Abstract
Glaucoma, a complex heterogenous disease, is the leading cause for optic nerve-related blindness worldwide. Primary open angle glaucoma (POAG) is the most common subset and by the year 2020 it is estimated that approximately 60 million people will be affected. MYOC, OPTN, CYP1B1 and WDR36 are the important candidate genes. Nearly 4% of the glaucoma patients have mutation in any one of these genes. Mutation in any of these genes causes disease either directly or indirectly and the severity of the disease varies according to position of the genes. We have compiled all the related mutations and SNPs in the above genes and developed a database, to help access statistical and clinical information of particular mutation. This database is available online at http:bicmku.in:8081/glaucoma The database, constructed using SQL, contains data pertaining to the SNPs and mutation information involved in the above genes and relevant study data.
Availability
The database is available for free at http:bicmku.in:8081/glaucoma
Keywords: WD repeats 36 (WDR36), Optineurin (OPTN), Cytochrome P450 1 B chain (CYP1B1), Myocilin (MYOC) Single Nucleotide polymorphism (SNP), Structured Query language (SQL)
Background
Glaucoma is a disease in which the optic nerve is damaged, leading to progressive irreversible loss of vision [1]. It is often, but not always, associated with increased pressure of the fluid in the eye. Primary open angle glaucoma is a neurodegenerative disorder characterized by progressive excavation of the optic disc with corresponding loss of peripheral vision, and is frequently associated with elevated intraocular pressure (IOP).
Genes that are associated with Glaucoma are MYOC, CYP1B1, WDR36 and OPTN. MYOC gene maps to the GLC1A locus at 1q24-q25. The gene has three exons, encoding myocilin, 504-amio acid polypeptide, which has an N-terminal leucine zipper domain and a C-terminal olfactomedin-like domain [2]. CYP1B1 gene maps to the GLC3A locus at 2p21 which contains three exons, encoding cytochrome P450, 543 aminoacid polypeptide. OPTN gene maps to GLC1E locus at 10p15-p14 and contains 3 noncoding exons in the 5'-untranslated region and 13 exons that code for a 577-amino acid protein. It plays a neuroprotective role in the eye and optic nerve. WDR36 gene maps to locus at 5q21.3-q22.1. It contains 23 exons that code for 951 amino acid protein. Mutation in this gene accounts for 1% of glaucoma.
Methodology
Data collection and curation
A literature search was conducted using PubMed from that all the available data collected till date. Search term included myocilin, MYOC, CYP1B1, OPTN, WDR36, glaucoma. The information has also been collected from the genecards database [3], myocilin database (www.myocilin.com) [4]. Data have been curated manually.
Data Management
Glaucoma database runs on MySql server as back end tool with Perl CGI in the front end. The database has more than 500 entries. To ensure flexibility and ease of management, the database was constructed using the MySQL database. The database contains tables with the information being stored in an organised way. The total number of control and glaucomatous case subjects recruited, as well as the number of case and control subjects identified as carrying a particular variant in the each gene is also recorded. The variant, phenotypic field, ethnicity, age, IOP and Number of patients for each identified gene is recorded in different tables.
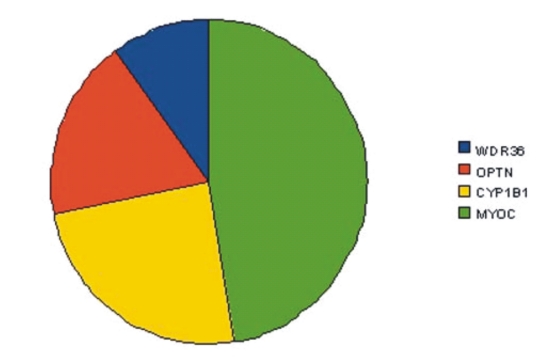
The database contains information relating the mutations and SNPs involved in the disease. The disease-causing mutation in the gene is coded as a field in the database and as a drop-down menu that allows the selection of the particular mutation. The information is used to determine the genomic location (nucleotide index), and its corresponding site in the gene (e.g., promoter,exon,introns etc). The Study type button allows the user to know about the phenotypic status of the selected mutation. There is a separate column named candidate gene where major genes such as MYOC, CYP1B1, OPTN, and WDR36 are displayed and Figure 1 shows the gene distribution pattern in the database. There is also a similar column named associate genes which contains the information stored in the associate gene table in the MySql database. The phenotype-genotypic information of the respective gene can be viewed by clicking the link provided. The database also presents the user with other associative genes involved in the glaucoma with their genomic location and phenotype. This database provides clinical information of the particular mutation which will help to predict severity of the disease for clinicians and Figure 2shows snapshot of the Database.
Figure 1.
The distribution of variants identified in gene. About 40% of disease causing variants were identified in CYP1B1, 18% of the variants were identified in OPTN and 10% of the variants were identified in WDR36 gene.
Figure 2.
Screenshot of the main page of the website.
Website Structure
A hypertext mark-up language (HTML) script, which uses minimal embedded script so as to eliminate browser-browser variation, was written for the website. Using the heading bar, it is easy to navigate from the homepage to other information such as physiology, overview, gallery etc., Perl programming along with CGI script is used to extract the user specified information from the MySql database. Phenotype and mutation data have been compiled initially from the published literature.
Utility and Future direction
The database provides the user with the SNP and mutation information for the major genes involved in the disease glaucoma. It also provides the user with the other associated genes which are indirectly involved in the disease glaucoma. The database aims to provide the user with the SNP information of the major genes thus providing the researcher useful information. A web server can be developed to predict mutation severity.
Acknowledgments
We would like to thank Aravind Eye Hospital, Madurai, Tamilnadu, India for the encouragement and Department of Biotechnology (DBT) for the support by providing the Center of Excellence in Bioinformatics facility.
Footnotes
Citation:Rangachari et al, Bioinformation 5(9): 398-399 (2011)
References
- 1.HA Quigley, et al. Br J Ophthalmol. 2006;90(3):262. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.P Sundaresan, et al. Molecular Vision. 2003;9:606. [PubMed] [Google Scholar]
- 3.I Peter, et al. GeneCardsTM. 2002;11(1542) [Google Scholar]
- 4.W Alex, et al. Human Mutation. 2008;29(2):207. doi: 10.1002/humu.20634. [DOI] [PubMed] [Google Scholar]