Abstract
Differentiation often requires conversion of analogue signals to a stable binary output through positive feedback. Hedgehog (Hh) signalling promotes myogenesis in the vertebrate somite, in part by raising the activity of muscle regulatory factors (MRFs) of the Myod family above a threshold. Hh is known to enhance MRF expression. Here we show that Hh is also essential at a second step that increases Myod protein activity, permitting it to promote Myogenin expression. Hh acts by inducing expression of cdkn1c (p57Kip2) in slow muscle precursor cells, but neither Hh nor Cdkn1c is required for their cell cycle exit. Cdkn1c co-operates with Myod to drive differentiation of several early zebrafish muscle fibre types. Myod in turn up-regulates cdkn1c, thereby providing a positive feedback loop that switches myogenic cells to terminal differentiation.
Keywords: muscle, Cdkn1c, zebrafish, Hedgehog, myod, myog, p57kip2
INTRODUCTION
Positive feedback helps commit cells to a differentiation step. A classical example is suggested by Myod auto-regulation during myogenesis (Weintraub, 1993). Myod is a transcription factor required for timely differentiation of certain muscle fibre populations in mice and zebrafish (Hammond et al., 2007; Hinits et al., 2009; Kablar et al., 1997; Maves et al., 2007; Sabourin et al., 1999; Yablonka-Reuveni et al., 1999). Myod reporter genes are down-regulated in myod null mice (Chargé et al., 2008; Kablar et al., 2003; Kablar et al., 1997), suggesting that positive feedback by Myod acts at the level of myod gene transcription. Indeed, Myod expression peaks early in differentiation of cultured myoblasts (Halevy et al., 1995). However, knockdown of Myod protein in zebrafish does not appear to decrease myod mRNA (Hinits et al., 2009; Maves et al., 2007). Thus, although Myod is essential for myogenesis in animals lacking the related Myf5/Mrf4 proteins (Kassar-Duchossoy et al., 2004; Rudnicki et al., 1993), whether Myod auto-regulation is required for muscle cell terminal differentiation is unclear.
MRF activation and cell cycle exit are two key steps in terminal myoblast differentiation (Andres and Walsh, 1996; Halevy et al., 1995). Studies in cell culture reveal that Myod can help drive cell cycle exit in multiple ways. One route is through activation of the cyclin-dependent kinase inhibitor Cdkn1a/p21Cip1, a protein that regulates cell cycle exit in G1 through its action on several CDKs (Nagahama et al., 2001). However, in vivo evidence that Cdkn1a is important for myogenesis is weak because cdkn1a null mice are viable and fertile (Deng et al., 1995). cdkn1a is a member of the Cip/Kip family of CDK inhibitors, which include cdkn1b/p27Kip1 and cdkn1c/p57Kip2 (Nagahama et al., 2001). Although null mutations of cdkn1b or cdkn1c are also viable, double mutants for cdkn1a;cdkn1c show, among other defects, a severe reduction in muscle differentiation (Fero et al., 1996; Yan et al., 1997; Zhang et al., 1997; Zhang et al., 1999). Why myogenesis fails in these mutants is unknown. Beyond mice, the function of the Cdkn1 family in myogenesis has only been analysed in Xenopus laevis, where the Cdkn1 gene p27Xic1 is required for myogenesis and can cooperate with Myod (Vernon and Philpott, 2003). Whether and how Cdkn1 proteins cooperate with Myod in embryonic myogenesis is unknown.
To understand the common themes of vertebrate myogenesis, we and others have recently begun to analyse the function of Myod and other MRFs in the zebrafish (Hammond et al., 2007; Hinits et al., 2009; Maves et al., 2007). As in mice, either Myod or Myf5 is required for early myogenesis in the somite (Hammond et al., 2007; Rudnicki et al., 1993). Distinct populations of muscle fibres in the zebrafish somite require distinct extrinsic signals in order to express MRFs and undergo terminal differentiation (Barresi et al., 2000; Blagden et al., 1997; Chong et al., 2007; Coutelle et al., 2001; Groves et al., 2005; Hammond et al., 2007; Hirsinger et al., 2004; Lewis et al., 1999; Ochi et al., 2008). For example, Hedgehog (Hh) signals from the ventral midline are required for proper myf5 and myod expression and slow muscle formation by adaxial cells (Barresi et al., 2000; Coutelle et al., 2001; Lewis et al., 1999; Ochi et al., 2008; Schauerte et al., 1998). Strikingly, however, MRF expression is initiated normally in the absence of Hh, suggesting that Hh is necessary for MRF maintenance, not initial induction (Ochi et al., 2008). Thus, expression of both myod and myf5 mRNAs is not sufficient to initiate an auto-regulatory loop maintaining MRF expression in vivo. Why not?
Here we show that a second action of Hh on muscle precursor cells is to activate expression of cdkn1c/p57kip2, initiating a positive feedback loop that stabilizes Myod protein and permits it to activate myogenin expression and drive muscle terminal differentiation. Cdkn1c does not act by promoting cell cycle exit. These findings reveal a role of Cdkn1c in terminal differentiation that goes beyond its known function in other cells during cell cycle exit.
Materials and methods
Zebrafish lines and maintenance
Null mutant lines smob641 (Varga et al., 2001), myf5hu2022 (Hinits et al., 2009) and shhatbx392 (Schauerte et al., 1998) were maintained on King’s wild type background. Staging and husbandry were as described previously (Westerfield, 1995).
In situ mRNA hybridization, immunohistochemistry and Western analysis
In situ mRNA hybridization for myf5, myod, myog, mrf4, eng2a and prdm1 was as described previously (Hinits et al., 2009). Additional probes were hsp90a, cdkn1a (Lee et al., 2008), cdkn1b (IMAGE 7002450), cdkn1bl (IMAGE 6799784) and cdkn1c (IMAGE 6892669). Embryos for immunohistochemistry were fixed in 4% PFA for 30 min and stained as described (Blagden et al., 1997; Groves et al., 2005). Primary antibodies used were MyHC (A4.1025) (Blagden et al., 1997), slow MyHC (F59) (Devoto et al., 1996), zebrafish Myod (Hammond et al., 2007), Myogenin (Hinits et al., 2009) and ß-Tubulin (Amersham N357). Myod antibody was absorbed against 8-24 hpf methanol-fixed embryos before use. HRP- (Vector) or Alexa dye-conjugated (Invitrogen) secondary antibodies were used with Citifluor mountant (Agar). Confocal images were collected on a Zeiss LSM510. Western analysis was as described (Hinits et al., 2009).
Embryo Manipulations
Embryos were injected with MOs described previously to myod (2-4 ng), myf5 (2-4 ng), myog (1-2 ng) (Hinits et al., 2009) and cdkn1c ATG MO (1-2 ng) (Shkumatava and Neumann, 2005), Park MO (3-5 ng) (Park and Chung, 2001) and 5′ MO (1-2 ng 5′tcaatgccgtgagccgacgtttgtt3′). Controls were vehicle or, in Fig. 6D, irrelevant mismatch MO 5′tgcttgatcatcctgagacaggcag3′. Cyclopamine (200 μM in fish medium) or vehicle control was added at 30-50% epiboly to embryos whose chorions had been punctured with a 30G hypodermic needle. BrdU treatment was performed as described (Appel et al., 2001). RNA (100 pg) was made with Ambion Megascript kit from pSP64T-shha or pßUT3 containing full length zebrafish myod subcloned into SacI/SalI sites and injected into embryos at 1-2 cell stage.
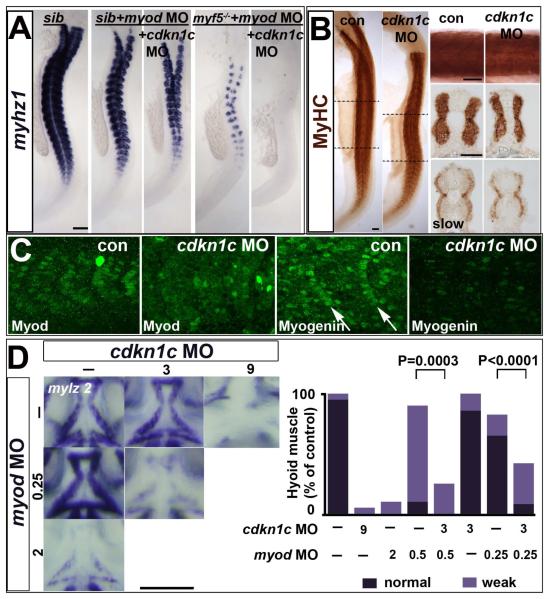
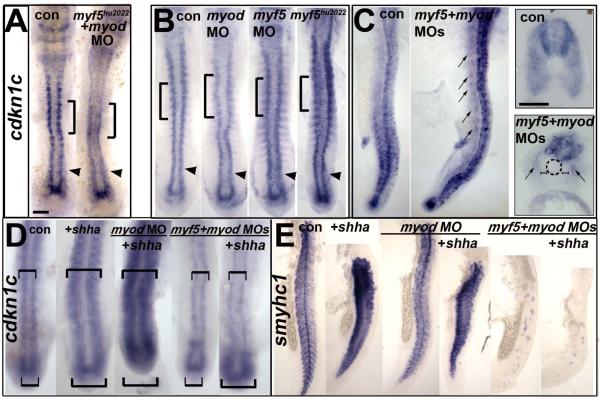
Figure 6. Cdkn1c drives fast myogenesis through Myod.
In situ mRNA hybridization (A,D) or protein immunodetection (B,C) at 24 hpf (A-C; lateral flatmount) or 72 hpf (D, dorsal flatmount) of embryos injected with cdkn1c MO. (A). Cdkn1c MO exacerbated somitic fast muscle loss induced by myod MO, and ablated residual fast muscle in mutant myf5hu2022;myod knockdown. Bar: 100 μm (B). Cdkn1c MO alone diminishes fast muscle differentiation, without reducing slow muscle. Dashed lines indicate positions of upper and lower transverse sections. Bars: flatmounts 100 μm, somite lateral zoom and sections 50 μm (C). Cdkn1c MO diminished Myod immunoreactivity in nuclei of nascent fast muscle and Myogenin immunoreactivity in more mature somites, particularly the expression at the posterior somite border (arrows). Bar: 50 μm. (D). Cdkn1c MO (9 ng) reduced ventral head myogenesis in a similar manner to myod MO (2 ng). Low doses of cdkn1c MO (3 ng) or myod MO (0.25 ng) that alone had little effect, synergised to reduce myogenesis in intermandibular, interhyoideus and hyohoideus muscles. Bar: 100 μm. Chart quantifies the extent of ventral hyoid muscle defects after injection of the indicated ng of each MO/embryo, with differences tested by Χ2 test. All embryos were classified as having either ‘normal’, ‘weak’ or ‘no’ mylz2 mRNA in the three stated muscles. ‘Normal’ embryos (dark bars) had strong mylz2 expression, as shown in control panel. ‘Weak’ embryos (light bars) had mylz2 expression visibly below the control range, as shown in the 2 ng myod MO panel. ‘No’ embryos entirely lacked mylz2 expression, as in the 9 ng cdkn1c MO panel, are shown as white space above the bars. Data shown has n = 20 for each condition from within a single experiment.
Full length zebrafish cdkn1c from IMAGE:6892669 was subcloned into the XbaI site of hsp70-4-MCS-IRES-mGFP6 (Hinits et al., 2007), adding a 5′ myc-tag and XbaI linkers with primers: 5′TCTTCTAGAATGCAGAAGCTGATCTCAGAGGAGGACCTGATGGCAAACGTGGACGTATC3′ and 5′TCTTCTAGATCATCTAATAGTTTTACGT3′ and sequence verified. DNA encoding this or Xenopus p28Kix1, the closest cdkn1c homologue (Habermann et al., 2004), under hsp70/4 heatshock control (Yamaguchi et al., 2005) was injected at 1 cell stage. Heat shock 39°C was applied at 5-12 ss for 1 hour and rescue analysed 4-6 somite stages later. For analysis of whether an embryo was rescued or not, embryos were sorted into those with and without detectable MyHC. All, or in larger experiments a sample, of MyHC positive embryos were flatmounted and individually scored as either ‘strong’ or ‘weak’ based on the number and intensity of MyHC positive adaxial fibres. The data shown are the fraction of ‘strong’ embryos out of the entire sample across all experiments. Controls shown are heat-shock without vector injection, but also included vector injection without heat-shock. Smob641 mutants were identified by myod mRNA pattern.
RESULTS
Myod protein accumulation is regulated by Hedgehog
Myf5 and Myod act together in adaxial cells to drive slow myogenesis (Hammond et al., 2007; Hinits et al., 2009; Maves et al., 2007). Initiation of myf5 and myod expression in adaxial cells is independent of Hh (Ochi et al., 2008). However, smoothened (smob641) mutant embryos, which lack an essential component of the Hh signaling pathway, fail to maintain myod or myf5 expression in adaxial cells and do not form adaxial-derived slow muscle fibres (Barresi et al., 2000). The same result arises in embryos treated with cyclopamine (cyA), a drug that inhibits all Hh signalling and prevents slow myogenesis (Barresi et al., 2001). In contrast, sonic hedgehog a mutants (shhatbx392, sonic you) contain significant, albeit reduced, quantities of slow muscle, probably because two other Hh genes, shhb and ihha, are expressed in ventral midline tissues prior to the 5 som stage (5 ss) (Currie and Ingham, 1996; Ekker et al., 1995; Lewis et al., 1999). Surprisingly, we noticed that smob641 and shhatbx39 mutants have an almost indistinguishable failure of myf5 and myod mRNA maintenance in slow cells (Fig. 1A; see Supplementary Table S1 for numbers of embryos analysed and outcome(s) of all experiments). The sole difference is that, after an indistinguishable decline in expression in anterior presomitic mesoderm (PSM), the differentiated slow muscle cells in shhatbx392 mutant somites retain low levels of re-accumulated myod transcripts.
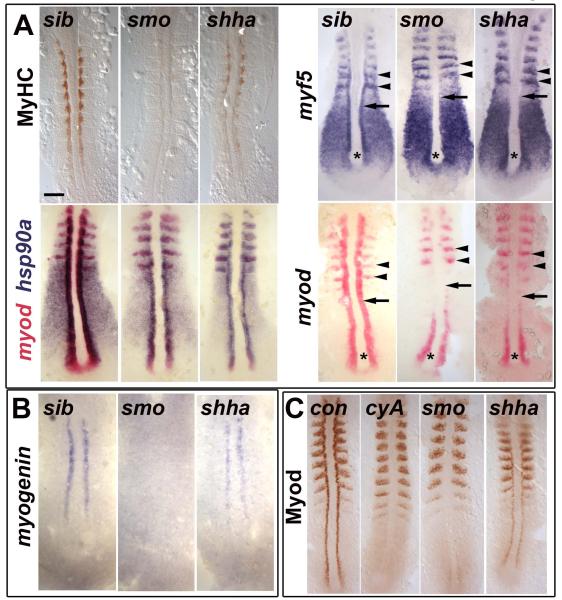
Figure 1. Myod protein level correlates with slow myogenesis in Hh signalling mutants.
In situ mRNA hybridization for myf5, myod, myog and hsp90a and immunodetection of Myod and Myosin Heavy Chain (MyHC) in dorsal flatmounts of 5 ss stage control, cyclopamine-treated (cyA), smob641 and shhatbx392 mutant embryos. Mutants were identified by significant reduction in one or both markers in ~25% of lay (shhatbx392: 9/41; smob641 14/50). Anterior to top. Bar: 100 μm. (A). Absence of MyHC in smob641 but only reduction in shhatbx392 mutants, despite indistinguishable loss of myod and myf5 mRNAs in adaxial cells in anterior pre-somitic mesoderm (arrows). Note that myf5 and myod expression in tailbud (asterisk) and presumptive fast muscle precursors (black arrowheads) is little affected by Hh manipulation. Shhatbx392 differs from smob641 only in expression of myod in differentiating adaxial muscle in somites (white arrowheads). (B). Myog mRNA is present in adaxial cells of shhatbx392 but not smob641 mutants. (C). Myod protein is absent in adaxial cells of cyA and smob641 embryos but weakly present in shhatbx392 mutant. Control embryos were either shhatbx392 siblings or untreated wild type.
Adaxial cells are not lost (Coutelle et al., 2001; Hirsinger et al., 2004). In cyA-treated, smob641 and shhatbx392 embryos, adaxial cells retain strong hsp90a expression throughout the adaxial cell region (Fig. 1A). Similarly, adaxial cells, which are unlabelled by S-phase marker BrdU in wild type, remain unlabelled in cyA or smob641 embryos (Fig S1). In 31 cyA or 15 smob641 embryos, none of the approximately 480 adaxial cells per 8 ss embryo were BrdU labelled. Similarly, no Brdu labelling was observed in adaxial cells at later stages (data not shown). Thus, differential adaxial cell survival or proliferation does not account for the absence and presence of slow muscle in cyA/smob641 and shhatbx392 mutants, respectively.
The formation of slow fibres in shhatbx392 mutants correlates with myogenin (myog) expression. shhatbx392 mutants express myog mRNA, whereas smob641 or cyA embryos do not (Fig. 1B and data not shown). Myod protein was undetectable in adaxial cells of smob641 and cyA embryos, although expression in presumed fast muscle precursors in the lateral somite is normal. In contrast, shhatbx392 embryos have significantly more Myod immunoreactivity in adaxial cell nuclei in PSM, although still much less than their siblings (Fig. 1C). We conclude that weak residual Hh signalling in shhatbx392 mutants promotes accumulation of Myod protein and myog expression, which might account for the residual slow myogenesis.
Myod promotes myogenin expression
As slow muscle formation correlates with MRF accumulation in adaxial cells, we asked whether Myod and/or Myog proteins drive adaxial slow muscle differentiation. Injection of wild type embryos with an antisense morpholino oligonucleotide (MO) to myod, which we have previously shown to knockdown Myod protein (Hammond et al., 2007), prevents myog mRNA accumulation in adaxial cells at 5 ss (Fig. 2A). This delays, but does not prevent, slow fibre formation, because of the presence of Myf5 (Hinits et al., 2009). Thus, in nascent adaxial cells Myod is rate limiting for myog expression and muscle differentiation.
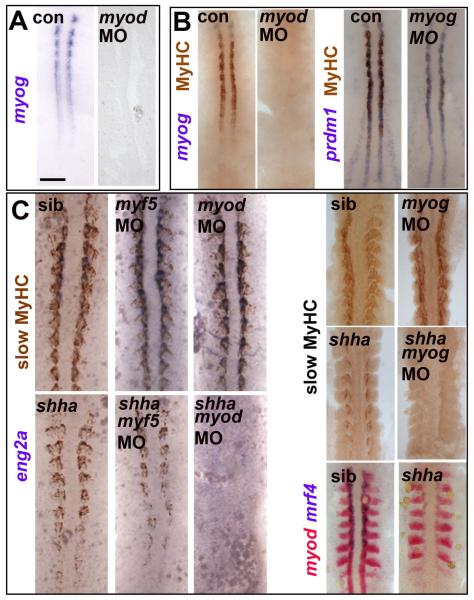
Figure 2. Myod-driven myogenin expression permits slow fibre formation in shha mutant.
Myog mRNA or MyHC accumulation in uninjected, myod MO- or myog MO-injected embryos from wild type or shhatbx/+ in-cross. Dorsal flatmounts of 5 ss (A,B) or 10-15 ss (C) embryos. Bar: 100 μm. (A). Myog mRNA is ablated by myod MO injection into wild type. (B) Myod and myog MOs each delay adaxial myogenesis. (C) Myod and myog, but not myf5, MOs ablate adaxial myogenesis in shhatbx392 mutants (21/79), but not from siblings.
We next tested whether Myogenin is also required for slow fibre formation. Two distinct myog MOs knockdown Myogenin protein and delay slow muscle differentiation (Hinits et al., 2009). However, myog MOs do not ultimately prevent slow myogenesis (Fig. 2C). Therefore, Myod-driven myog expression hastens slow muscle formation, possibly by raising overall MRF activity.
Myod and Myogenin are essential for slow myogenesis in shha mutant
We tested the role of MRFs in shhatbx392 mutants, a situation in which MRF activity may be limiting after failure of myf5 and myod mRNA maintenance (Fig. 1A). Injection of either myod or myog MOs into a shhatbx392/+ incross ablates the residual slow muscle normally observed in mutants (Fig. 2C). Myf5 MO has no such effect. Mrf4 is not expressed early in shhatbx392 mutants, and therefore cannot drive slow fibre formation (Hinits et al., 2007). Thus, by driving myog expression, Myod accounts for the presence of slow fibres in shhatbx392 mutants. As Hh signalling is essential for adaxial slow myogenesis and yet the accumulation of myod and myf5 mRNAs are indistinguishable between smob641 and shhatbx392 mutants, some other action of weak Hh signalling must account for the activation of Myod and induction of myog.
Cdkn1c is a Hh-dependent myogenic regulator present in adaxial cells
We hypothesized that residual Hh signalling promotes Myod stability and/or activity. In the retina, Shha drives terminal differentiation of neurons through Cdkn1c, also known as p57Kip2 (Shkumatava and Neumann, 2005). Loss of Cdkn1 activity leads to failure of murine myogenesis (Zhang et al., 1999). Zebrafish cdkn1c is expressed in adaxial cells throughout their differentiation, as well as in the central nervous system and the base of the notochord (Fig. S2; Park et al., 2005). Cdkn1c mRNA is essentially eliminated from adaxial cells, but not from the notochord, by Hh blockade (Fig. 3A). Conversely, Shha over-expression up-regulates Cdkn1c (Fig. 3A). In contrast, shhatbx392 mutants have reduced but significant levels of adaxial cdkn1c mRNA; expression is near normal as cells leave the tailbud, but is much below the control level in more anterior adaxial cells of nascent somites (Fig. 3A). The correlation of residual cdkn1c mRNA with residual Myod protein (compare Figs 1C and 3A) makes it an excellent candidate Myod regulator.
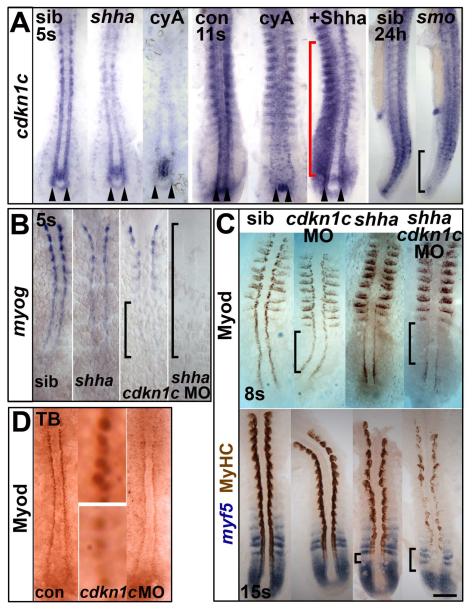
Figure 3. Hh causes Cdkn1c to stabilize Myod.
(A). Cdkn1c mRNA is Hh-dependent in adaxial cells. Shhatbx392 or smob641 mutation or cyA treatment reduces cdkn1c mRNA. Note that loss was proportional to reduction in Hh signal (arrowheads and black bracket). Shha mRNA injection caused unilateral somitic cdkn1c up-regulation (red bracket). (B,C). Cdkn1c 5′ MO injection into embryos from a shhatbx392/+ incross reduced adaxial myog mRNA (B), Myod or MyHC proteins (C). Note delay in myog mRNA and reduction in Myod protein (brackets). (D). Cdkn1c ATG MO injection reduces adaxial Myod at tailbud stage. Bar: 100 μm.
Cdkn1c promotes Myod accumulation in adaxial cells
We used three different cdkn1c MOs, two previously shown to knockdown Cdkn1c in brain and retina (Park et al., 2005; Shkumatava and Neumann, 2005), and were able to reduce myog mRNA in shhatbx392 mutant embryos and delay it in siblings (Fig. 3B). Cdkn1c MO greatly decreased slow muscle differentiation in shhatbx392 mutants, but siblings were not noticeably affected (Fig. 3C lower panel). We conclude that Cdkn1c promotes slow fibre terminal differentiation when MRF levels are limiting.
Cdkn1c acts downstream of myod mRNA accumulation. Cdkn1c MOs did not reduce myod or myf5 mRNA levels in the adaxial region, although myod expression in differentiated slow fibres within somites of shhatbx392 mutants was diminished, paralleling the reduction in slow fibre differentiation (Figs 3C and S3A). Cdkn1c has been suggested both to stabilize Myod protein through its effect on CDK2 activity and to enhance Myod DNA binding through direct protein:protein interaction (Reynaud et al., 2000; Reynaud et al., 1999). Cdkn1c MO-injected wild type embryos have reduced Myod immunoreactivity in adaxial nuclei, which is not apparent in the more numerous lateral fast muscle precursors (Fig. 3C,D). Consistent with this, Western analysis failed to reveal a significant alteration in level or size of Myod protein in 24 hpf embryos (Fig. S3B). Cdkn1c MO-injected shhatbx392 mutant (cdkn1c−;shhatbx392) embryos had Myod in the tailbud region, but it was reduced and did not persist in numerous adaxial cells in more anterior PSM, as it did in control shhatbx392 mutants (Fig. 3C upper panel). In smob641 mutants, however, even less residual Myod protein was detected than in cdkn1c−;shhatbx392, paralleling their lower formation of residual slow fibres (Fig. 1C). Thus, in adaxial cells, Cdkn1c promotes Myod protein accumulation in vivo.
MRFs maintain Cdkn1c expression in adaxial cells
In mice, Cdkn1c co-operates with Cdkn1a to drive myogenesis (Zhang et al., 1999). Among the three additional zebrafish cdkn1 genes, cdkn1a (p21Cip1), cdkn1b and cdkn1b-like (both p27Kip1 homologues), none are significantly expressed in adaxial cells, although cdkn1b mRNA is observed in the lateral somite, where fast muscle forms (Fig. S4). Thus, Cdkn1c appears to be the major Cdkn1 involved in zebrafish adaxial myogenesis.
Mammalian Cdkn1a and Xenopus p27Xic1 are Myod targets that mediate cell cycle withdrawal (Halevy et al., 1995; Vernon and Philpott, 2003; Wang and Walsh, 1996). Knockdown of Cdkn1c did not lead to proliferation of adaxial cells (none of the ~480 adaxial cells in any of 62 morphant embryos, Fig. S1, Table S1). We therefore tested whether MRFs were required to maintain cdkn1c expression. When both Myf5 and Myod proteins were depleted in the same embryo, adaxial cdkn1c mRNA was reduced in PSM and essentially ablated in somites (Fig. 4A). Nevertheless, significant adaxial cdkn1c mRNA was present in PSM adaxial cells flanking the tailbud (Fig. 4A). Loss of myf5 alone did not diminish cdkn1c mRNA, but myod MO reduced cdkn1c mRNA, particularly in rostral somites (Fig. 4B). Conversely, when Myod was over-expressed by mRNA injection, ectopic cdkn1c and myog mRNAs and slow myogenesis were observed in both somitic and head mesoderm (Fig. S5). Thus, MRFs drive cdkn1c expression, implicating Cdkn1c in a positive feedback loop whereby MRF activity induces Cdkn1c, which then enhances MRF protein activity leading to terminal differentiation.
Figure 4. Cdkn1c expression depends on Hh in tailbud and on MRFs anteriorly.
MO-injected wild type (B-D) or myf5hu2022 incross embryos (A,B) co-injected with shha mRNA (D,E) analysed for cdkn1c mRNA in 12-15 ss dorsal (A,B,D) or 24 hpf lateral flatmount, dorsal to right (C,E). Anterior is to top. Bars: flatmounts 100 μm, sections 50 μm. (A-C). Adaxial cdkn1c mRNA accumulation initiates independent of MRF activity (arrowheads), but fails to be maintained (bracket). Myod MO reduces cdkn1c expression in older somites (brackets) but tailbud is unaffected (arrowheads). Myf5 and myod knockdown ablates somite cdkn1c mRNA except in small groups of cells lateral to the un-migrated adaxial cells (arrows). Neural, cloacal and transient notochordal expression is unaffected. (D). Over-expression of Shha promotes cdkn1c up-regulation in anterior PSM and somite in contorls and after Myod knockdown, but not in myf5+myod double morphants (upper brackets). Note that Shha up-regulates cdkn1c in the lateral tailbud of myf5+myod double morphants (lower brackets), but this is not maintained in anterior PSM. (E). Over-expression of Shha promotes ectopic smyhc1 expression at 24 hpf in control or myod morphants, but not in myf5+myod double morphant embryos.
Hh promotes Cdkn1c expression specifically in tailbud
To distinguish better between a direct effect of Hh on cdkn1c expression and an indirect effect via MRF activity, we next knocked down Myod activity in embryos expressing ectopic Shha. Shha could still induce ectopic cdkn1c and ectopic slow fibres, showing that Myod was not essential (Fig. 4D,E). This finding raised the possibility that Myf5, which is also induced by ectopic Shha (Coutelle et al., 2001), might help mediate cdkn1c up-regulation. Knockdown of both Myf5 and Myod does not prevent Shha-driven up-regulation of cdkn1c mRNA in the lateral tailbud (Fig. 4D), confirming that nascent adaxial cells in tailbud regions do not require MRF activity, but that Hh signalling acts independently to promote cdkn1c expression. Strikingly, however, this ectopic cdkn1c mRNA is not maintained as tailbud cells enter the anterior PSM, and ectopic slow fibres do not form at 24 hpf, demonstrating that MRF activity is essential for the maintenance of Cdkn1c and slow myogenesis in the lateral PSM region (Fig. 4D,E).
Cdkn1c rescues adaxial myogenesis in the absence of Hedgehog signalling
We next asked whether Cdkn1c can act in the absence of Hh signalling. A notable, but little emphasized, characteristic of both smob641 mutant and cyA-treated embryos, is the presence of rare residual slow muscle fibres, that form prior to the generation of secondary slow fibres (Barresi et al., 2001; Hirsinger et al., 2004)(Fig. 5). We dub this process ‘escape’, and asked whether it depends on Cdkn1c. Knockdown of Cdkn1c in smob641 prevents most escape, whether assayed at 15 ss or 24 hpf (Fig. 5A,C). This suggests that Cdkn1c enhances slow myogenesis in the absence of Hh signalling, presumably through MRF-driven positive feedback.
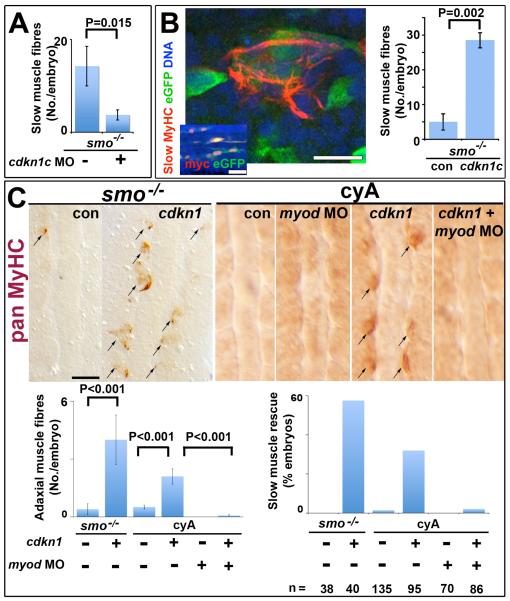
Figure 5. Cdkn1c rescue of slow myogenesis is Myod-dependent.
Slow MyHC (A,B confocal sections) or pan MyHC (C, dorsal flatmount) in embryos lacking Hh signalling. (A). Cdkn1c MO injection reduces residual slow muscle fibres in 24 hpf smob641 mutants. (B). Mosaic over-expression of zebrafish cdkn1c induced by heat-shock of hs70/4:cdkn1cIRESeGFP at 12 ss rescues slow MyHC at 18 ss. Inset shows coexpression of myc-Cdkn1c and eGFP. eGFP mosaicism was ≤5%, which corresponds to ~36 somitic adaxial cells at 18 ss. Bar: 10 μm; inset 50 μm. (C). Mosaic Xenopus cdkn1 expression rescues adaxial muscle in smob641 mutant or cyA-treated embryos at 10-12 ss. Cdkn1-driven rescue is prevented by myod knockdown. Charts show rescue of slow fibres (left, mean±sem; Wilcoxon significance test) and fraction of embryos showing rescue (right; n = total embryos). Note that mosaic expression in ~10% of adaxial cells would mark ~33 somitic slow fibres per embryo at this stage. Bar: 50 μm.
A stringent test of the ability of Cdkn1c to promote MRF activity is to rescue adaxial myogenesis in embryos that entirely lack Hh signalling and fail to maintain MRF mRNAs. We induced zebrafish or Xenopus Cdkn1c in either cyA embryos or a smob641/+ incross by heat shock at 8-15 ss and assayed myogenesis at 12-18 ss (Fig. 5B,C). Isolated adaxial cells in smob641 or cyA embryos expressing Cdkn1c and eGFP frequently differentiated into muscle, whereas control injected or un-heat-shocked adaxial cells did not (Fig. 5B,C). Rescued cells were restricted to anterior somites but less than half accumulated detectable eGFP, perhaps due to the short period post-heat-shock (Fig. 5B). No such rescue was observed in cyA-treated heat-shocked embryos injected with myod MO in addition to the cdkn1c heat-shock construct (Fig. 5C). As adaxial cells in smob641 or cyA embryos do not proliferate despite failing to differentiate (Fig. S1), Cdkn1c does not simply act by forcing cell cycle exit. We conclude that Cdkn1c activity is sufficient to rescue adaxial myogenesis in the absence of Hh, presumably by promoting MRF activity. Thus, by promoting Myod accumulation from the otherwise transiently induced myod mRNA, Cdkn1c triggers a positive feedback loop that rescues adaxial myogenesis.
Cdkn1c drives fast fibre differentiation
Cdkn1c is expressed in fast myogenic cells from mid-somitogenesis until at least 24 hpf (Figs 3A and S2). Reduction of MRF activity severely curtails cdkn1c expression throughout the somites, including the precursors of fast cells, but does not inhibit neural expression (Fig. 4C). Myf5 and Myod knockdown leaves small groups of cells in the medial region of each somite retaining cdkn1c mRNA (Fig. 4C). These are the Hh-dependent residual fast muscle fibres that escape MRF knockdown, as shown by their expression of myhz1 mRNA (Hinits et al., 2009). Knockdown of Cdkn1c entirely ablates these residual fast fibres, suggesting that Cdkn1c activity enhances the low residual activity of Myod in myf5;myod morphants or myf5 mutants injected with myod MO (Fig. 6A and data not shown). Moreover, loss of Cdkn1c enhances the loss of fast muscle caused by myod MO alone (Fig. 6A). Therefore, Cdkn1c can also contribute to differentiation of fast fibres.
To test for the function of Cdkn1c in otherwise un-manipulated fast muscle, we injected cdkn1c MOs into embryos and analysed expression of fast muscle markers. Little change in myhz1 mRNA or MyHC immunoreactivity was apparent in cdkn1c morphants when viewed in wholemount, yet sectioning at 24 hpf revealed a reduction in fast muscle area (Fig. 6B). No obvious change in accumulation of any MRF mRNA was detected in cdkn1c morphants, but Myod and Myogenin proteins were reduced in specific cell groups (Figs 6C and data not shown). Thus, Cdkn1c is required for efficient fast muscle differentiation, and appears to act on MRFs downstream of their transcription.
Myod and Cdkn1c interact genetically
To test the hypothesis that Myod and Cdkn1c act in the same pathway to drive muscle differentiation we looked for genetic interactions. Loss of Myod function in zebrafish leads to ablation of head myogenesis (Hinits et al., 2009). Cdkn1c is expressed in head muscle anlagen at 48 hpf, just as differentiation commences (Fig. S2). Cdkn1c morphant embryos showed a similar reduction in head myogenesis, which was particularly marked in the ventral hyoid muscles (Fig. 6D). Co-injection of low doses of cdkn1c MO and myod MO, which alone had no significant effect on myogenesis, led to depletion of head muscle, particularly the hyohyoideus and interhyoideus (Fig. 6D). Thus, cdkn1c is epistatic to myod and the genetic enhancement of myod loss of function shows that Cdkn1c and Myod operate in the same pathway.
DISCUSSION
The current work makes three major points. First, that Cdkn1c functions to promote both slow and fast myogenesis in the zebrafish. Second, that muscle terminal differentiation is driven by a positive feedback loop, with Cdkn1c stabilizing and activating Myod which, in turn, maintains cdkn1c expression. Third, that in slow muscle Hh drives cdkn1c expression leading to active Myod, myog expression and differentiation. Our work shows how a cell cycle regulator functions beyond the cell cycle to regulate terminal differentiation in vivo.
Cdkn1c and myogenesis
Our data show that Cdkn1c synergizes genetically with Myod to promote myogenesis. Although a major role of Cdkn1c elsewhere is to inhibit the G1/S transition, we find that Cdkn1c is not essential for cell cycle exit in slow muscle precursors: cdkn1c mRNA is lost in Hh signalling blockade and yet adaxial cells do not enter S-phase. Moreover, Cdkn1c knockdown does not prevent adaxial cell cycle exit, consistent with the absence of mRNA encoding cyclin D or E homologues in adaxial cells (www.zfin.org). Cdkn1c not only inhibits CDKs, but can also bind to a variety of other cellular proteins including Myod, PCNA, Skp2, Nr4a2 and LIMK-1 (www.hprd.org). It is highly likely that Cdkn1c promotes myogenesis through Myod. Firstly, we show that loss of Cdkn1c function mildly phenocopies loss of Myod. Second, Cdkn1c promotes accumulation of Myod immunoreactivity in adaxial cells. Third, Cdkn1c can rescue slow myogenesis only if Myod is present. Fourth, as has previously been shown, in cultured cells Cdkn1c can directly bind and stabilize Myod and may additionally enhance Myod activity by preventing CDK-dependent phosphorylation that otherwise leads to Myod degradation (Reynaud et al., 2000; Reynaud et al., 1999). Although zebrafish are not amenable to similar biochemical analyses, taken together, these data argue strongly that Cdkn1c acts in zebrafish myogenesis by promoting Myod activity.
The role of Cdkn1c in myogenesis appears to be ancient. In both mice and zebrafish, cdkn1c is highly expressed in muscle tissues and, while not essential for all myogenesis, contributes to muscle terminal differentiation (Yan et al., 1997; Zhang et al., 1997). Surprisingly, a cdkn1c gene has not been identified in Xenopus laevis, reptiles or birds. The lack of slow adaxial muscle in X. laevis trunk parallels this loss of the cdkn1c present in ancestral amphibia (Grimaldi et al., 2004; Habermann et al., 2004). Nevertheless, the unique amphibian Cdkn1 family member p27Xic1 drives myogenesis in X. laevis, implying functional conservation of Cdkn1 action in muscle (Vernon and Philpott, 2003). In chicken, Cdkn1c function may also have been taken by Cdkn1b (p27Kip1), which can promote muscle differentiation in cell culture (Leshem et al., 2000; Messina et al., 2005). We observe zebrafish cdkn1b expression specifically in fast muscle precursors, followed by cdkn1c. This suggests that each has a role in successive phases of the terminal differentiation process, as recently observed in murine pituitary development (Bilodeau et al., 2009). Mouse Cdkn1c is reported to accumulate in a subset of muscle nuclei (Zhang et al., 1997), reminiscent of the differential expression and role of cdkn1c in subsets of zebrafish myogenic cells. We wonder whether differing requirement for Cdkn1 function may reflect a fundamental distinction within vertebrate muscle cell populations.
In zebrafish, Cdkn1c is required for a subset of Myod-dependent head and lateral somitic muscles. Myod acts together with Myog in these muscles, independent of a Myf5 requirement, which we have termed the lateral mode of myogenesis (Hinits et al., 2009). In mice, Cdkn1c and Myod are also required in subsets of lateral somite and head muscles. Only certain somite-derived and head myogenic cells require MyoD, so that defects in Myod nulls appear transient (Kablar et al., 1997). Similarly, cdkn1c null mice lack lateral somite-derived abdominal muscle, yet some recover and survive (Zhang et al., 1997). The transient nature of the murine defects have prevented thorough analysis to date, but the similarities between species suggest an ancient function for Myod and Cdkn1c in lateral somitic myogenic mode.
We find that Cdkn1c co-operates with Myod to drive myog expression. However, Myogenin itself has a relatively minor role in early zebrafish myogenesis, contrasting with its major role in mice (Hasty et al., 1993; Nabeshima et al., 1993). The importance of Myogenin in mouse may relate to the greater myogenic role of Mrf4 (Kassar-Duchossoy et al., 2004; Rawls et al., 1998), and the more complex involvement of Cdkn1 family members. Murine Cdkn1a is a Myod target gene that is also expressed in early myogenesis and during terminal differentiation of myoblasts. Cdkn1a is redundant with cdkn1c for cell cycle exit. The phenotype of loss of function of both Cdkn1s is strikingly similar to a myogenin null (Halevy et al., 1995; Parker et al., 1995; Zhang et al., 1999). Although mouse myogenin expression is not entirely Cdkn1-dependent, these correlations demand further study.
Like cdkn1c, cdkn1a is also an ancient vertebrate gene, but is not detectably expressed in embryonic zebrafish muscle, suggesting it lacks a function in myogenesis, perhaps because of the limited proliferation in this species. Alternatively, murine Cdkn1a may have been recruited into myogenesis during the evolution in mammals of imprinting at the syntenically-conserved Cdkn1c region (Dunzinger et al., 2007). Imprinting represses paternal Cdkn1c, which might, without Cdkn1a, have prevented proper muscle differentiation in the fetus. As mammalian Cdkn1a now drives myoblast cell cycle exit, Cdkn1c may primarily regulate differentiation in mice, as in zebrafish adaxial cells. On the other hand, our data do not eliminate the possibility that Cdkn1c may regulate both cell cycle exit and differentiation in other zebrafish muscle cell types. Strikingly, repression of human CDKN1C in region 11p15 is implicated in Beckwith-Weidemann Syndrome (BWS), and polymorphisms in CDKN1C are associated with increased risk of atherosclerosis and myocardial infarction (Rodriguez et al., 2007; Zhang et al., 1997). The finding that loss of Cdkn1c disrupts cranial and lateral somitic myogenesis suggests that CDKN1C may contribute to the overgrowth of tongue muscle in BWS. Our data raise the question of whether Cdkn1c dysfunction contributes to other muscle disease, which often targets specific cranial or limb muscle groups.
Cdkn1c and Myod positive feedback loop
How does Cdkn1c promote Myod activity? It has been proposed that CDKs phosphorylate and inactivate Myod in proliferating myoblasts (Reynaud et al., 1999). In post-mitotic cells, similarly, CDK activity may decrease Myod activity and/or stability. Our data show that adaxial Cdkn1c promotes Myod accumulation, but is not required for cell cycle exit. So Cdkn1c may both confirm cell cycle exit, through inhibition of CDK activity, and also drive cells to terminal differentiation through the combined effects of decreased Myod phosphorylation and direct Myod binding by Cdkn1c, both of which stabilize Myod (Reynaud et al., 2000; Reynaud et al., 1999). Our data do not, however, rule out a contribution of other mechanisms by which Cdkn1c might enhance Myod activity. Indeed, we have been unable to quantify any stabilization or altered phosphorylation of Myod in vivo after Cdkn1c manipulation, although this may be due to the presence of Myod in multiple cell populations.
We find that Myod activity is also required to maintain adaxial cdkn1c expression. In cultured murine cell lines kept in differentiation conditions, Myod has been shown to regulate Cdkn1c indirectly when Cdkn1a is absent (Bean et al., 2005; Figliola et al., 2008; Vaccarello et al., 2006). Our finding that Cdkn1c promotes early zebrafish muscle differentiation in the absence of cdkn1a expression suggests that similar mechanisms operate in vivo. We propose a model in which positive feedback between Cdkn1c and Myod drives terminal differentiation (Fig. 7). The model contains a striking symmetry: two extracellular signals (Fgf and Hh) each initiate one half of a positive feedback system. Each initial response (MRF and cdkn1c expression, respectively) then acts to maintain and amplify the other. Ultimately, this leads to an irreversible commitment to terminal differentiation. Such a mechanism likely explains the correlations observed between myod and cdkn1c expression in various mammalian systems (Abou-Khalil et al., 2009; Bigot et al., 2008).
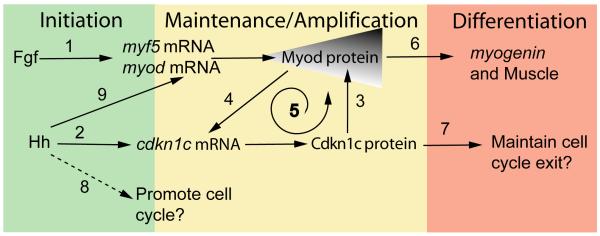
Figure 7. Model of Myod:Cdkn1c Positive Feedback.
Adaxial MRF expression initiates independent of Hedgehog (Hh) signals, through Fgf (1). Hh initiates cdkn1c expression (2) leading to gradual accumulation of Myod protein (3) that maintains cdkn1c expression (4), creating a positive feedback loop (5) that triggers muscle differentiation (6). A possible additional role of Cdkn1 is maintenance of cell cycle exit (7). A proliferative effect of Hh in myogenesis has been reported in mouse myoblasts and chick somite cells, as indicated by the dashed line (8). However, all available evidence argues against a proliferative effect of Hh in zebrafish myogenesis. Instead, Hh apparently independently maintains MRF mRNA in anterior PSM (9).
Cdkn1c and the role of Hedgehog in myogenesis
Positive feedback is a well-known mechanism for converting an analogue signal to a digital output; in this case, terminal differentiation. In zebrafish adaxial myogenesis, the level of Hh signaling regulates such a binary switch. We observed that, in smob641 or cyA-treated embryos, which lack cdkn1c mRNA, myod mRNA is present in the tailbud but Myod protein generally does not accumulate. However, very rare cells do succeed in progressing to terminal differentiation. Such ‘escape’ is Myod-dependent and presumably, therefore, occurs in cells that accumulate enough Myod without the aid of normal Cdkn1c activity. We could ablate these residual slow fibres with cdkn1c MO. At an intermediate level of Hh signaling (in shhatbx392 mutants), a low level of cdkn1c mRNA accumulates. This Cdkn1c drives increase in Myod protein, initially without altering myod mRNA. The consequence is that about half the usual number of superficial slow muscle fibers form but, consistent with a binary switch model, those that do form express slow markers and undergo normal differentiation and migration. When Cdkn1c is experimentally reduced in shha mutants a further parallel reduction in adaxial Myod and residual slow muscle occurs. In the wild type condition, it seems that Cdkn1c is part of a system of genetic redundancy in terminal differentiation that establishes the correct number of slow muscle fibers. Both Hh and Cdkn1c promote fast muscle differentiation in myf5+myod morphant zebrafish (Hinits et al., 2009), suggesting that these proteins also cooperate in fast myogenesis. Hh-driven Cdkn1c expression appears to control the binary decision to cell cycle exit in the retina (Shkumatava and Neumann, 2005). It remains to be determined how widespread is the role of Hh and Cdkn1 in such analogue-to-digital conversion.
Hh signaling is a potent regulator of myogenesis, but has been described to both promote myoblast proliferation and drive terminal differentiation (Elia et al., 2007; Li et al., 2004). Susceptibility of Cdkn1 genes to Hh signaling may regulate a cell’s response to Hh. Indeed, whereas cdkn1c expression is Hh-dependent in the muscle and retina, it is Hh-independent in notochord. Within muscle, situations in which Cdkn1 is not activated by Hh may favour proliferation. We have no evidence that Hh ever promotes proliferation of myogenic cells in zebrafish; its induction of Cdkn1c correlates with terminal differentiation. This suggests that, if Hh can drive Cdkn1 expression, promotion of terminal differentiation results. Our findings suggest that Cdkn1 function in myogenesis may form a node at which other signals can regulate Hh response.
That Cdkn1c over-expression alone can rescue slow myogenesis in smob641 mutant or cyA-treated embryos is a dramatic demonstration of its central role in slow fibre formation. On the other hand, we did not observe ectopic slow fibres when Cdkn1c was over-expressed in lateral somitic cells, indicating that Cdkn1c is not responsible for determining the slow fate. At present, it is unclear if the action of Cdkn1c on Myod is sufficient to rescue slow fibres, or if other molecular targets are also involved. For example, adaxial expression of prdm1, which encodes a chromatin remodelling factor required for proper slow myogenesis, is Hh-dependent but MRF-independent (Baxendale et al., 2004; Hinits et al., 2009). It will be interesting to know if prdm1 expression is also rescued by Cdkn1c. We note that Cdkn1c is insufficient to rescue all adaxial cells in which it is expressed. It is possible that Cdkn1c is required in a specific time window. Alternatively, only the adaxial cells with the highest level of residual myod mRNA may be rescued by Cdkn1c.
Our data show that cdkn1c sensitivity to Hh diminishes as the PSM matures. When MRF activity is blocked, to prevent the positive feedback loop we describe, cdkn1c is still Hh-responsive in tailbud, but not in anterior PSM. The molecular basis of this change in Hh sensitivity is unclear. It is known that, later in development, Hh signaling acts directly on lateral somitic cells to regulate dermomyotome behaviour (Feng et al., 2006). Although Fig. 4C shows that much cdkn1c expression in the lateral somite is MRF dependent, it will be important to determine how directly and independently Hh regulates MRFs and cdkn1c in dermomyotome.
Cdkn1c is one of the most highly up-regulated genes when murine myoblasts or muscle stem cells enter quiescence (Fukada et al., 2007). It seems likely that, in this context, Cdkn1c helps block the cell cycle but can not drive terminal differentiation, possibly due to a lack of MyoD. In human myoblasts, the parallel failure of MRF and Cdkn1c accumulation has been suggested to underlie replicative ageing (Bigot et al., 2008). It will be important to determine how cellular context controls the functioning of the positive feedback loop we have described.
Conclusion
Understanding the molecular mechanisms driving a committed cell to undergo terminal differentiation remains a fundamental problem in developmental and cell biology, with implications for stem cell science, regenerative medicine and cancer. Since the Weintraub lab’s classic series of studies on MyoD auto-regulation, controlled positive feedback by MyoD has provided a paradigm in developmental biology. Subsequent work, however, has failed to prove that direct positive feedback of MyoD protein on the Myod gene has a significant role in either commitment to myogenesis or terminal differentiation. Here, we resurrect Weintraub’s idea in new molecular clothes. We show how a major cell cycle regulator, Cdkn1c (p57Kip2), helps drive terminal differentiation in vertebrate skeletal muscle in vivo by establishing a positive feedback loop with MyoD at the level of Myod protein activity, not myod transcription (Fig. 7). The work therefore fills a gap in the understanding of MyoD function in muscle differentiation; it shows that indirect positive feedback by Myod drives myogenesis.
Supplementary Material
Figure S1: Hh signalling does not drive cell cycle exit of adaxial cells.
Embryos from a smob641/+ incross, treated with cyA or injected with cdkn1c MO were exposed to BrdU (or not) at ~6 ss at 4°C for 20 min, transferred to Embryo Medium at 28.5°C for 30 min, and BrdU was detected by immunohistochemistry at ~8 som. (A). Smob641 mutants were genotyped by in situ hybridization for myod and separated prior to BrdU staining. As a control, omission of BrdU prevented all nuclear labeling. Note the lack of BrdU in smob641 mutant adaxial cells lacking myod mRNA, but its presence in more anterior notochord, consistent with Fucci results showing cells in S/G2/M in maturing notochord (Sugiyama, M., Sakaue-Sawano, A., Iimura, T., Fukami, K., Kitaguchi, T., Kawakami, K., Okamoto, H., Higashijima, S.I., and Miyawaki, A. 2009. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A 106: 20812-20817.) (B). hsp90a mRNA reveals adaxial cell nuclei. Note the lack of BrdU+ nuclei in the hsp90a-containing adaxial cells and in the notochord/myod focal plane in the high magnifications at right, irrespective of genotype/treatment. Bars: 25 μm.
Figure S2: Cdkn1c mRNA accumulation in slow and fast muscle development.
Wholemount in situ mRNA hybridization for zebrafish cdkn1c in dorsal (A,B,L; anterior to top) or lateral (G,M; anterior to top, dorsal to right) flatmount or transverse cryosections at the levels indicated by dashed lines (C-F, H-K, N-R; dorsal to top). Bar: 100 μm (A). At 5 som, Cdkn1c mRNA is detected in adaxial cells, the base of the notochord and the overlying neural plate. (B-F). By 15 som, expression continues in basal notochord, adaxial cells and differentiated slow muscle, commences in fast muscle precursors in rostral somites and in ventrolateral regions near the tailbud and becomes regionalized in ventral and lateral spinal chord. (G-K). At 25 som, expression becomes widespread in rostral somites, persists in neural tube, adaxial cells and posterior notochord and is high in cloacal region. (L-R). By 24 hpf, cdkn1c mRNA has complex neural expression reminiscent of differentiating neurons, is abundant in fast and slow muscle, persists in cloaca but is declining in posterior notochord. (S-V). Cdkn1c (S,T) and mylz2 myosin (U,V) mRNA in head muscle anlagen at 48 hpf in ventral (S,U) or lateral (T,V) view, anterior up, dorsal to right. Note hyoid muscle anlagen (arrow) and bilateral precursors overlying the differentiated fibers (arrowheads). (T). Cdkn1c mRNA in fin muscle masses at 48 hpf. ad adaxial cells, cl cloaca, dc diencephalon, fm fast muscle precursors, hb hindbrain, m mouth, nc notochord, np neural plate, nt neural tube, pn posterior notochord, sm slow muscle, ss superficial somite, tc telencephalon, tm tegmentum, y yolk.
Figure S3. Cdkn1c knockdown does not affect myod mRNA in presomitic mesoderm. (A). In situ mRNA hybridization for myod alone (left panel) or myod and myog (right panel) in a shhatbx392/+ incross injected with cdkn1c MOs or vehicle-control. Note the reduced numbers of myog-expressing cells in shha;cdkn1c MO embryo. Dorsal view of flatmounts at 8 som, anterior to top. (B). Western analysis of Myod in 24 hpf embryos injected with cdkn1c MO. A single major band of Mr ~44 000 is not reduced relative to ß-Tubulin loading control (Mr ~55 000). When averaged over three experiments on 16-24 hpf embryos, no significant down-regulation of Myod band was detected after correction for protein loading. Bars: 25 μm.
Figure S4. Cdkn1b is expressed in the developing lateral somite.
In situ hybridization of 15 ss embryos for cdkn1a, cdkn1b and cdkn1b-like, also showing cdkn1c for comparison. Dorsal view flatmounts, anterior to top. Boxed areas are magnified at right. Cdkn1a mRNA is not detected above background. Cdkn1b mRNA appears in the lateral region of each somite shortly after its formation. Cdkn1bl mRNA is barely detectable above background in somites. In contrast, cdkn1c mRNA is readily detected in notochord, muscle and CNS. Bars: 50 μm.
Figure S5. Myod RNA injection drives ectopic myogenesis and cdkn1c up-regulation. Embryos injected with RNA encoding Myod were analysed at 15 ss immunohistochemically for MyHC (A,C) and by mRNA in situ hybridization for myog (B), prdm1 (C) and cdkn1c (D). Dorsal flatmounts, anterior to top. Bar: 100 μm. (A). Unilateral ectopic muscle in somitic and head mesodermal regions. (B). Myod represses myog mRNA in somites, probably due to premature differentiation, and induces ectopic myog mRNA the head region. (C). Ectopic myogenesis is not accompanied by ectopic prdm1 expression. (D). Two examples of diffuse low level ectopic cdkn1c mRNA up-regulation (brackets) after myod RNA injection.
Acknowledgments
Thanks to Phil Ingham, Ichiro Masai and Tom Hawkins for reagents. SMH is an MRC Scientist with Programme Grant support.
References
- Abou-Khalil R, Le Grand F, Pallafacchina G, Valable S, Authier FJ, Rudnicki MA, Gherardi RK, Germain S, Chretien F, Sotiropoulos A, Lafuste P, Montarras D, Chazaud B. Autocrine and paracrine angiopoietin 1/tie-2 signaling promotes muscle satellite cell self-renewal. Cell Stem Cell. 2009;5:298–309. doi: 10.1016/j.stem.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J. Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel B, Givan LA, Eisen JS. Delta-notch signaling and lateral inhibition in zebrafish spinal cord development. BMC Dev Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi MJ, D’Angelo JA, Hernandez LP, Devoto SH. Distinct mechanisms regulate slow-muscle development. Curr Biol. 2001;11:1432–1438. doi: 10.1016/s0960-9822(01)00428-6. [DOI] [PubMed] [Google Scholar]
- Barresi MJ, Stickney HL, Devoto SH. The zebrafish slow-muscle-omitted gene product is required for hedgehog signal transduction and the development of slow muscle identity. Development. 2000;127:2189–2199. doi: 10.1242/dev.127.10.2189. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The b-cell maturation factor blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Bean C, Salamon M, Raffaello A, Campanaro S, Pallavicini A, Lanfranchi G. The ankrd2, cdkn1c and calcyclin genes are under the control of myod during myogenic differentiation. J. Mol. Biol. 2005;349:349–366. doi: 10.1016/j.jmb.2005.03.063. [DOI] [PubMed] [Google Scholar]
- Bigot A, Jacquemin V, Debacq-Chainiaux F, Butler-Browne GS, Toussaint O, Furling D, Mouly V. Replicative aging down-regulates the myogenic regulatory factors in human myoblasts. Biol. Cell. 2008;100:189–199. doi: 10.1042/BC20070085. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J. Distinct developmental roles of cell cycle inhibitors p57kip2 and p27kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Mol. Cell. Biol. 2009;29:1895–1908. doi: 10.1128/MCB.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden CS, Currie PD, Ingham PW, Hughes SM. Notochord induction of zebrafish slow muscle mediated by sonic hedgehog. Genes Dev. 1997;11:2163–2175. doi: 10.1101/gad.11.17.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chargé SB, Brack AS, Bayol SA, Hughes SM. Myod- and nerve-dependent maintenance of myod expression in mature muscle fibres acts through the drr/prr element. BMC Dev Biol. 2008;8:5. doi: 10.1186/1471-213X-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SW, Nguyet LM, Jiang YJ, Korzh V. The chemokine sdf-1 and its receptor cxcr4 are required for formation of muscle in zebrafish. BMC Dev Biol. 2007;7:54. doi: 10.1186/1471-213X-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutelle O, Blagden CS, Hampson R, Halai C, Rigby PW, Hughes SM. Hedgehog signalling is required for maintenance of myf5 and myod expression and timely terminal differentiation in zebrafish adaxial myogenesis. Dev Biol. 2001;236:136–150. doi: 10.1006/dbio.2001.0193. [DOI] [PubMed] [Google Scholar]
- Currie PD, Ingham PW. Induction of a specific muscle cell type by a hedgehog-like protein in zebrafish. Nature. 1996;382:452–455. doi: 10.1038/382452a0. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21cip1/waf1 undergo normal development, but are defective in g1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Devoto SH, Melancon E, Eisen JS, Westerfield M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development. 1996;122:3371–3380. doi: 10.1242/dev.122.11.3371. [DOI] [PubMed] [Google Scholar]
- Dunzinger U, Haaf T, Zechner U. Conserved synteny of mammalian imprinted genes in chicken, frog, and fish genomes. Cytogenet Genome Res. 2007;117:78–85. doi: 10.1159/000103167. [DOI] [PubMed] [Google Scholar]
- Ekker SC, Ungar AR, Greenstein P, von Kessler DP, Porter JA, Moon RT, Beachy PA. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr. Biol. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- Elia D, Madhala D, Ardon E, Reshef R, Halevy O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of mapk/erk and pi3k/akt pathways. Biochim. Biophys. Acta. 2007;1773:1438–1446. doi: 10.1016/j.bbamcr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Feng X, Adiarte EG, Devoto SH. Hedgehog acts directly on the zebrafish dermomyotome to promote myogenic differentiation. Dev Biol. 2006;300:736–746. doi: 10.1016/j.ydbio.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, Kaushansky K, Roberts JM. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Figliola R, Busanello A, Vaccarello G, Maione R. Regulation of p57(kip2) during muscle differentiation: Role of egr1, sp1 and DNA hypomethylation. J. Mol. Biol. 2008;380:265–277. doi: 10.1016/j.jmb.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Fukada S, Uezumi A, Ikemoto M, Masuda S, Segawa M, Tanimura N, Yamamoto H, Miyagoe-Suzuki Y, Takeda S. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- Grimaldi A, Tettamanti G, Martin BL, Gaffield W, Pownall ME, Hughes SM. Hedgehog regulation of superficial slow muscle fibres in xenopus and the evolution of tetrapod trunk myogenesis. Development. 2004;131:3249–3262. doi: 10.1242/dev.01194. [DOI] [PubMed] [Google Scholar]
- Groves JA, Hammond CL, Hughes SM. Fgf8 drives myogenic progression of a novel lateral fast muscle fibre population in zebrafish. Development. 2005;132:4211–4222. doi: 10.1242/dev.01958. [DOI] [PubMed] [Google Scholar]
- Habermann B, Bebin AG, Herklotz S, Volkmer M, Eckelt K, Pehlke K, Epperlein HH, Schackert HK, Wiebe G, Tanaka EM. An ambystoma mexicanum est sequencing project: Analysis of 17,352 expressed sequence tags from embryonic and regenerating blastema cdna libraries. Genome Biol. 2004;5:R67. doi: 10.1186/gb-2004-5-9-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by myod. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- Hammond CL, Hinits Y, Osborn DP, Minchin JE, Tettamanti G, Hughes SM. Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev Biol. 2007;302:504–521. doi: 10.1016/j.ydbio.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Bradley A, Morris JH, Edmondson DG, Venuti JM, Olson EN, Klein WH. Muscle deficiency and neonatal death in mice with targeted mutation in the myogenin gene. Nature. 1993;364:501–506. doi: 10.1038/364501a0. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Carvajal JJ, Rigby PW, Hughes SM. Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle. Gene Expr Patterns. 2007;7:738–745. doi: 10.1016/j.modgep.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development. 2009;136:403–414. doi: 10.1242/dev.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsinger E, Stellabotte F, Devoto SH, Westerfield M. Hedgehog signaling is required for commitment but not initial induction of slow muscle precursors. Dev Biol. 2004;275:143–157. doi: 10.1016/j.ydbio.2004.07.030. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Tajbakhsh S, Rudnicki MA. Myf5 and myod activation define independent myogenic compartments during embryonic development. Dev Biol. 2003;258:307–318. doi: 10.1016/s0012-1606(03)00139-8. [DOI] [PubMed] [Google Scholar]
- Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. Myod and myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- Kassar-Duchossoy L, Gayraud-Morel B, Gomes D, Rocancourt D, Buckingham M, Shinin V, Tajbakhsh S. Mrf4 determines skeletal muscle identity in myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- Lee KC, Goh WL, Xu M, Kua N, Lunny D, Wong JS, Coomber D, Vojtesek B, Lane EB, Lane DP. Detection of the p53 response in zebrafish embryos using new monoclonal antibodies. Oncogene. 2008;27:629–640. doi: 10.1038/sj.onc.1210695. [DOI] [PubMed] [Google Scholar]
- Leshem Y, Spicer DB, Gal-Levi R, Halevy O. Hepatocyte growth factor (hgf) inhibits skeletal muscle cell differentiation: A role for the bhlh protein twist and the cdk inhibitor p27. J. Cell. Physiol. 2000;184:101–109. doi: 10.1002/(SICI)1097-4652(200007)184:1<101::AID-JCP11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Lewis KE, Currie PD, Roy S, Schauerte H, Haffter P, Ingham PW. Control of muscle cell-type specification in the zebrafish embryo by hedgehog signalling. Dev Biol. 1999;216:469–480. doi: 10.1006/dbio.1999.9519. [DOI] [PubMed] [Google Scholar]
- Li X, Blagden CS, Bildsoe H, Bonnin M-A, Duprez D, Hughes SM. Hedgehog can drive terminal differentiation of amniote slow skeletal muscle. BMC Dev Biol. 2004;4:9. doi: 10.1186/1471-213X-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maves L, Waskiewicz AJ, Paul B, Cao Y, Tyler A, Moens CB, Tapscott SJ. Pbx homeodomain proteins direct myod activity to promote fast-muscle differentiation. Development. 2007;134:3371–3382. doi: 10.1242/dev.003905. [DOI] [PubMed] [Google Scholar]
- Messina G, Blasi C, La Rocca SA, Pompili M, Calconi A, Grossi M. P27kip1 acts downstream of n-cadherin-mediated cell adhesion to promote myogenesis beyond cell cycle regulation. Mol. Biol. Cell. 2005;16:1469–1480. doi: 10.1091/mbc.E04-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y, Hanaoka K, Hayasaka M, Esumi E, Li S, Nonaka I, Nabeshima Y.-i. Myogenin gene disruption results in perinatal lethality owing to severe muscle defect. Nature. 1993;364:532–535. doi: 10.1038/364532a0. [DOI] [PubMed] [Google Scholar]
- Nagahama H, Hatakeyama S, Nakayama K, Nagata M, Tomita K. Spatial and temporal expression patterns of the cyclin-dependent kinase (cdk) inhibitors p27kip1 and p57kip2 during mouse development. Anat Embryol (Berl) 2001;203:77–87. doi: 10.1007/s004290000146. [DOI] [PubMed] [Google Scholar]
- Ochi H, Hans S, Westerfield M. Smarcd3 regulates the timing of zebrafish myogenesis onset. J. Biol. Chem. 2008;283:3529–3536. doi: 10.1074/jbc.M708594200. [DOI] [PubMed] [Google Scholar]
- Park CW, Chung JH. Age-dependent changes of p57(kip2) and p21(cip1/waf1) expression in skeletal muscle and lung of mice. Biochim. Biophys. Acta. 2001;1520:163–168. doi: 10.1016/s0167-4781(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J. Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SB, Eichele G, Zhang P, Rawls A, Sands AT, Bradley A, Olson EN, Harper JW, Elledge SJ. P53-independent expression of p21cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- Rawls A, Valdez MR, Zhang W, Richardson J, Klein WH, Olson EN. Overlapping functions of the myogenic bhlh genes mrf4 and myod revealed in double mutant mice. Development. 1998;125:2349–2358. doi: 10.1242/dev.125.13.2349. [DOI] [PubMed] [Google Scholar]
- Reynaud EG, Leibovitch MP, Tintignac LA, Pelpel K, Guillier M, Leibovitch SA. Stabilization of myod by direct binding to p57(kip2) J. Biol. Chem. 2000;275:18767–18776. doi: 10.1074/jbc.M907412199. [DOI] [PubMed] [Google Scholar]
- Reynaud EG, Pelpel K, Guillier M, Leibovitch MP, Leibovitch SA. P57(kip2) stabilizes the myod protein by inhibiting cyclin e-cdk2 kinase activity in growing myoblasts. Mol. Cell. Biol. 1999;19:7621–7629. doi: 10.1128/mcb.19.11.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez I, Coto E, Reguero JR, Gonzalez P, Andres V, Lozano I, Martin M, Alvarez V, Moris C. Role of the cdkn1a/p21, cdkn1c/p57, and cdkn2a/p16 genes in the risk of atherosclerosis and myocardial infarction. Cell Cycle. 2007;6:620–625. doi: 10.4161/cc.6.5.3927. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. Myod or myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- Sabourin LA, Girgis-Gabardo A, Seale P, Asakura A, Rudnicki MA. Reduced differentiation potential of primary myod-/- myogenic cells derived from adult skeletal muscle. J. Cell Biol. 1999;144:631–643. doi: 10.1083/jcb.144.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauerte HE, van Eeden FJ, Fricke C, Odenthal J, Strahle U, Haffter P. Sonic hedgehog is not required for the induction of medial floor plate cells in the zebrafish. Development. 1998;125:2983–2993. doi: 10.1242/dev.125.15.2983. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Neumann CJ. Shh directs cell-cycle exit by activating p57kip2 in the zebrafish retina. EMBO Rep. 2005;6:563–569. doi: 10.1038/sj.embor.7400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarello G, Figliola R, Cramerotti S, Novelli F, Maione R. P57kip2 is induced by myod through a p73-dependent pathway. J. Mol. Biol. 2006;356:578–588. doi: 10.1016/j.jmb.2005.12.024. [DOI] [PubMed] [Google Scholar]
- Varga ZM, Amores A, Lewis KE, Yan YL, Postlethwait JH, Eisen JS, Westerfield M. Zebrafish smoothened functions in ventral neural tube specification and axon tract formation. Development. 2001;128:3497–3509. doi: 10.1242/dev.128.18.3497. [DOI] [PubMed] [Google Scholar]
- Vernon AE, Philpott A. A single cdk inhibitor, p27(xic1), functions beyond cell cycle regulation to promote muscle differentiation in xenopus. Development. 2003;130:71–83. doi: 10.1242/dev.00180. [DOI] [PubMed] [Google Scholar]
- Wang J, Walsh K. Resistance to apoptosis conferred by cdk inhibitors during myocyte differentiation. Science. 1996;273:359–361. doi: 10.1126/science.273.5273.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. The myod family and myogenesis: Redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book - a guide for the laboratory use of zebrafish (danio rerio) University of Oregon Press; 1995. [Google Scholar]
- Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking myod. Dev Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, Masai I. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing wnt and notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Yan Y, Frisen J, Lee MH, Massague J, Barbacid M. Ablation of the cdk inhibitor p57kip2 results in increased apoptosis and delayed differentiation during mouse development. Genes Dev. 1997;11:973–983. doi: 10.1101/gad.11.8.973. [DOI] [PubMed] [Google Scholar]
- Zhang P, Liegeois NJ, Wong C, Finegold M, Hou H, Thompson JC, Silverman A, Harper JW, DePinho RA, Elledge SJ. Altered cell differentiation and proliferation in mice lacking p57kip2 indicates a role in beckwith-wiedemann syndrome. Nature. 1997;387:151–158. doi: 10.1038/387151a0. [DOI] [PubMed] [Google Scholar]
- Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. P21(cip1) and p57(kip2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Hh signalling does not drive cell cycle exit of adaxial cells.
Embryos from a smob641/+ incross, treated with cyA or injected with cdkn1c MO were exposed to BrdU (or not) at ~6 ss at 4°C for 20 min, transferred to Embryo Medium at 28.5°C for 30 min, and BrdU was detected by immunohistochemistry at ~8 som. (A). Smob641 mutants were genotyped by in situ hybridization for myod and separated prior to BrdU staining. As a control, omission of BrdU prevented all nuclear labeling. Note the lack of BrdU in smob641 mutant adaxial cells lacking myod mRNA, but its presence in more anterior notochord, consistent with Fucci results showing cells in S/G2/M in maturing notochord (Sugiyama, M., Sakaue-Sawano, A., Iimura, T., Fukami, K., Kitaguchi, T., Kawakami, K., Okamoto, H., Higashijima, S.I., and Miyawaki, A. 2009. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc Natl Acad Sci U S A 106: 20812-20817.) (B). hsp90a mRNA reveals adaxial cell nuclei. Note the lack of BrdU+ nuclei in the hsp90a-containing adaxial cells and in the notochord/myod focal plane in the high magnifications at right, irrespective of genotype/treatment. Bars: 25 μm.
Figure S2: Cdkn1c mRNA accumulation in slow and fast muscle development.
Wholemount in situ mRNA hybridization for zebrafish cdkn1c in dorsal (A,B,L; anterior to top) or lateral (G,M; anterior to top, dorsal to right) flatmount or transverse cryosections at the levels indicated by dashed lines (C-F, H-K, N-R; dorsal to top). Bar: 100 μm (A). At 5 som, Cdkn1c mRNA is detected in adaxial cells, the base of the notochord and the overlying neural plate. (B-F). By 15 som, expression continues in basal notochord, adaxial cells and differentiated slow muscle, commences in fast muscle precursors in rostral somites and in ventrolateral regions near the tailbud and becomes regionalized in ventral and lateral spinal chord. (G-K). At 25 som, expression becomes widespread in rostral somites, persists in neural tube, adaxial cells and posterior notochord and is high in cloacal region. (L-R). By 24 hpf, cdkn1c mRNA has complex neural expression reminiscent of differentiating neurons, is abundant in fast and slow muscle, persists in cloaca but is declining in posterior notochord. (S-V). Cdkn1c (S,T) and mylz2 myosin (U,V) mRNA in head muscle anlagen at 48 hpf in ventral (S,U) or lateral (T,V) view, anterior up, dorsal to right. Note hyoid muscle anlagen (arrow) and bilateral precursors overlying the differentiated fibers (arrowheads). (T). Cdkn1c mRNA in fin muscle masses at 48 hpf. ad adaxial cells, cl cloaca, dc diencephalon, fm fast muscle precursors, hb hindbrain, m mouth, nc notochord, np neural plate, nt neural tube, pn posterior notochord, sm slow muscle, ss superficial somite, tc telencephalon, tm tegmentum, y yolk.
Figure S3. Cdkn1c knockdown does not affect myod mRNA in presomitic mesoderm. (A). In situ mRNA hybridization for myod alone (left panel) or myod and myog (right panel) in a shhatbx392/+ incross injected with cdkn1c MOs or vehicle-control. Note the reduced numbers of myog-expressing cells in shha;cdkn1c MO embryo. Dorsal view of flatmounts at 8 som, anterior to top. (B). Western analysis of Myod in 24 hpf embryos injected with cdkn1c MO. A single major band of Mr ~44 000 is not reduced relative to ß-Tubulin loading control (Mr ~55 000). When averaged over three experiments on 16-24 hpf embryos, no significant down-regulation of Myod band was detected after correction for protein loading. Bars: 25 μm.
Figure S4. Cdkn1b is expressed in the developing lateral somite.
In situ hybridization of 15 ss embryos for cdkn1a, cdkn1b and cdkn1b-like, also showing cdkn1c for comparison. Dorsal view flatmounts, anterior to top. Boxed areas are magnified at right. Cdkn1a mRNA is not detected above background. Cdkn1b mRNA appears in the lateral region of each somite shortly after its formation. Cdkn1bl mRNA is barely detectable above background in somites. In contrast, cdkn1c mRNA is readily detected in notochord, muscle and CNS. Bars: 50 μm.
Figure S5. Myod RNA injection drives ectopic myogenesis and cdkn1c up-regulation. Embryos injected with RNA encoding Myod were analysed at 15 ss immunohistochemically for MyHC (A,C) and by mRNA in situ hybridization for myog (B), prdm1 (C) and cdkn1c (D). Dorsal flatmounts, anterior to top. Bar: 100 μm. (A). Unilateral ectopic muscle in somitic and head mesodermal regions. (B). Myod represses myog mRNA in somites, probably due to premature differentiation, and induces ectopic myog mRNA the head region. (C). Ectopic myogenesis is not accompanied by ectopic prdm1 expression. (D). Two examples of diffuse low level ectopic cdkn1c mRNA up-regulation (brackets) after myod RNA injection.