alternate DSB repair pathways such as NHEJ for survival. In direct contradiction to this prediction, we found that disabling NHEJ diminished the genomic instability and lethality of PARP inhibition in HR-deficient cells rather than exacerbating it. Our results extend the growing body of literature that has connected NHEJ to genomic instability after exposure to chemotherapeutic agents. In a recent study, disabling NHEJ was shown to reverse the DNA-repair defects and chromosomal instability of FANCD2 mutants exposed to platinum cross-linking agents (11). Moreover, ablation of 53BP1, a molecule recently demonstrated to facilitate NHEJ-mediated DSB repair (38) in addition to its other roles (39), also rescued the genotoxicity of DNA-damaging agents in a BRCA1 background (34, 40). These earlier studies provide support for a model in which unrestricted NHEJ could induce genomic instability and eventual lethality in HR-deficient cells.
Because of the error-prone nature of NHEJ, the interplay between HR and NHEJ has important implications for genomic stability. Our findings are consistent with the observation that competition between these two DSB repair pathways occurs at sites of DNA damage (11, 18). In particular, we demonstrate that BRCA2-deficient PEO1 cells are hypersensitive to both PARP1 catalytic inhibition and siRNA depletion, and this effect is reversed by disabling NHEJ. Coupled with the observation that this behavior was also seen in BRCA1-deficient and ATM-deficient cell lines, our findings strongly implicate NHEJ as a process that contributes to the toxicity of PARP inhibitors in HR-deficient cells. It is worth emphasizing that the necessity for active NHEJ for PARP inhibitor synthetic lethality was demonstrated through multiple different approaches that diminish NHEJ through either genetic (knockdown of Ku80 or Artemis or the use of DNA-PKcs–deficient cells) or pharmacologic (small-molecule inhibition of DNA-PKcs) means.
In summary, a variety of genetic and pharmacologic approaches indicate a critical role for NHEJ in the synthetic lethality of PARP inhibition and HR deficiency. Our findings support a model (Fig. 1E) in which PARP inhibition induces aberrant activation of NHEJ in HR-deficient cells, and this activation is responsible for the ensuing genomic instability and eventual lethality. PARP inhibition is being extensively investigated as a method of exploiting genetic lesions in cancer cells (3–8), with promising results in clinical trials (41, 42). Despite the early success of PARP inhibitors in the treatment of BRCA-deficient cancers, many BRCA-deficient tumors resist this therapy. Recent phase 2 trials of the PARP inhibitor olaparib describe objective responses of 33% in BRCA-deficient ovarian cancers (41) and 41% in BRCA-deficient breast cancers (42). Although remarkable, these results fall short of regressions observed with other targeted therapies, which have tumor response rates of 50–70% (43). The more limited response of BRCA-deficient tumors to PARP inhibitors raises the possibility that factors in addition to HR deficiency play a role in sensitivity of BRCA-deficient tumors to PARP inhibition. To this end, our findings predict that BRCA-deficient tumors with low NHEJ activity might be less responsive to PARP inhibitors.
Materials and Methods
Reagents and Antibodies.
Reagents were purchased from the following companies: ABT-888 (veliparib) and etoposide from Enzo Life Sciences, 6-TG from Sigma-Aldrich, and HindIII and I-SceI from New England Biolabs. The DNA-PK inhibitor, AZ12594248/KU60648 (a water-soluble analog of NU7441), and ATM inhibitor, KU55933, were kindly provided by KuDOS Pharmaceuticals. Antibodies and their suppliers are provided in SI Materials and Methods.
Cell Culture and siRNA Transfections.
Cell culture conditions, including methods used to perform siRNA- or shRNA-mediated knockdown and subsequent clonogenic assays (44), are described in SI Materials and Methods.
NHEJ Assay.
The end-joining reporter plasmid pEGFP-Pem1-Ad2 (provided by E. Hendrickson, University of Minnesota, Minneapolis, MN) (Fig. 3A) was used as previously described (16, 30, 31). Further details are provided in SI Materials and Methods.
Cytogenetics.
Cell harvest and metaphase slide preparation were performed for metaphase analysis as previously described (45). One hundred nonbanded metaphases from each cell line were analyzed and scored for radial formations as well as major and minor breakage according to the International System for Human Cytogenetic Nomenclature. Images of cells with breakage were captured with a CytoVision Imaging System (Genetix).
HPRT Mutagenesis Assays.
HPRT mutagenesis was performed as described previously (46). Detailed descriptions of this assay are provided in SI Materials and Methods.
Immunoblotting and Immunofluorescence Microscopy.
Detailed descriptions of protein preparation, immunoblotting, and confocal microscopy are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Z. Lou, E. Hendrickson, F. Couch, J. Chen, L. Karnitz, D. Toft, G. Poirier, J. Sorace, and C. Segovis for reagents and/or advice. We also thank the Mayo Clinic Cytogenetics and Flow Cytometry Shared Resources for experimental assistance and Deb Strauss for editorial help. This work was supported in part by National Institutes of Health Grants P50 CA136393 and T32 GM072474.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013715108/-/DCSupplemental.
References
- 1.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferraris DV. Evolution of poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors. From concept to clinic. J Med Chem. 2010;53:4561–4584. doi: 10.1021/jm100012m. [DOI] [PubMed] [Google Scholar]
- 3.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 4.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 5.Mendes-Pereira AM, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sourisseau T, et al. Aurora-A expressing tumour cells are deficient for homology-directed DNA double strand-break repair and sensitive to PARP inhibition. EMBO Mol Med. 2010;2:130–142. doi: 10.1002/emmm.201000068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williamson CT, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weston VJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 9.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 10.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 11.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 12.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J Biol Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 13.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- 15.Gagné J-P, et al. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochegger H, et al. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saberi A, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol Cell Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair(Amst) 2003;2:955–969. doi: 10.1016/s1568-7864(03)00118-6. [DOI] [PubMed] [Google Scholar]
- 20.Sakai W, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69:6381–6386. doi: 10.1158/0008-5472.CAN-09-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottipati P, et al. Poly(ADP-ribose) polymerase is hyperactivated in homologous recombination-defective cells. Cancer Res. 2010;70:5389–5398. doi: 10.1158/0008-5472.CAN-09-4716. [DOI] [PubMed] [Google Scholar]
- 22.Penning TD, et al. Discovery of the Poly(ADP-ribose) polymerase (PARP) inhibitor 2-[(R)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-4-carboxamide (ABT-888) for the treatment of cancer. J Med Chem. 2009;52:514–523. doi: 10.1021/jm801171j. [DOI] [PubMed] [Google Scholar]
- 23.Chan DW, et al. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002;16:2333–2338. doi: 10.1101/gad.1015202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiff T, et al. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 25.Hingorani M, et al. Inhibition of repair of radiation-induced DNA damage enhances gene expression from replication-defective adenoviral vectors. Cancer Res. 2008;68:9771–9778. doi: 10.1158/0008-5472.CAN-08-1911. [DOI] [PubMed] [Google Scholar]
- 26.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 27.Uematsu N, et al. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 29.Drouet J, et al. Interplay between Ku, Artemis, and the DNA-dependent protein kinase catalytic subunit at DNA ends. J Biol Chem. 2006;281:27784–27793. doi: 10.1074/jbc.M603047200. [DOI] [PubMed] [Google Scholar]
- 30.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. DNA end joining becomes less efficient and more error-prone during cellular senescence. Proc Natl Acad Sci USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fattah F, et al. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6:e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377–5388. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verkaik NS, et al. Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur J Immunol. 2002;32:701–709. doi: 10.1002/1521-4141(200203)32:3<701::AID-IMMU701>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 34.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goggins M, et al. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 36.Tomlinson GE, et al. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 1998;58:3237–3242. [PubMed] [Google Scholar]
- 37.Ziv Y, et al. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
- 38.Orsburn B, et al. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Mol Cell Biol. 2010;30:2341–2352. doi: 10.1128/MCB.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: Function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 40.Bouwman P, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet. 2010;376:245–251. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 42.Tutt A, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 43.Chan SL, Mok T. PARP inhibition in BRCA-mutated breast and ovarian cancers. Lancet. 2010;376:211–213. doi: 10.1016/S0140-6736(10)61119-1. [DOI] [PubMed] [Google Scholar]
- 44.Karnitz LM, et al. Gemcitabine-induced activation of checkpoint signaling pathways that affect tumor cell survival. Mol Pharmacol. 2005;68:1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- 45.Fletcher J. Current Protocols in Human Genetics. New York: Wiley; 2001. Metaphase harvest and cytogenetic analysis of solid tumor cultures; pp. 10.3.1–10.3.10. [DOI] [PubMed] [Google Scholar]
- 46.Hashimoto H, Chatterjee S, Berger NA. Mutagenic activity of topoisomerase I inhibitors. Clin Cancer Res. 1995;1:369–376. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.