Abstract
After immunization or infection, activation-induced cytidine deaminase (AID) initiates diversification of immunoglobulin (Ig) genes in B cells, introducing mutations within the antigen binding V regions (somatic hypermutation, SHM) and double-strand DNA breaks (DSBs) into switch (S) regions, leading to antibody class switch recombination (CSR). We asked if during B cell activation, AID also induces DNA breaks at genes other than IgH genes. Using a non-biased genome-wide approach, we have identified hundreds of reproducible AID-dependent DSBs in mouse splenic B cells shortly after induction of CSR in culture. Most interestingly, AID induces DSBs at sites syntenic with sites of translocations, deletions, and amplifications found in human B cell lymphomas, including within the oncogene B cell lymphoma11a (bcl11a)/evi9. Unlike AID-induced DSBs in Ig genes, genome-wide AID-dependent DSBs are not restricted to transcribed regions, and frequently occur within repeated sequence elements, including CA-repeats and non-CA tandem repeats, and SINEs.
Introduction
During an immune response, both the Ig V region genes and heavy chain constant (C) region genes diversify in order to increase the effectiveness of the antibody response. AID initiates CSR by deaminating cytosines, converting them to uracils, which are excised by Uracil DNA glycosylase (UNG), leaving abasic sites that are nicked by AP endonucleases, forming single-strand breaks (SSBs) (Guikema et al., 2007; Rada et al., 2002; Stavnezer et al., 2008). Nearby SSBs (on opposite DNA strands) form DSBs required for the deletional recombination occurring during CSR. AID initiates CSR by deaminating cytosines in special tandemly repeated switch (S) region sequences (ranging from 1 – 12 kb in length), which have numerous repeats of a preferential target motif for AID, WGCW (where W=A or T), and which are located upstream of each of the IgH constant region genes. AID has been shown to initiate mutations at several non-Ig genes in B cells in Peyer’s patches, where Ig variable region genes have undergone multiple rounds of SHM (Liu et al., 2008; Pasqualucci et al., 2001; Shen et al., 1998). However, it is unknown if during activation of B cells to undergo CSR, DSBs, in addition to mutations, are induced at non-IgH sites in the genome. This is important because DSBs can lead to chromosomal translocations, chromosomal amplifications and deletions. Although several oncogenes involved in B lymphomas and myelomas have been shown to undergo translocation to S regions in the IgH locus, it has not been demonstrated that these breaks are due to AID activity.
Results and Discussion
Detection of AID-dependent Nbs1-binding sites in the IgH locus of B cells induced to undergo CSR in culture
To identify AID-dependent DSBs throughout the genome, we used chromatin immunoprecipitation (ChIP) to detect Nbs1-binding sites. Nbs1 is a component of the Mre11-Nbs1-Rad50 (MRN) complex, which binds within 1 kb of DSBs immediately after their formation (Berkovich et al., 2007; Difilippantonio and Nussenzweig, 2007; You et al., 2009). Importantly, Nbs1 has been shown to co-localize with IgH genes in B cells undergoing CSR, indicating that it binds to AID-induced DSBs (Petersen et al., 2001). AID-dependent S region DSBs are detectable by ligation-mediated PCR (LM-PCR) approximately 2 days after addition of lipopolysaccharide (LPS), BlyS/BAFF, and anti-IgD dextran (anti-δ-dex) to splenic B cell cultures (Schrader et al., 2005). Using these conditions, we stimulated wild-type (WT) and aid−/− B cells and performed Nbs1-ChIP, followed by whole-genome tiling array hybridization analysis (ChIP-chip) to identify AID-dependent DSBs. Suppl Fig S1 diagrams the experimental plan, which is detailed in Experimental Procedures. The immunoprecipitated DNA probes used for the experiments were first tested by quantitative PCR, which showed that anti-Nbs1 specifically precipitates Sμ sequences from WT cells but not from aid−/− cells, and does not precipitate Cμ sequences (Fig 1A,B). Cμ is not targeted by AID-induced mutations or DSBs (Peled et al., 2008; Schrader et al., 2005).
Fig 1. ChIP-qPCR and IgH ChIP-chip experiments show AID-dependent binding of Nbs1 to Ig Sμ sequences in splenic B cells treated for 48 hrs with LPS+anti-δ dextran.
A. Quantitative PCR analysis of Nbs1-immunoprecipitated DNA used to prepare probes for the ChIP-chip experiments. Means of Sμ and Cμ signals (+SEM) relative to input DNA signals of 4 independent experiments are shown. Each individual experiment consisted of 4–6 individual ChIPs from the same cell culture, and independent experiments were performed using separately isolated splenic B-cells. B. Quantitative PCR analysis of the Nbs1-ChIP samples (after whole genome amplification, WGA), which were used to hybridize to the arrays. The Sμ and Cμ signals are reported relative to signals obtained from an identical quantity of input DNA, also amplified by WGA. C. ChIP-chip results, prepared using Nimblescan, showing Nbs1 and RNA Pol II signals across a 100-kb segment of the IgH locus from aid−/− and WT cells induced to switch. Y-axis shows log2 ratio of the ChIP signal vs. input, X-axis shows genomic coordinates of the presented results. Results from 5 ChIP-chip experiments are shown. From top to bottom: 2 Nbs1 ChIP-chip experiments with aid−/− cells, 2 ChIP-chip Nbs1 experiments with WT cells and 1 representative RNA Pol II ChIP-chip experiment. Significance relative to false discovery rate calculated by Nimblescan is indicated by the color. The lowest panel shows the density of WGCW AID hotspots across this region, indicating the abundance of these motifs within the three S regions in this segment. The Y-axis indicates density in 500 bp relative to genome average = 1. Red bars indicate 1st percentile; orange, 2nd–5th percentile; yellow, 6th–15th percentile; green, 16th–30th percentile; light blue, 31st–50th percentile; dark blue, 51st–80th percentile; black, 81st–100th percentile.
Nbs1 signals are detected at S regions in the Ig locus in WT B cells but not in aid−/− cells activated for CSR
Fig 1C shows the Nbs1 ChIP-chip peak signals across a 100-kb segment of the IgH locus from aid−/− cells in the top two panels and from WT littermates in the third and fourth panels. The 5th panel presents representative results from one of two ChIP-chip experiments for RNA polymerase II, showing that the IgH genes are highly transcribed. No obvious differences were found in the pattern of RNA polymerase II binding in WT versus aid−/− cells, with the exception of the exons deleted in the aid gene, which were not detected in aid−/− (data not shown). The lowest panel shows the locations of the highly preferred AID target motif (WGCW) across this region. There are no Nbs1-signals in ChIP’ed material from aid−/− cells across this entire segment, whereas Nbs1 binds reproducibly at Sμ and Sγ1 in WT cells. Although the cells used for these experiments were induced to switch to IgG3, Nbs1-binding was detected at Sγ3 in only one of the two experiments, which is consistent with data showing that fewer DSBs are detected by ligation-mediated (LM)-PCR in the Sγ3 region than in Sμ regions (Schrader et al., 2005; Wu and Stavnezer, 2007). This is not due to repeat masking of the arrays, as the Sγ3 region is well-represented on the arrays. However, across the Sμ core tandem repeats the probes on the arrays are more distantly spaced, leading to underestimation of signals here. Nbs1 binding at Sγ1 is likely detected because there is considerable switching to IgG1 (~8%) in these cultures, and the Sγ1 region is unusually long, thus providing a larger AID target than Sγ3. Fig 1C demonstrates the specificity of the Nbs1-ChIP-chip assay for detecting AID-dependent DSBs, and is in agreement with results of ligation-mediated (LM)-PCR experiments, indicating that AID-dependent DSBs are present in IgH S regions in mouse B cells induced to switch in culture (Schrader et al., 2005).
AID-dependent Nbs1-binding sites are detected at many non-IgH sites, including the B cell oncogene, bcl11a
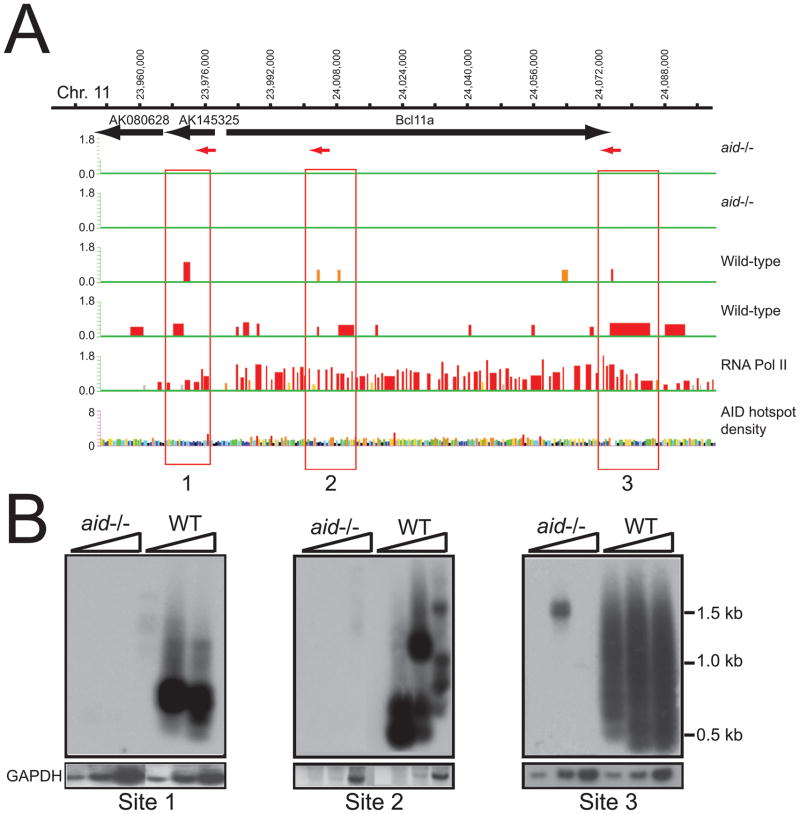
In addition to the Ig Sμ region, we identified 364 reproducible AID-dependent Nbs1-binding sites having a p<0.05, and 36 sites with p<0.0001 (Tables S1, S2). Interestingly, we found Nbs1-ChIP signals across a 146-kb segment that includes the bcl11a gene (Fig 2). In this region, 3 reproducible Nbs1-binding sites were detected in WT cells, but not in aid−/− cells. To confirm that these Nbs1 signals mark DSBs, we performed LM-PCR using primers within or near each of the 3 Nbs1-binding sites shown in Fig 2A. The DNA was treated with T4 DNA polymerase (T4 Pol) to detect staggered DSBs or mock treated to detect only blunt DSBs. Southern blots of the LM-PCR products confirm the induction of DSBs at these sites in B cells activated for CSR (Fig 2B). To provide further evidence that DSBs occur at the sites indicated by the Nbs1 ChIP signals, we cloned and sequenced 37 bcl11a LM-PCR products from WT cells. 89% of the breaks occurred at G:C bp, consistent with having been instigated by AID activity and similar to our results for S region DSBs (Schrader et al., 2005; Stavnezer et al., 2008) (Table S3). Further confirming the specificity of these DSBs, the 11 DSB segments cloned from aid−/− cells did not occur preferentially at G:C bp.
Figure 2. Nbs1 and Pol II ChIP-chip across the bcl11a locus.
A. Upper two panels show Nbs1 ChIP-chip signals across a 146-kb segment as described for Fig 1A. Sites boxed in red: 1, and 3 have significance of p<0.05 (Tamalpais) (Bieda et al., 2006), and correspond to p05_ ID_79 and _80 in Table S1. Site 2 does not achieve significance using the Tamalpais program, although site 2 has AID-dependent DSBs (see Fig 2B). Significance calculated by Nimblescan relative to false discovery rate is indicated by the color, as in Fig 1C. In addition, yellow bars correspond to p>0.1–<0.2; grey: >0.2–<0.4. B. LM-PCR blots show AID-dependent DSBs at each of the 3 Nbs1-binding sites shown in panel A. Primer locations and directionality are indicated with red arrows in A, and sequences are in Table S5. For Site 2, primer for Site 2a (left) is used, although the sequences in Table S3 were obtained using primers for both sites 2a and 2b. The assays for sites 1 and 2 were performed without the addition of T4 DNA Pol, which detects blunt DSBs, and for site 3 with the addition of T4 Pol, which detects both blunt and staggered DSBs. Lower panel shows gapdh signal amplified to control for template input.
The bcl11a locus and surrounding region of mouse chromosome 11 is syntenic to human chromosome 2, at a region that is frequently translocated or amplified in human B cell lymphoma and leukemia (Lenz et al., 2008). The DSB site located 5′ of bcl11a (site 1) corresponds to a site where translocations with Ig Sγ regions have been mapped in 4 cases of human B cell chronic lymphocytic leukemia (B-CLL) that consequently express high levels of nuclear Bcl11a (Satterwhite et al., 2001). Bcl11a is a transcriptional repressor and associates with Bcl6, both of which are normally expressed in germinal centers. Earlier, Bcl11a was identified (evi9) in mice in a screen for dominant transforming oncogenes (Li et al., 1999). Furthermore, the bcl11a gene is amplified and over-expressed in 20% of Non-Hodgkin’s lymphoma, in at least 50% of primary mediastinal B cell lymphoma (PMBL), and 50% of primary Hodgkin’s lymphoma cases (Satterwhite et al., 2001; Weniger et al., 2006). DSBs can lead to amplifications of large chromosome segments by breakage-fusion-bridge cycles (Difilippantonio et al., 2002).
We determined whether other AID-dependent Nbs1-binding sites and DSBs are found in the mouse genome at sites syntenic for chromosomal regions reported to be amplified or deleted in human DLBCL (Lenz et al., 2008), and found that many have AID-dependent Nbs1-binding sites and DSBs, as confirmed by LM-PCR. Suppl Figs S2-S11 present maps of ChIP-chip data for ~100 kb segments showing several other Nbs1-binding sites verified by LM-PCR, several of which were confirmed by sequencing LM-PCR products, as indicated in the figure legends. Figs S2-S8 show p<0.05 sites, and Fig S9-S11 show p<0.0001 sites. For each site, we provide a genome map of the surrounding region and the syntenic region in humans. Shown are sites located near: pten, a gene frequently deleted in germinal center B cell-like (GCB)-DLBCL (Fig S2); jak2, frequently amplified in all 3 subtypes of DLBCL, especially in PMBL (Fig S4); Ppp1R13, and fosB, amplified in activated B cell like (ABC)- and GCB-DLBCL (Fig S8). See the Fig legends for additional information. LM-PCR confirmed DSBs in 15 out of 18 (83%) AID-dependent non-IgH Nbs1-binding sites. The LM-PCR results are indicated in Tables S1 and S2.
AID-dependent DSBs were not detected in the c-myc gene
Surprisingly, we did not detect AID-dependent Nbs1-signals (p<0.05) in the c-myc locus. Also, by LM-PCR we detect a comparable low frequency of DSBs in the c-myc locus in both aid−/− and WT cells, either 5′ of the gene or in the intron between exons 1 and 2, where translocations between IgH and c-myc genes most frequently occur (data not shown). Although c-myc-IgH translocations have been shown to be AID-dependent (Ramiro et al., 2004; Robbiani et al., 2008; Takizawa et al., 2008), and AID is required for germinal center derived B cell lymphomas in mice (Pasqualucci et al., 2008), AID-dependent DSBs in the c-myc gene in cells expressing physiological levels of AID have not been detected. However, when AID is over-expressed by retroviral transduction in mouse splenic B cells, chromosomal breaks are induced in the c-myc gene and also near the gene encoding miR-142 (Robbiani et al., 2009). We did not detect AID-dependent Nbs1 signals near or within the miR-142 gene in our experiments. It has been shown that a low frequency of AID-dependent mutations occur in the c-myc gene in msh2−/−ung−/− germinal center cells, but these are repaired in an error-free manner in WT cells (Liu et al., 2008), which might explain our inability to detect AID-dependent DSBs.
Characterization of the AID targets
Presence of WGCW motifs
We interrogated the non-IgH AID target sites for recurrent sequence motifs. AID is known to preferentially deaminate dC in the context of the WRC sequence motif, and the preferred hotspot is WGCW (Martomo et al., 2004). We found a highly significant (p<0.001) association of the WGCW motif with the Nbs1-binding sites (Suppl Fig S12), but no significant enrichment of the more ubiquitous WRC or the more restricted AGCT motif. The mean enrichment is modest (1.4-fold), and not all of the Nbs1-binding sites have the WGCW motif enriched, consistent with the fact that the preference of AID for the WGCW sequence is not absolute, and many other dC’s are targeted by AID (Bransteitter et al., 2004; Peled et al., 2008).
Many AID-dependent DSBs occur at non-transcribed sites
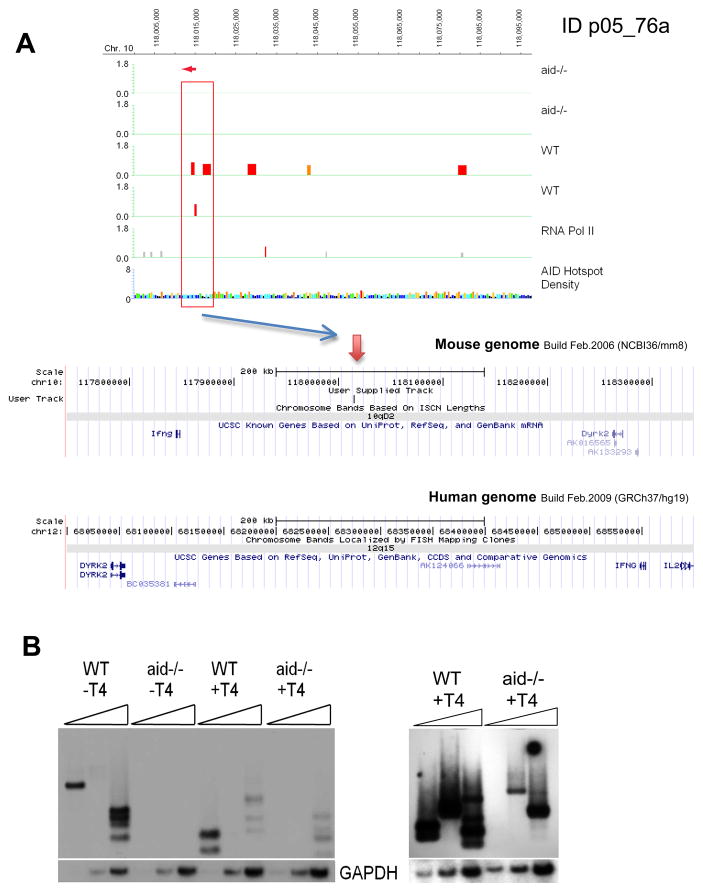
Surprisingly, only 50% of the p<0.05 Nbs1-binding sites and 68% of the p<0.0001 sites are transcribed in activated splenic B cells, as determined by RNA Pol II ChIP-chip (Tables S1, S2). As indicated in the tables, several of the AID-dependent Nbs1 peaks are in transcribed regions that are not within annotated genes; also indicated are peaks that are within genes or located within 10 kb of annotated genes. Importantly, we find that many of the peaks are in non-transcribed intergenic regions. Fig 3 presents an example of AID-dependent Nbs1-binding sites in an intergenic region between the dyrk2 and IFNγ genes, two genes that are frequently amplified in GCB-DLBCL. Other examples of sites in non-transcribed regions are shown in Suppl Figs S3, 4, 5, 7. The finding that only 50 of the p<0.05 and 68% of the p<0.0001 AID-dependent Nbs1 sites are transcribed by Pol II is unexpected, since only transcribed Ig S regions and V genes are targeted by AID. Furthermore, the substrate for AID is single-strand (ss) DNA, which is generated by transcription and by R-loops that are dependent on transcription (Dickerson et al., 2003; Shen and Storb, 2004; Stavnezer et al., 2008; Yu et al., 2003). One possibility is that these non-transcribed sites could be targeted by AID during DNA replication, when DNA is transiently single-stranded. Although AID-dependent DSBs in the Sμregion are only detected in G1 phase and not in S or G2/M phases (Petersen et al., 2001; Schrader et al., 2007), this might be due to locus-specific regulation (Lundgren et al., 1995).
Fig 3.
A: ChIP-chip results for ID p05_76a on chromosome 10. Upper panel: reproducible AID-dependent Nbs1-binding site indicated by red box. Lower panel: mouse genomic region including the peak site on chromosome 10. This region is syntenic to the region on chromosome arm 12q in human, which is amplified in 12.5% of GCB (Lenz et al., 2008). The human map is inverted relative to the mouse map in order to maintain the convention of having the centromere to the left. B. LM-PCR results for this site using site-specific primer OS218, indicated with a red arrow in A. Three DSBs were cloned and confirmed by sequencing.
Three types of repeat elements are highly enriched at AID-dependent Nbs1-binding sites
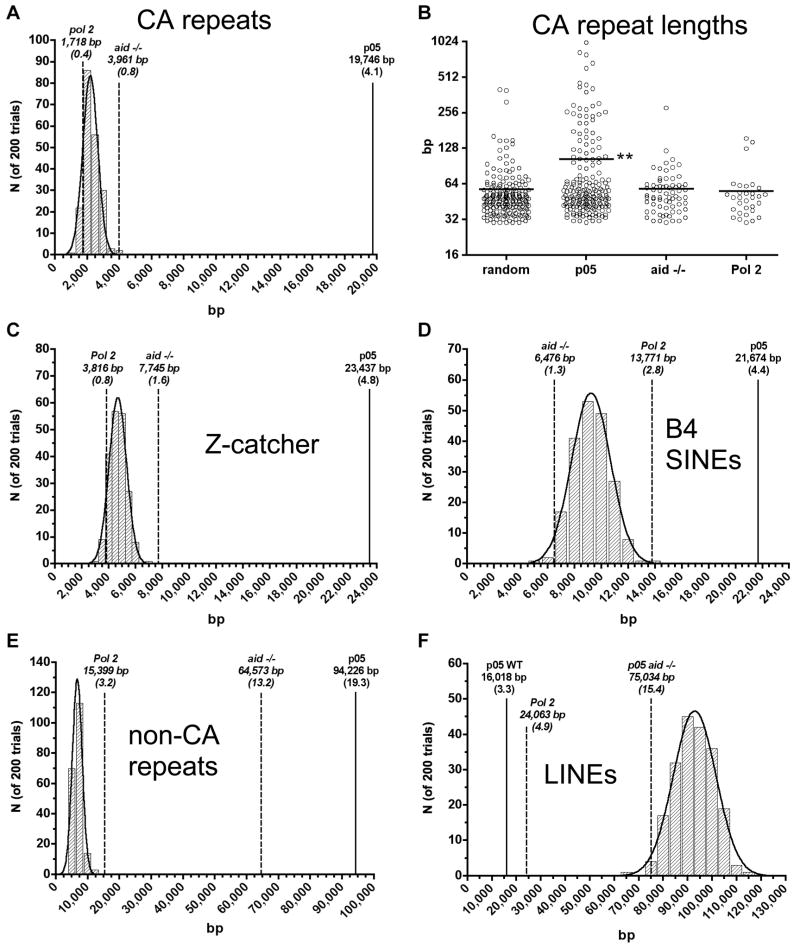
We did not find any consensus transcription factor binding motifs consistently enriched at the AID-dependent Nbs1-binding sites, although several other types of sequences are highly significantly associated with these sites. Tandem CA repeats are highly enriched: 41% of the p<0.05 sites, and 32% of the p<0.0001 sites contain CA repeats at least 30 bp in length. Tables S1 and S2 indicate the sequence elements associated with the AID-dependent Nbs1-binding sites. The enrichment in CA repeat content of the AID-dependent Nbs1 p<0.05 sites relative to random genomic sequences of similar length and chromosome distribution is 9.0-fold (p<0.005), and 5.0-fold relative to AID-independent Nbs-1 sites (Fig 4A). Further supporting the importance of CA repeats for AID-targeting, the average length of the CA repeats at the AID-dependent Nbs1-binding sites is twice as long as CA repeats in randomly chosen sequences, or in Nbs1-binding sites in aid−/− cells, or in Pol II-binding sites (Fig 4B). This alternating purine-pyrimidine sequence is capable of forming Z DNA, a left-handed helix, whereas B DNA is a right-handed helix (Nordheim and Rich, 1983). As the CA repeat forms unstable Z DNA, it transitions between Z and B DNA, forming ss DNA during the transition (Ho, 1994), possibly explaining the ability of AID to attack these regions. Also, two extruded bases are present at the junction between Z and B DNA (Ha et al., 2005). Using Z-catcher, a program that calculates the likelihood of Z DNA formation at a given genomic sequence, we found that ~55% of the AID-dependent Nbs1-sites could potentially form Z DNA at a superhelical density typical of nuclear DNA (Kramer and Sinden, 1997; Li et al., 2009) (Tables S1, S2). For the p<0.05 sites, this is 5.2-fold enriched relative to random genomic sequences (Fig 4C). Although nucleosome-binding precludes Z DNA formation, it was previously shown by use of a transfected Z DNA binding protein domain (ADAR1) that Z DNA is often present in non-transcribed chromosomal regions (Li et al., 2009). CA repeats and Z DNA potential are found in both transcribed and non-transcribed AID-dependent Nbs1-binding sites.
Fig 4. Enrichment of repeats and predicted Z DNA.
(A) CA repeat content of AID-dependent Nbs1 p<0.05 binding sites (solid line) is shown compared to that of length-matched aid−/− Nbs1 and Pol II-binding sites (dashed lines), along with the frequency distribution of CA repeat content in 200 trials of randomly drawn genomic intervals having the same lengths and chromosomal distribution as the AID-dependent Nbs1 p<0.05 sites. Numbers in parentheses are the percent of total bp in CA repeats in the the 365 AID-dependent Nbs1 p<0.05 sites (487,526 bp), or aid−/− or Pol 2 sites, normalized to 487,526 bp. (B) Lengths of individual CA repeats present in the AID-dependent Nbs1, aid−/− Nbs1, Pol II, or random sites. Horizontal lines indicate the respective means (**; p< 0.001, pairwise t test for p<0.05 vs. random, aid−/− Nbs1, and Pol II). (C-F) As in (A), except predictions of Z DNA content by the program Z-catcher (C), B4 SINE (D), non-CA tandem repeat (E), and LINE (F) content, respectively.
Specific members of the short-interspersed-element (SINE) family are also enriched within the AID-dependent Nbs1-binding sites. B4 elements are 2.4-fold enriched relative to randomly selected genomic sequences, and ID SINEs are 1.8-fold enriched. The combined frequency of these elements at the p<0.05 sites is 33% (Table S1). Importantly, SINEs are not enriched at Nbs1-sites found in aid−/− cells (Fig 4D). SINEs have evolved from tRNA genes, and some are transcribed by RNA Pol III. Their transcription is induced by DNA damaging agents and by viral infection, i.e. conditions of stress (Rudin and Thompson, 2001; Walters et al., 2009; White et al., 1990), and we speculate perhaps also by LPS. This might allow AID targeting. Alternatively, due to their relationship to tRNA genes, they might form secondary structures that generate small segments of ss DNA at the ends of hairpins. However, in a search for short palindromic sequences, which theoretically would form hairpins, we did not detect enrichment at the AID-dependent Nbs1-binding sites.
We also found that 44% of the AID-dependent Nbs1-binding sites contain highly significant (p<0.005) enrichment of non-CA tandem DNA repeats (using Tandem Repeat Finder, set at score ≥100) (Benson, 1999). The repeat units vary from 4 to 50 bp in length, with an average of 23 repeats (Table S4). The average length of the repeat region is 501 bp, and the identity between repeats averages 85%. Note that only repeats within the Nbs1-binding region are counted; repeats that extend beyond the Nbs1-binding regions are not included, so these numbers are underestimates. The enrichment of non-CA repeats was not observed for Pol II-binding sites (Fig 4E). Short tandem repeats are found at unstable sites in the human genome, and are often associated with genetic diseases. This is thought to be due to the fact that they can be amplified or deleted due to slipped misalignment during DNA replication (Bzymek and Lovett, 2001; Castel et al., 2010). We hypothesize that if misalignment occurs during DNA replication, and if AID is present in nuclei during S phase, this could generate ss DNA regions that could serve as AID substrates. It is also possible that DNA misalignment during replication could lead to DSBs independent of AID, which would explain our finding that non-CA tandem repeats are also enriched at AID-independent Nbs1-binding sites. However, a significantly greater proportion of AID-dependent Nbs1-binding sites than AID-independent sites contain these tandem repeats (44% vs. 27%; χ2, p<0.0001). These results provide direct evidence that tandem repeat elements lead to DSBs at multiple genomic sites. A total of 91% of the p<0.05 and 97% of the p<0.0001 AID-dependent Nbs1-binding sites are either transcribed, have one or more of the repetitive elements discussed above, or have the potential to form Z DNA.
As discussed above, AID-dependent DSBs in the Sμ region are only detected during G1 phase (Schrader et al., 2007). AID might also deaminate transcriptionally active non-Ig genes and also Z DNA regions during G1 phase. However, it is difficult to understand how the non-CA tandem repeat elements would create ss DNA for AID targeting during G1 phase. Thus, these results suggest that AID might be sufficiently active during S phase to deaminate ss DNA sites created by misalignment during DNA replication.
Whether AID instigates DSBs at non-Ig genes during G1 and/or S phase, it is likely that some of the lesions are repaired during S phase. Recently it was demonstrated that numerous AID-dependent γH2AX foci and chromosomes broken at non-IgH loci are present in activated B cells lacking XRCC2, a protein essential for homologous recombination (Hasham et al. 2010). These cells also showed cell cycle arrest during S phase. These results suggest that at least some of the AID-dependent DSBs detected at non-Ig sites are repaired during S phase by a mechanism involving homologous recombination.
Simple tandem repeats, and less frequently, interspersed repeats, are over-represented among false positive peaks in ChIP-chip experiments (Johnson et al., 2008); however, our evidence indicates that the observed association of repeat sequences with AID-dependent Nbs1 binding sites is genuine. The enrichments for CA repeats and B4 SINEs are specific to the AID-dependent Nbs1 ChIP-chip peaks and not observed for Pol II and aid−/− Nbs1 peaks detected using the same experimental protocol and informatic criteria (Fig 4A,D). Likewise, although non-CA tandem repeats are enriched in both AID-dependent and AID-independent Nbs1 binding sites, the fraction of AID-dependent Nbs1-binding sites containing non-CA tandem repeats is significantly greater than that of the AID-independent Nbs1 sites, and there is no observed enrichment of these tandem repeats at Pol II binding sites. We indicate in Tables S1 and S2 all of the AID-dependent Nbs1-binding sites found in regions of the genome that are segmentally duplicated, as these are also associated with false positive calls (Johnson et al., 2008). Of reproducible AID-dependent Nbs1-binding sites, 9.6% of p<0.05 and 13.5% of p<0.0001 sites map to segmental duplications, higher than found in randomly drawn intervals (6.2%). This is as expected if a given IP’d sequence has homology to more than one genomic site. This problem does not affect the other types of repeated sequences we have analyzed above, because highly repeated sequences are masked from the arrays, and CA repeats and SINES, while present in the IP’d DNA, are detected indirectly by hybridization to adjacent unique sequences.
Interestingly, LINEs (long interspersed elements), derived from retroviruses, are greatly under-represented (5.8-fold) within the AID-dependent Nbs1 binding sites relative to randomly drawn sequences (p<0.005) (Fig 4F). This does not appear to be due to an under-representation of LINE probes on the microarrays, as the LINE content in aid−/− Nbs1 binding sites is similar to that expected from random sequences. LINEs are also under-represented at the Pol II binding sites (3.8-fold, p<0.005), suggesting they are located in repressed/heterochromatic chromosome regions. These data suggest that although the AID-dependent Nbs1-binding sites are not all transcribed, they are more likely to be found in active rather than repressed chromatin regions.
Common fragile sites (CFS) occur in chromosomal regions that are difficult to replicate and are generally found in large transcriptionally active genes (Helmrich et al., 2006). They are identified as gaps or as DSBs in metaphase chromosomes when cells are under replicative stress. Two of the AID-dependent Nbs1 sites (IDs p05_522 and 523) reside within one of the 8 CFS that have been identified in the mouse (Wwox). Since none of the other mouse CFS have AID-dependent Nbs1-binding, this result is difficult to interpret. As chromosome breaks are found at CFS, it is possible that DSBs at CFS are often AID-independent and would be filtered from our listings of AID-dependent Nbs1 sites.
Although AID can deaminate 5meC (Morgan et al., 2004), none of the AID-dependent Nbs1-binding sites we identified occur at CpG islands, as defined by the UCSC genome browser. This might be due to the low efficiency of 5meC deamination compared to unmethylated dC (Bransteitter et al., 2003; Larijani and Martin, 2007), and is consistent with the finding that deamination of 5meC by AID is repaired in an error-free manner in zebrafish, resulting in unmethylated dC (Rai et al., 2008). We asked if the Nbs1-binding sites have a higher frequency of CpG dinucleotides than the genome average, but found they do not (data not shown), although the overall G:C content in the sites is higher than the genome average (46.4% in AID-dependent Nbs1 sites vs 41.7% in the random sequences).
Conclusions
We demonstrate here that physiological levels of AID induce DSBs at reproducible sites throughout the genome in WT mouse B cells induced to undergo CSR. Importantly, several of the AID-induced DSBs occur at sites syntenic with regions that undergo translocations or are amplified or deleted in human B cell lymphomas (Lenz et al., 2008), indicating that AID-induced DNA damage might be the basis of several types of genomic alterations frequently observed in human B cell lymphomas. Of interest, we show that the gene encoding Bcl11a, which is frequently over-expressed and amplified in DLBCL and in Hodgkin’s lymphoma (HL), sustains AID-dependent DNA damage in activated mouse splenic B cells. Previously, the bcl11a gene was shown to be translocated to Sγ in sporadic cases of aggressive B-CLL. Most DLBCL cases have undergone either normal or aberrant switch recombination events, consistent with their derivation from B cells that have undergone or are undergoing CSR.
Although we detected 364 reproducible AID-dependent non-Ig Nbs1-binding sites (p<0.05), there were many other sites that have Nbs1-signals in only one of two experiments, as can be seen in all the figures. By reporting only reproducible AID-dependent Nbs1-binding sites, it is highly likely that we are underestimating the true numbers of genome-wide DSBs due to AID activity in B cells undergoing CSR. For example, Sγ3 signals were only detected in one of two experiments, despite the fact that in both experiments, B cells switched to IgG3 (10–15% of cells). This could be due to infrequent AID targeting and/or to very rapid repair of the DSBs, so that at any one moment a small fraction of cells will have Sγ3 DSBs (Schrader et al., 2007; Schrader et al., 2005; Wu and Stavnezer, 2007). Note that in the Nbs1-ChIP-chip experiments, no S region other than Sμ achieved a signal with a significance of p<0.05, although it is clear that downstream S regions are genuine AID targets in switching B cells. This is not due to repeat-masking of the arrays, as the oligonucleotide probes extend across the S regions. We hypothesize that in cells with normal functioning DNA repair, the vast bulk of DSBs will be transient, due to being rapidly repaired before cell replication, which occurs very rapidly in these cultures.
Recent results using the Hi-C conformation capture assay show that transcriptionally active and non-transcribed genes each co-localize in distinct nuclear compartments (Lieberman-Aiden et al., 2009). Thus, our finding that AID targets are present in both transcribed and non-transcribed loci indicates that AID targets do not all co-localize within nuclei. Although evidence from studying SHM in a mouse Ig light chain gene transgene suggests that a binding site for transcription factors encoded by the E2A gene is essential for targeting AID to these genes (Michael et al., 2003; Tanaka et al., 2010), we did not find any evidence for association of the E2A consensus motif with the non-IgH AID-dependent Nbs1-binding sites. This might be consistent with the low frequency of AID targeting to these non-IgH sites. On the other hand, this result clearly indicates that the E2A motif is not required for AID targeting. We also did not find any other known transcription factor-binding motif enriched at these sites.
Our data suggest that the deamination of genome-wide sites by AID is not due to specific targeting, for example, directed by a transcription factor(s), but rather that AID might non-specifically deaminate a wide variety of sites, with a preference for regions that are transcribed, or able to form Z DNA, or have tandem repeats, or contain B4 or ID SINEs within them. Perhaps these types of elements form ss DNA at some time during the cell cycle. However, it is also possible that the repeat sequences found at the AID-dependent Nbs1-binding sites and DSBs are not only involved in AID targeting but also important for converting SSBs into DSBs, and/or cause delayed repair. This last possibility is suggested by the finding that when the two Nbs1-ChIP-chip experiments are analyzed individually, the CA repeats are less highly enriched (although still highly significant) than in the reproducible p<0.05 peaks. This reduced enrichment in the individual experiments suggests that the reproducible Nbs1 signals are likely to occur at DSBs that are more slowly/less efficiently repaired, therefore more likely to be reproducibly detected than the average AID-induced DSB.
Experimental Procedures
Antibodies
Rabbit anti-Nbs1 antibody was obtained from Abcam (cat # ab32074 clone Y112), anti-RNA Pol II antibody (clone CTD4H8), specific for both unphosphorylated and phosphorylated Pol II, was obtained from Upstate Corp. (cat # 05–623).
Mice
AID-deficient mice were obtained from T. Honjo (Kyoto University, Kyoto, Japan) (Muramatsu et al., 2000), and were backcrossed to C57BL/6. Mice were housed in the Institutional Animal Care and Use Committee-approved specific pathogen-free facility at the University of Massachusetts Medical School. The mice were bred and used according to the guidelines from University of Massachusetts Animal Care and Use committee. For each experiment, splenic B cells from WT mice and aid−/− littermates were analyzed.
Splenic B-cell isolation and culture
Single-cell suspensions were prepared from spleens of 6 to 12 week old mice as described previously (Schrader et al., 1999). Cultures contained LPS (50 μg/ml; Sigma-Aldrich, St. Louis, MO) human BLyS (100 ng/ml; Human Genome Sciences, Rockville, MD) and anti δ-dextran (25 ng/ml; FinaBio, Rockville, MD).
Genomic DNA preparation and LM-PCR
After culture for 2 days, viable cells were isolated by flotation on Lympholyte (Cedar Lane, Ontario, Canada). Cells were imbedded in low melt agarose plugs, and DNA isolated as described (Schrader et al., 2005). For linker ligation, 50 μl 1X ligase buffer was added to the plugs, which were then heated to 62°C to melt the agarose. 20 μl DNA (about 200,000 cell equivalents) was added to 2 μl T4 DNA ligase (2 Weiss units, MBI Fermentas, Hanover, MD), 10 μl ds annealed linker in 1X ligase buffer, 3 μl 10x ligase buffer and 30 μl dH20 and incubated overnight at 18°C. Linker was prepared as described (Schrader et al., 2005). Ligated DNA samples were heated at 70°C for 10 min, diluted 5x in dH20 and then assayed for gapdh DNA by PCR to adjust DNA input prior to LM-PCR. The gene-specific primers (Integrated DNA Technologies, Coralville, IA) were used in conjunction with linker primer (LMPCR.1) to amplify DNA breaks in Sμ and other Nbs1 peak sites. All oligonucleotides used in this study are listed in Table S5. Site-specific primers directed in each direction within each AID-dependent Nbs1 site chosen for analysis were identified using Primer3Plus. The primers were then tested for the ability to detect AID-dependent LM-PCR signals. Three-fold dose titrations of input DNA (1.5, 4.5 and 13.5 μl; ~920, 2770, and 8000 cell equivalent, respectively) were amplified by HotStar Taq (Qiagen) using a touchdown PCR program (Schrader et al., 2005)(35 cycles after touchdown). PCR products were electrophoresed on 1.25% agarose gels and blotted onto nylon membranes (GeneScreen Plus, Perkin Elmer, Waltham, MA). Blots were hybridized with a gene-specific oligonucleotide probe, end-labeled with [γ32P]-ATP at 42°C overnight and washed at 55°C with 2X SSC/0.1% SDS.
Chromatin Immunoprecipitation
ChIP was performed according to a modified version of the Upstate Corp protocol. In brief: B-cells cultured with LPS, anti-δ dextran and BLyS for 2 days were harvested and live cells were isolated by centrifugation on Lympholyte-M (Cedarlane Laboratories, Burlington NC). Cells were washed in PBS and fixed for 10 min at 37°C with 1% formaldehyde. Fixation was quenched by incubating with glycine at a final concentration of 125 mM for 5 minutes at room temperature. Fixed cells were then washed 3 times with PBS and the cell pellet was stored at −80°C. To perform ChIP, the pellet was thawed in Lysis buffer as per the Upstate protocol in the presence of protease and phosphatase inhibitors, and then sonicated 20 times for 10 sec bursts using a VibraCell (Sonics) sonicator at a medium setting. Aliquots containing 2 million cell equivalents of lysate were incubated overnight at 4°C with a mixture of Protein A/Protein G Dynabeads (Invitrogen) coupled with either specific antibody or normal IgG control. Beads were then washed according to the Upstate protocol and DNA was de-crosslinked and eluted by overnight incubation in elution buffer at 65°C. Samples were treated with Proteinase K and RNase A (final concentrations of 125 μg/ml) at 55°C overnight. DNA was precipitated in ethanol after phenol/chloroform extraction and resuspended in Tris-EDTA (10/0.1) pH 7 buffer.
ChIP-chip
ChIP-chip was performed following the protocol provided by Nimblegen Corp. Samples were amplified using the WGA2 kit (Sigma Corp). WGA was performed according to the provided protocol, except the first fragmentation step was omitted. ChIP samples were amplified undiluted, whereas input control samples were amplified at a starting concentration of 1–2 ng/μl. After amplification, samples were labeled with Cy5 nonamers (TriLink Biotechnologies) using random primers and linear amplification with Klenow fragment of E. coli DNA Polymerase I (eBioscience Biotechnologies). Labeled samples were hybridized along with Cy3-labeled input control DNA to whole genome tiling arrays provided by Nimblegen (catalog no. 05340659001), washed with buffers provided by Nimblegen, and scanned using a glass microarray scanner (Agilent). Scan files were processed using the software packages Ringo (Toedling et al., 2007), Nimblescan, MA2C (Song et al., 2007), and Tamalpais (Bieda et al., 2006). Nimblescan and Signalmap were used to prepare the ChIP-chip data figures. The final Nbs1 peak sets (Suppl Tables S1, S2) were obtained using Tamalpais (Bieda et al., 2006; Waterman et al., 1987) modified to work for 2.1 million probe arrays. In short, the number of consecutive probes above threshold values required for peak calling was increased to account for the increased number of probes, as compared to the 384,000 probes for which the software was designed. We used thresholds between the 1st and 15th percentile of highest signal intensity. For a site to achieve a significance of p<0.0001, 6 contiguous probes must show signal at or above the 1st percentile of signal intensity, while 13 consecutive probes are required at the 15th percentile threshold. For a significance p<0.05, 4 to 9 contiguous probes must have signal above the thresholds. AID-dependent Nbs1 binding sites were defined as Nbs1 ChIP-chip peaks found in both biological replicates using WT cells and neither of two experiments using aid−/− cells. RNA Pol II signals were processed using MA2C, inputting data from two replicate experiments and setting the peak detection threshold at FDR ≤5%. Peak sets from experiments with different antibodies and/or cell genotypes were compared using MS Access (Microsoft) or the UCSC Table Browser (http://genome.ucsc.edu).
Three segments of the Nbs1-ChIP arrays failed in one experiment, so we have no results for these regions. These segments are: chr 2: 1– 68,000,000 bp; chr 13: 56,000,000–121,000,000; chr 14: 1–26,000,000.
Bioinformatics analyses
Searches for sequence motifs were performed using Emboss (Rice et al., 2000), MEME (Bailey and Elkan, 1994), and CEAS (Ji et al., 2006). Interspersed repeats were detected using RepeatMasker (A.F.A. Smit, R. Hubley & P. Green RepeatMasker at http://repeatmasker.org) version open-3.2.8 in default mode (Crossmatch search engine), with RM database version 20090604. Counts of individual repeat types were parsed from the raw output files using custom Perl scripts. CA and non-CA tandem repeats were detected using Tandem Repeat Finder version 4.04 (Benson, 1999). For CA repeats, the minimum score was set at 60, and output files were filtered for repeats of CA, AC, TG, GT, or multiples thereof. For non-CA repeats, the minimum score was set at 100 and maximum repeat length at 50. Repeats consisting of ≥90% CA/GT were removed from the output, and where multiple repeats occurred in the same region of sequence, only the highest scoring repeat was retained. Potential Z DNA-forming sequences were detected using Z-catcher (Li et al., 2009) with the superhelicity cutoff set at −0.07. Statistical significance was assessed by running the various sequence analysis programs on 200 sets of randomly drawn DNA sequences having the same length and chromosomal distribution as the AID-dependent Nbs1 p<0.05 peak set (365 intervals). The same analyses were performed on 365 width-matched aid−/− Nbs1 and Pol II peaks, called using Tamalpais with the same parameters as for the WT Nbs1 p<0.05 peak set. Nbs1 binding sites present in segmental duplications were identified using the genomic SuperDups annotation table of the UCSC Genome browser (http://genome.ucsc.edu) and confirmed using BLAT (Kent, 2002).
Supplementary Material
Acknowledgments
We thank Drs. David LaPointe, Zhipeng Weng, and Jeffrey Bailey for advice on bioinformatics analyses and helpful discussions regarding repetitive sequences. We thank Erin K. Linehan for help with LM-PCRs, cloning, and statistics. This research was supported by a grant from NIH, RO1 AI23283 (J.S.), and by the Irvington Institute Fellowship Program of the Cancer Research Institute (J.E.J.G.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.molecularcell.com/com. Microarray data have been deposited into the GEO database (Series Accession # GSE24827).
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–690. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome research. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Calabrese P, Goodman MF. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J Biol Chem. 2004;279:51612–51621. doi: 10.1074/jbc.M408135200. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 2001;98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel AL, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nature Reviews Molecular Cell Biology. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. J Exp Med. 2003;197:1291–1296. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Nussenzweig A. The NBS1-ATM connection revisited. Cell cycle (Georgetown, Tex) 2007;6:2366–2370. doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med. 2007;204:3017–3026. doi: 10.1084/jem.20071289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- Hasham MG, Donghia NM, Coffey E, Maynard J, Snow KJ, Ames J, Wilpan RY, He Y, King BL, Mills KD. Widespread genomic breaks generated by activation-induced cytidine deaminase are prevented by homologous recombination. Nat Immunol. 2010;11:820–826. doi: 10.1038/ni.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome research. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PS. The non-B-DNA structure of d(CA/TG)n does not differ from that of Z-DNA. Proc Natl Acad Sci U S A. 1994;91:9549–9553. doi: 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Li W, Song J, Wei L, Liu XS. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 2006;34:W551–554. doi: 10.1093/nar/gkl322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Li W, Gordon DB, Bhattacharjee A, Curry B, Ghosh J, Brizuela L, Carroll JS, Brown M, Flicek P, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome research. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. BLAT--the BLAST-like alignment tool. Genome research. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer PR, Sinden RR. Measurement of unrestrained negative supercoiling and topological domain size in living human cells. Biochemistry. 1997;36:3151–3158. doi: 10.1021/bi962396q. [DOI] [PubMed] [Google Scholar]
- Larijani M, Martin A. Single-stranded DNA structure and positional context of the target cytidine determine the enzymatic efficiency of AID. Mol Cell Biol. 2007;27:8038–8048. doi: 10.1128/MCB.01046-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci U S A. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xiao J, Li J, Lu L, Feng S, Droge P. Human genomic Z-DNA segments probed by the Z alpha domain of ADAR1. Nucleic Acids Res. 2009;37:2737–2746. doi: 10.1093/nar/gkp124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr, Jenkins NA, Copeland NG. Leukaemia disease genes: large-scale cloning and pathway predictions. Nat Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- Lundgren M, Strom L, Bergqvist LO, Skog S, Heiden T, Stavnezer J, Severinson E. Cell cycle regulation of germline immunoglobulin transcription: potential role of Ets family members. Eur J Immunol. 1995;25:2042–2051. doi: 10.1002/eji.1830250736. [DOI] [PubMed] [Google Scholar]
- Martomo SA, Yang WW, Gearhart PJ. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J Exp Med. 2004;200:61–68. doi: 10.1084/jem.20040691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael N, Shen HM, Longerich S, Kim N, Longacre A, Storb U. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–242. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004;279:52353–52360. doi: 10.1074/jbc.M407695200. [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- Nordheim A, Rich A. The sequence (dC-dA)n X (dG-dT)n forms left-handed Z-DNA in negatively supercoiled plasmids. Proc Natl Acad Sci U S A. 1983;80:1821–1825. doi: 10.1073/pnas.80.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, 3rd, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nat Genet. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Petersen S, Casellas R, Reina-San-Martin B, Chen HT, Difilippantonio MJ, Wilson PC, Hanitsch L, Celeste A, Muramatsu M, Pilch DR, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID Is Required for c-myc/IgH Chromosome Translocations In Vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani DF, Bunting S, Feldhahn N, Bothmer A, Camps J, Deroubaix S, McBride KM, Klein IA, Stone G, Eisenreich TR, et al. AID produces DNA double-strand breaks in non-Ig genes and mature B cell lymphomas with reciprocal chromosome translocations. Mol Cell. 2009;36:631–641. doi: 10.1016/j.molcel.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Edelmann W, Kucherlapati R, Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J Exp Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol. 2007;179:6064–6071. doi: 10.4049/jimmunol.179.9.6064. [DOI] [PubMed] [Google Scholar]
- Schrader CE, Linehan EK, Mochegova SN, Woodland RT, Stavnezer J. Inducible DNA breaks in Ig S regions are dependent upon AID and UNG. J Exp Med. 2005;202:561–568. doi: 10.1084/jem.20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM, Peters A, Baron B, Zhu X, Storb U. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 1998;280:1750–1752. doi: 10.1126/science.280.5370.1750. [DOI] [PubMed] [Google Scholar]
- Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci U S A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Johnson WE, Zhu X, Zhang X, Li W, Manrai AK, Liu JS, Chen R, Liu XS. Model-based analysis of two-color arrays (MA2C) Genome biology. 2007;8:R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Ann Rev Immunol. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa M, Tolarova H, Li Z, Dubois W, Lim S, Callen E, Franco S, Mosaico M, Feigenbaum L, Alt FW, et al. AID expression levels determine the extent of cMyc oncogenic translocations and the incidence of B cell tumor development. J Exp Med. 2008;205:1949–1957. doi: 10.1084/jem.20081007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Shen HM, Ratnam S, Kodgire P, Storb U. Attracting AID to targets of somatic hypermutation. J Exp Med. 2010;207:405–415. doi: 10.1084/jem.20090821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toedling J, Skylar O, Krueger T, Fischer JJ, Sperling S, Huber W. Ringo--an R/Bioconductor package for analyzing ChIP-chip readouts. BMC bioinformatics. 2007;8:221. doi: 10.1186/1471-2105-8-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB life. 2009;61:831–837. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman MS, Gordon L, Arratia R. Phase transitions in sequence matches and nucleic acid structure. Proc Natl Acad Sci U S A. 1987;84:1239–1243. doi: 10.1073/pnas.84.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weniger MA, Pulford K, Gesk S, Ehrlich S, Banham AH, Lyne L, Martin-Subero JI, Siebert R, Dyer MJ, Moller P, Barth TF. Gains of the proto-oncogene BCL11A and nuclear accumulation of BCL11A(XL) protein are frequent in primary mediastinal B-cell lymphoma. Leukemia. 2006;20:1880–1882. doi: 10.1038/sj.leu.2404324. [DOI] [PubMed] [Google Scholar]
- White RJ, Stott D, Rigby PW. Regulation of RNA polymerase III transcription in response to Simian virus 40 transformation. Embo J. 1990;9:3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Stavnezer J. DNA polymerase beta is able to repair breaks in switch regions and plays an inhibitory role during immunoglobulin class switch recombination. J Exp Med. 2007;204:1677–1689. doi: 10.1084/jem.20070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilop A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.