Abstract
Stem cells are permanent residents of tissues and thought to be targets of cancer initiation. The frequent, and often early, upregulation of the FOXM1 transcription factor in the majority of human cancers suggests it may participate in the initiation of human tumorigenesis. However, this hypothesis has not been tested. Herein, we show that targeting the ectopic expression of FOXM1 to the highly clonogenic cells of primary human keratinocytes with stem/progenitor cell properties, but not to differentiating cells, caused clonal expansion in vitro. We show, using a functional 3D-organotypic epithelial tissue regeneration system, that ectopic FOXM1 expression perturbed epithelial differentiation generating a hyperproliferative phenotype reminiscent of that seen in human epithelial hyperplasia. Furthermore, transcriptional expression analysis of a panel of 28 epithelial differentiation-specific genes reveals a role for FOXM1 in suppression of epithelial differentiation. This study provides the first evidence that FOXM1 participates in an early oncogenic pathway that predisposes cells to tumorigenesis by expanding the stem/progenitor compartment and deregulating subsequent keratinocyte terminal differentiation. This finding reveals an important window of susceptibility to oncogenic signals in epithelial stem/progenitor cells prior to differentiation, and may provide a significant benefit to the design of cancer therapeutic interventions that target oncogenesis at its earliest incipient stage.
Keywords: epithelial progenitor/stem cells, organotypic culture, tissue regeneration, cancer initiation, epithelial keratinocyte differentiation
Introduction
FOXM1 directs a transcriptional network of genes that is necessary for cell cycle promotion and for the coordination of timely cell division and exit from the cell cycle (1, 2). Previous studies have demonstrated its role as a major regulator of G2/M phase of the cell cycle where it is required to ensure successful execution of the mitotic program and the maintenance of chromosome stability (3). In 2002, we provided the first evidence that directly linked FOXM1 with human cancer demonstrating that FOXM1 was abundantly expressed in human basal cell carcinomas and further identified this gene as a downstream target of the oncogenic transcription factor GLI1 (4). FOXM1 has since been listed amid the most common differentially expressed genes in the majority of human cancers (1, 2). Our finding that transcriptional upregulation of FOXM1 precedes malignancy in a number of solid human cancer types including oral, esophagus, lung, breast, kidney, bladder and uterus suggests a role for FOXM1 in tumor initiation (5). We have also found that ectopic FOXM1 expression induces genomic instability in primary human oral and skin cells (5, 6) which supports a role for FOXM1 in both malignant progression and metastatic invasion found in a number of human cancer types (1, 2). The fact that FOXM1 is implicated throughout early and late stages of cancer progression suggests a fundamental role for FOXM1 in both tumor initiation and maintenance. FOXM1 activity is dependent on oncogenic signaling downstream of Ras (7) and Cyclin D1 (2), the deregulation of which is often implicated in epithelial tumor initiation and promotion. As an important executor of oncogenic stimulation, the aberrant activation of FOXM1 in models of chemical induced carcinogenesis in mice is tumor promoting (2, 8, 9), while its ablation has strong tumor suppressive effects (10, 11). Although the tumorigenic role for FOXM1 has been attributed to aberrant cell cycle regulation, the mechanism of how its upregulation contributes to cancer initiation in human epithelial cells, if at all, remains unclear.
Squamous epithelia are continuously maintained by permanent stem cell populations that are responsible for homeostasis (12-14). During wound repair however, a different, slow cycling population of cells (regenerative stem/progenitor cells) is responsible for tissue regeneration (15) and much like stem cells, it is also capable of repopulating all the lineages of the tissue in which it resides. Both populations however, behave like clonogenic (Clonehi) populations of keratinocytes when expanded in vitro (12, 13). We therefore refer to these cells as stem/progenitor as there are currently no methods to isolate bona fide stem cells from human tissues (12-14). Although progenitor cells can self renew very efficiently in culture, they inevitably exit this compartment as a consequence of either differentiation (16, 17), senescence (17-20) or both, and then resemble transit amplifying cells (or paraclones) (Clonelo) in that they quickly exit the cell cycle and terminally differentiate (18). The highly self-renewing cells form colonies that have a high cloning efficiency on transfer or, in other words, have a high capacity for self renewal and have been termed holoclones (18). The progressive reduction in self renewal capacity leads to the formation of paraclones, which only form small or abortive colonies that quickly terminally differentiate. Numerous approaches have been used to separate Clonehi from Clonelo keratinocytes (17, 21-23) on the basis of cell surface expression signatures. This has facilitated the investigation into the response of such populations to various oncogenic stresses. In response to overexpression of mitogen-activated kinase (MAPK) (24) or constitutively active β-catenin (25), Clonehi keratinocytes retain their high clonogenic capacity when detached from integrin or expand in numbers, respectively. On the other hand, the overexpression of c-MYC depletes the Clonehi pool both in vitro (26) and in vivo (27). Furthermore, the Clonelo population can regain its clonogenic capacity by Adenovirus 5 E1A, which inactivates a variety of tumor suppressor pathways (28), but not by a single oncogene. In the in vivo setting, RAS oncogene can induce neoplasia when targeted to the stem/progenitor containing basal layer of the epidermis (29) while it can only produce regression prone papillomas when its expression is driven by differentiation specific promoters (29-31).
The upregulation of FOXM1 during tissue regeneration in response to injury (32), suggests that its physiological activation represents a signal which is required for the expansion of regenerative stem/progenitor cells for the replenishment of the epithelium. FOXM1 is an early event in human epithelial neoplasia (5), and in line with the current ‘cancer stem cell’ concept that malignancies are maintained by a subpopulation of cells that possess tumor initiation and self-renewal capacity, we hypothesised that the acquisition of aberrant FOXM1 gene expression in stem/progenitor cells may give rise to progeny cells inheriting excessive FOXM1 levels, as often found in many premalignant tissues (5) and malignant cancers (1, 2). To date, studies of FOXM1 as a cancer causing gene have been performed in the context of transformed and/or immortalized cell lines, undoubtedly providing invaluable insight into the molecular mechanisms mediated by FOXM1. However, due to the aberrant molecular background already present in these cell line systems, the role of FOXM1 in the earliest stages of neoplasia could not be delineated and it is not fully understood. Herein, we sought to investigate the role of FOXM1 (isoform B; FOXM1B) proto-oncogene, by examining its regulation and the effect of its overexpression, in normal primary human oral keratinocyte stem/progenitor cells as they represent the main targets of human malignancies.
Material & Methods
Clinical samples
The use of human tissue was approved by the relevant Research Ethics Committees at each institution. The present study involved 3 oral SCC tumor tissue explants primary cultures for flow cytometry and gene expression analysis, and 6 independent normal oral mucosa tissues donated by healthy volunteers with informed patient consent and ethical approval.
Cell culture
Primary normal human oral keratinocytes (NHOK) were established from clinically normal oral buccal or gingival tissue and were maintained in RM+ medium (5) and were grown on mitomycin (10 μg/ml for 4h) treated 3T3-feeder layers which were plated at a density of 1.8×104/cm2 24 hours prior to keratinocyte seeding. All established cell lines and tumor explants culture procedures used in this study have been characterised and cultured as described previously (5, 33). Retroviral transduction cell synchronization and cell clonogenic assay protocols are described in the Supplemental Data file.
Fluorescence activated cell sorting (FACS)
FACS was performed on BD FACSAria Cell-Sorter (BD Biosciences). Cells were washed once with cold PBS and resuspended in cold FACS buffer (PBS/5% FBS; 1% penicillin/streptomycin) followed by direct immunofluorescence for 15 minutes on ice with FIT-C conjugated anti-integrin β1 (10 μl antibody/106 cells, CD29-FITC, Abcam), PE conjugated anti-p75NTR (20 μl antibody/106 cells, CD271-PE, BD Biosciences) or anti-CD44 (1 μl antibody/106 cells BD Biosciences, clone G44.26) in 100 μl FACS buffer). Cells were centrifuged twice (800 rpm, 3 minutes) with one PBS wash in between and finally resuspended in cold FACS buffer containing 200 ng/ml DAPI dilactate (Sigma). Integrin β1, p75NTR or CD44 high/low(hi/lo) fractions were sorted by selecting the highest/lowest 20% of stained cells respectively. Random sorted controls were selected randomly from the total pool of DAPI negative cells. Keratinocytes were then plated through the automated cell dispenser onto 6-well plates at 50~1000 cells/cm2, containing pre-plated mitomycin C-treated 3T3-feeder layer (1.8×104/cm2) or in serum free medium.
Organotypic culture on de-epidermalized dermis
Organotypic keratinocyte cultures were performed as described (22). Tissue processing was carried out by Dr. Wesley Harrison at Centre for Cutaneous Research at the Blizard Institute of Cell and Molecular Sciences (BICMS), and tissue sections were cut at the Pathology Core Facility of BICMS. Organotypic culture on collagen matrix protocol is described in Supplementary Data file.
Immunoblotting
For protein extraction from keratinocytes growing on feeder layers, cells were incubated for 5 minutes in PBS:Versene (Gibco, Invitrogen) (1:1 ratio) to remove feeder cells prior to protein extraction. Protein extraction and separation on SDS-PAGE gels and immunoblotting was performed as previously described (5). All antibodies used are listed in the Supplementary Data file.
Immunofluorescence and confocal microscopy
Frozen tissue sections were fixed in ice cold methanol:acetone (1:1) for 10 minutes or in 4% formaldehyde (in PBS) for 20 minutes at RT and blocked by 10% goat serum (Sigma) in 0.025% Tween-20 (TBS) for 1 hour at RT. Incubation with primary antibody (in 1%BSA/TBS) was performed for 2 hours at RT or overnight at 4°C. Sections were washed twice in TBS prior to incubation with secondary antibody for 1 hour at RT. Finally, sections were washed twice in TBS and mounted with Shadon Immu-Mount™ (Thermo Scientific) solution containing DAPI dilactate (final concentration at 300 ng/ml). Paraffin embedded tissue sections were initially heated at 60°C for 8 minutes and were then deparaffinised in Xylene (3×5 minutes) and dehydrated (95% ethanol, 90% ethanol, 70% ethanol for 5 minutes each) prior to rehydrated in water (5 minutes). Antigen unmasking was performed by incubation in sub-boiling temperature for 20 minutes in antigen retrieval solution (pH 9.0; DakoCytomation). The sections were washed twice in TBS and permeabilised with TBS 0.2% Triton-X (Sigma) for 10 minutes at RT and were then processed as described above. Confocal and fluorescence microscopy were performed as described previously (22)
Real-time absolute quantitative RT-PCR (qPCR)
Absolute qPCR were performed according to previously established protocols involving standard curves for each gene (5). FOXM1 isoform-specific primers and reference genes (YAP1 and POLR2A) have been previously described and validated (5). Absolute qPCR were performed using the LightCycler 480 qPCR system (Roche Diagnostics Ltd, England, UK). Primer sequences are listed in Supplementary Table S1.
Results & Discussion
Of the three alternatively spliced isoforms, FOXM1B was found to be the key isoform involved in oncogenesis (1, 2). However, it remained unclear which isoform(s) was directly involved in the cell cycle. We addressed this question using both normal telomerase-immortalised human keratinocytes (N/TERT) and metastatic cervical carcinoma cells (HeLa), where FOXM1B was the only isoform found to be differentially expressed in cycling cells upon release from growth arrest (Supplementary, Fig. S1A, B; see Methods). Herein, we investigated the possibility that acquisition of aberrant FOXM1B expression in clonogenic stem/progenitor cells may be a fundamental oncogenic initiation mechanism which contributes to its widespread upregulation detected in most human cancers.
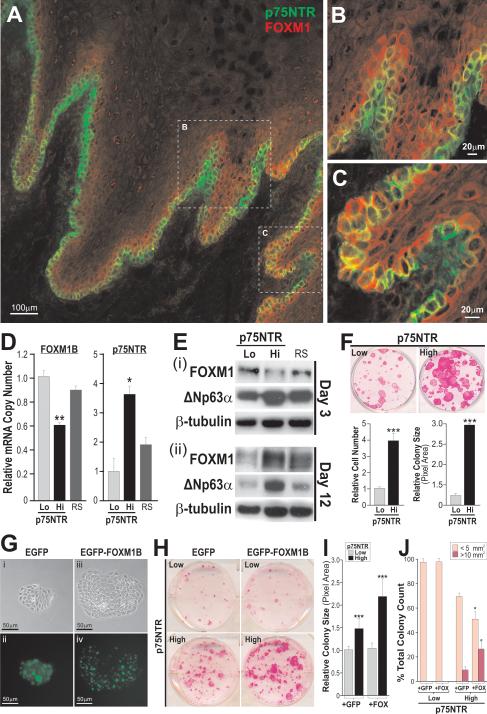
To test this hypothesis, we firstly investigated the expression profile of FOXM1 protein in human oral mucosa tissue. As the antibody for FOXM1 protein was unable to differentiate between isoforms we henceforth use the term FOXM1 whenever isoform-specific information was not available. Secondly, we examined the cellular and molecular effects of its ectopic expression in primary human oral mucosa keratinocyte stem/progenitor cells identified and isolated using a well established oral stem/progenitor cell marker p75NTR (a low affinity nerve growth factor receptor; also known as CD271, TNFRSF16, Gp80-LNGFR; NGFR) (23). In agreement, p75NTR expression was strictly basal, with regions of intense positivity at the tips of connective tissue papillae (Fig. 1A, B), whereas its expression was less intense in deep rete-ridges (Fig. 1C). In contrast, FOXM1 expression was found mainly in the more proliferative epibasal layers of the oral epithelium and occasionally detected in isolated basal cells (Fig. 1A, C). FOXM1 protein is mainly cytoplasmic within the normal mucosa (which has generally very low proliferative index), this being in agreement with the finding that inactive proteins are sequestered in the cytoplasm and that upon mitogenic stimulation, FOXM1 protein translocates into the nucleus (7). We have further shown that in normal primary oral keratinocytes, FOXM1 proteins were predominantly found in the cytoplasm whereas in oral SCC cell line UK1 (33), both cytoplasmic and nuclear FOXM1 proteins were ubiquitously detected (Supplemental Figure S1E) which is consistent with the FOXM1 protein expression pattern found in both premalignant dysplastic and oral SCC tumor tissues (5). In areas of intense p75NTR staining at the tips of connective tissue papillae, FOXM1 expression was almost undetectable (Fig. 1B) suggesting an inverse expression pattern between the p75NTR and FOXM1. To further investigate this expression pattern, we have FACS-isolated p75NTRlo (containing differentiating/non-clonogenic cells or paraclones) and p75NTRhi (containing both stem and early progenitor cells which are clonogenic or are able to form holoclones in vitro) from early passage (P1) primary normal human oral keratinocytes and performed gene expression (Fig. 1D, E) and clonogenicity experiments (Fig. 1F and Supplementary Fig. S2). Although integrin β1, an epidermal stem cell marker (16), was found to be co-expressed with p75NTR (Supplementary Fig. S2A), the integrin β1hi/p75NTRhi double-sorted cells showed only marginal advantage in clonogenic potential over single-sorted p75NTRhi cells (Supplementary Fig. S2C-F), indicating that cells expressing p75NTR alone represent the majority of clonogenic stem/progenitor cells. In agreement with the inverse expression pattern found in oral mucosa tissue above, p75NTRhi cells were found to express significantly lower levels of FOXM1B mRNA (Fig. 1D) and protein (Fig. 1E-I) compared to p75NTRlo or randomly sorted keratinocytes. Although both FOXM1B and FOXM1C are transcriptionally active (32), the reduced mRNA expression was detected only for FOXM1B isoform (Fig. 1D), but not isoform C (Supplementary Fig S1C). Given the proliferation-specific role of FOXM1B, we reasoned that the lower FOXM1B level was due to the quiescent nature of epithelial stem/progenitor cells in vivo and in vitro (17, 34, 35), as well as to the slow cycling status of p75NTRhi keratinocytes in vitro (36) and in vivo (23). Expression analysis of human epidermal keratinocyte stem cells in vitro revealed that the maintenance of their quiescent state is partly attributed to a negative regulator of c-MYC, namely LRIG1 (37). As there is evidence that FOXM1 is a downstream target of c-MYC (38), it is possible that LRIG1-induced suppression of c-MYC indirectly contributed to the observed repression of FOXM1 in oral stem/progenitor cells (p75NTRhi). The upregulation of FOXM1 protein in cells immediately above the basal cell layer (Fig. 1A-C) led us to hypothesise that p75NTRhi cells may give rise to progeny with high clonogenic potential when maintained in prolonged culture (Fig. 1F). Indeed, the role of FOXM1 in progenitor compartment expansion became apparent following prolonged culture (12 days) where FOXM1 was significantly upregulated in p75NTRhi cells and conversely, its expression diminished in p75NTRlo cells (Fig. 1E-ii). In both cultures at day 3 and 12 (Fig. 1E), p75NTRhi cells retained high levels of another epithelial stem cell marker ΔNp63α (20), indicating that FOXM1 is induced specifically during the expansion of p75NTRhi cells with high clonogenic potential. In contrast, FOXM1 expression was suppressed in the differentiating p75NTRlo cells. Indeed, FOXM1 and involucrin (IVL, a keratinocyte differentiation marker) expression were found to be inversely correlated in early and late passage p75NTRlo cells (Supplementary Fig. S3A) and another differentiation marker cytokeratin 4 (KRT4) was found to be suppressed in p75NTRhi cells compared to p75NTRlo cells (Supplementary Fig. S3B). These findings further illustrate that FOXM1 expression is upregulated during the expansion of clonogenic stem/progenitor cells prior to the onset of epithelial differentiation. In agreement, during embryonic development, the expression of FOXM1 coincides with the transient proliferation of neural precursors preceding terminal differentiation (39), while in adult epithelial tissues in mice, its expression is confined to the most proliferative layers (32).
Figure 1.
A, Immunohistochemistry staining of p75NTR (green) and FOXM1 (red) in normal human oral mucosa epithelium. B and C, Magnified views of tips of connective tissue papilla and deep-rete ridges, respectively, (dotted boxes) in A are shown here where co-expression (yellow) of p75NTR and FOXM1 are found predominantly in the suprabasal layer and also in isolated cells within the basal cell layer. D, Absolute qPCR analysis of FOXM1B and p75NTR endogenous mRNA expression levels in p75NTR-sorted primary oral keratinocytes three days after flow sorting. RS, random sorted populations. All values are representative of three independent experiments. Bars represent the mean ± SEM of triplicate samples.*P≤0.05, **P≤0.005, ***P≤0.001. E, Endogenous FOXM1 and ΔNp63α protein expression patterns in p75NTR-sorted primary oral keratinocytes following 3 (i) or 12 (ii) days in culture. β-tubulin was blotted for protein loading control. F, Clonogenecity assays for p75NTR-sorted primary oral keratinocytes. Primary human oral keratinocytes were FACS sorted according to p75NTR levels (low or high) and were plated at equal densities in 6 well plates (n=6). Cells were then allowed to clonally expand for 12 days after which time they were either trypsinised to obtain cell number values, or were stained with rhodamine B for colony visualisation and the quantification of colony growth. G, p75NTRhi cells were transduced with either (i, ii) pSIN-EGFP or (iii, iv) pSIN-EGFP-FOXM1B and individual keratinocyte colonies were examined under bright and fluorescence (FIT-C) microscopy following 3T3-feeder removal. H, Clonogenic assays performed on p75NTRlo and p75NTRhi oral keratinocytes following transduction with either pSIN-EGFP or pSIN-EGFP-FOXM1B. I, Quantification of the average colony area of p75NTRlo and p75NTRhi oral keratinocytes transduced with either construct. Each bar represents mean ± SEM values obtained from n=9 replicates performed in two individual primary human oral keratinocyte strains. J. Graphical representation of the colony-size distribution according to surface area (mm2) that emerged 15 days after transduction. A total of 200-500 colonies were counted. *P<0.05, **P<0.01, ***P<0.001.
Many epithelial malignancies, including oral squamous cell carcinoma, are thought to arise from premalignant epithelial progenitors (40). To understand why FOXM1 is abundantly expressed in epithelial premalignancies (5) and majority of human cancers (1, 2), we investigated the possibility that FOXM1 may be an initiating step that confers ectopic proliferation in keratinocyte stem/progenitor cells leading to the expansion of potentially premalignant progenies. To test this, FACS-sorted p75NTRlo and p75NTRhi cells were each retrovirally transduced with either pSIN-MCS (empty vector), pSIN-EGFP control, or pSIN-EGFP-FOXM1B. pSIN-MCS and pSIN-EGFP transduced cells gave identical experimental results (data not shown). Individual p75NTRhi keratinocyte colonies expressing FOXM1B showed an altered cellular morphology where cells exhibited higher nuclear to cytoplasmic ratio while colonies appeared more compact and relatively larger in size (Fig. 1G, compare panels i and iii). EGFP was homogenously expressed throughout the colony (Fig. 1G-ii), whereas EGFP-FOXM1B protein levels were heterogeneous with increased expression towards the periphery of each colony (Fig. 1G-iv). In order to establish whether ectopic expression of FOXM1B confers any functional effects, clonogenic assays were performed on primary keratinocytes derived from all subsets (p75NTRlo/hi +EGFP/+FOXM1B). The p75NTRlo cells showed limited growth capacity regardless of the construct with which they have been transduced, consistent with a population containing differentiating/non-clonogenic cells. However, upregulation of FOXM1B in p75NTRhi cells significantly enhanced clonal expansion over control p75NTRhi+EGFP cells (Fig. 1H-J). p75NTRhi+FOXM1B cells showed a ~2.6-fold increase (compared to p75NTRhi+EGFP) in the percentage of large cell colonies representing self-renewing holoclones (stem/progenitor cell-derived; >10mm2), and a concomitant reduction in the percentage of smaller/abortive colonies (18) (differentiating/non-clonogenic cell-derived; <5mm2, Fig. 1J). Hence, the elevation of FOXM1B expression in p75NTRhi, but not p75NTRlo keratinocytes, increased the intrinsic clonogenic capacity of primary keratinocytes. This suggests that the onset of epithelial differentiation renders cells refractory to FOXM1B-induced proliferation. In order to gain proliferative advantage, we therefore hypothesised that ectopic FOXM1B expression promotes stem/progenitor cell expansion by perturbing the onset of terminal differentiation.
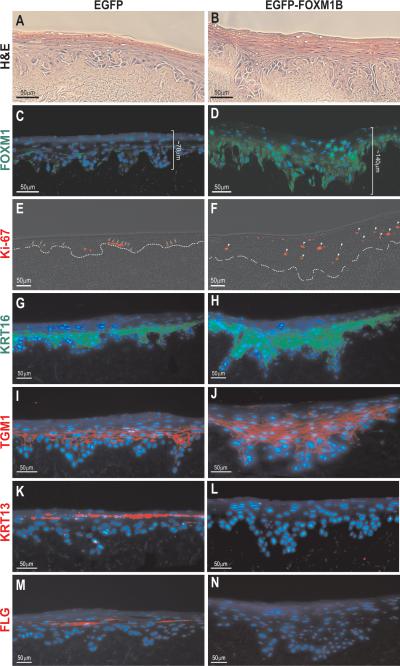
To address this, we performed 3D-organotypic epithelial regeneration model system using EGFP or EGFP-FOXM1B transduced primary human oral keratinocytes according to our previously established method (22). In support, EGFP-FOXM1B-transduced oral keratinocytes grown in the organotypic cultures showed a hyperproliferative phenotype by producing thicker cell layers compared to control cultures using EGFP-transduced cells (Fig. 2, compare between panels A/B and C/D). FOXM1 protein was detected at low levels in the basal and proliferative epithelial layers in the control EGFP organotypics (Fig. 2C). In contrast, FOXM1 protein was abundantly expressed in both the basal and suprabasal layers of the FOXM1B organotypics (Fig. 2D). As expected, immunoreactivity of the proliferation marker Ki-67 was strictly confined to the basal proliferative layer of the control EGFP organotypics (Fig. 2E). In contrast, keratinocytes of the suprabasal layers of FOXM1B organotypics retained ectopic FOXM1B expression, which correlated with increased Ki-67 immunoreactivity (Fig. 2F and Supplemental Fig. S4F and S4G) indicating that they remained proliferative despite their locations within the upper spinous layers which should otherwise host non-proliferative differentiated cells. Only a subset of FOXM1 expressing cells are Ki-67 positive and this is consistent with the staining patterns of FOXM1 and Ki-67 in oral mucosa tissue (Supplementary Fig. S4E) and basal cell carcinoma (4), suggesting the involvement of FOXM1 in processes other than cell proliferation alone. Single-cell expression analysis of human epidermal stem cells has revealed that the ablation of EGFR (growth factor receptor) by Lrig1 (stem cell marker) is required for the maintenance of stem cell quiescence, while it is also suggested that loss of Lrig1 may trigger stem cell expansion and epithelial hyperproliferation through EGFR activation (37). In agreement with this model, EGFR was found to be significantly upregulated in FOXM1B organotypics (Supplementary Fig. S4A, B) and this further confirms the role for FOXM1B in progenitor compartment expansion resulting in epithelial hyperproliferation. The expansion of the suprabasal layers suggests that FOXM1B perturbed the balance between stem/progenitor cell renewal and the commitment to terminal differentiation. We therefore hypothesized that the FOXM1B-induced hyperproliferation may intricately impact on epithelial differentiation.
Figure 2.
Characterisation of FOXM1B-induced hyperproliferation phenotype using 3D-Organotypic epithelial tissue regeneration model system. Haematoxylin and eosin stained organotypic cultures of human oral mucosa derived from primary human oral keratinocytes retrovirally transduced with either A, pSIN-EGFP or B, pSIN-EGFP-FOXM1B. Adjacent sections of respective organotypic tissues were stained with C and D, FOXM1, E and F, Ki-67 (arrows and arrow heads indicate expression of Ki-67 in basal and suprabasal cells, respectively; dotted line demarcates the basement membrane), G and H, cytokeratin 16 (KRT16), I and J, transglutaminase-1 (TGM1), K and L, cytokeratin 13 (KRT13), M and N, filaggrin (FLG). Nuclear DNA was stained with DAPI (blue).
To this end, we investigated the expression profile of cytokeratin 16 (KRT16), which is a marker of hyperproliferative conditions such as wound healing and its upregulation has been shown to delay differentiation (42), and transglutaminase-1 (TGM1), which is specifically expressed in the suprabasal layers of the oral epithelium (43). Both markers were significantly upregulated in FOXM1B organotypics (Fig. 2G-J) indicating a sustained keratinocyte hyperproliferation. Furthermore, cytokeratin 19 (KRT19), which is a marker for pre-neoplastic oral epithelia (44) and a target of Gli1 in hair follicle stem cells that is upregulated in basal cell carcinoma (45), was also upregulated in our FOXM1B-organotypics (Supplementary Fig. S4C, D). This expression pattern, along with the inappropriate mitotic activity of keratinocytes within the upper suprabasal layers, suggests that FOXM1B-expressing cells have acquired a defective terminal differentiation. Indeed, immunostaining with early and late keratinocyte differentiation markers cytokeratin 13 (KRT13; Fig. 2K, L) and filaggrin (FLG; Fig. 2M, N) respectively, showed complete absence in the FOXM1B organotypics. Similar results were also obtained from organotypic cultures using collagen gels (data not shown).
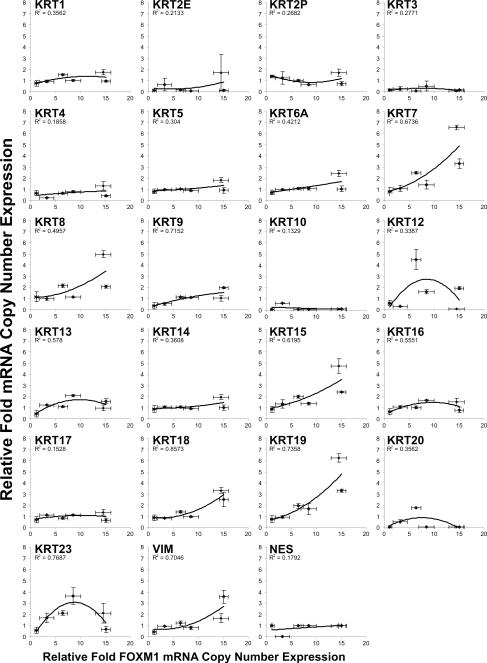
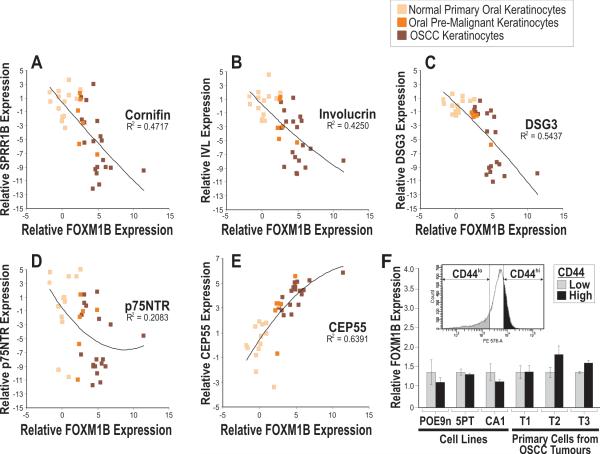
To obtain a more comprehensive understanding of the differentiation characteristics in FOXM1B-expressing oral keratinocytes, we have measured the mRNA levels using absolute qPCR on a panel of 21 classical cytokeratin genes (KRT1, 2E, 2P, 3, 4, 5, 6A, 7, 8, 9, 10, 12, 13, 14, 15, 16, 17, 18, 19, 20 and 23) and 2 mescenchymal-specific genes (VIM and NES) in primary oral keratinocytes transduced with varying doses of FOXM1B in culture (Fig. 3). FOXM1B was found to dose-dependently transactivate a number of simple basal keratin genes including KRT7, 8, 18, 15 and 19. Keratins 15 and 19 are basal-cell-specific markers in stratified epithelia including skin and oral mucosa (46-48). Expression of these keratins has also been associated with the clonogenic stem/progenitor cells of the bulge region in hair follicles that are responsible for maintenance and regeneration of the epidermis (49, 50). Keratins 7, 8 and 18 are simple or mixed-epithelia-specific keratins that are normally not expressed in stratified epithelia but they are induced during carcinogenesis (48). Induction of simple epithelial-specific and stem-cell specific keratins indicates that ectopic FOXM1 expression activated a defective differentiation pathway - a common pathological feature found in epithelial malignancies (41). High levels of FOXM1B expression (Fig. 3) was found to activate VIM which may represent a signal for epithelial-mescenchymal transition (51). Furthermore, genes involved in keratinocyte differentiation, such as cornifin (SPRR1B; Fig. 4A), involucrin (IVL; Fig. 4B) and desmoglein 3 (DSG3; Fig. 4C), were found to be inversely correlated with FOXM1B mRNA expression across a large panel of human oral keratinocytes consisting of 14 primary normal human oral keratinocytes, 6 oral premalignant and 19 oral squamous cell carcinoma cell lines. In agreement with expression patterns found in oral mucosa (Fig. 1A-C) and primary oral keratinocytes (Fig. 1D, E-i), p75NTR expression was also found to be inversely correlated with FOXM1B across this cell line panel (Fig. 4D) supporting a tumor suppressor role for p75NTR (52-54). Given that upregulation of p75NTR inhibits cell proliferation (54) and regulates epithelial differentiation (55), the inverse expression between FOXM1B and p75NTR indicates deregulation of cell proliferation and differentiation pathways during malignant progression. A previously characterized FOXM1B target CEP55 (5), as expected, correlated positively with FOXM1B expression (Fig. 4E). Collectively, these results indicate that the progressive upregulation of FOXM1B and downregulation of keratinocyte differentiation genes may be intricately associated not only during normal epithelial renewal, but also during malignant progression.
Figure 3.
Transcriptional regulation of keratinocyte differentiation genes in primary human oral keratinocytes transduced with varying levels of FOXM1B. Each data point represents mean ± S.E.M from triplicate determinations of 6 independent primary oral keratinocyte strains. Second-order polynomial regression analysis was performed to obtain the R2 coefficient of determination value which indicates the degree of correlation between each gene with FOXM1B.
Figure 4.
Absolute quantitative RT-PCR analysis of FOXM1B mRNA expression and A, cornifin (SPRR1B); B, involucrin (IVL); C, desmoglein 3 (DSG3); D, p75NTR and E, CEP55 across a cell panel consisting of 14 normal primary oral keratinocytes, 6 oral premalignant (SVpgC2a, DOK, OKF6/T, POE9n, D19 and D20) and 19 oral SCC cell lines (SCC9, SCC25, UK1, CALH2, SCC15, VB6, SqCC/Y1, CaDe12, 5PT, SCC4, H357, SVFN1-8). Second-order polynomial regression analysis was performed to obtain the R2 coefficient of determination value which indicates the degree of correlation between two genes. F, Relative FOXM1B mRNA expression levels in FACS-sorted CD44lo and CD44hi cells from cell lines (POE9n, 5PT and CA1) and three independent primary cells extracted from oral SCC tumor tissues (T1-T3), each performed in triplicates. Inset histogram shows respective CD44lo and CD44hi cell populations harvested for qPCR analysis.
Our finding that a defined genetic hit, such as FOXM1 upregulation, induces the sustained expansion of normal stem/progenitor cells led us to question whether putative ‘cancer stem cells’ express different levels of FOXM1 compared to the rest of the tumor cell population. To test this, we investigated the endogenous expression levels of FOXM1 using absolute qPCR in ‘cancer stem cells’ which were FACS-isolated from one premalignant oral dysplastic cell line (POE9n), two established oral SCC cell lines (5PT and CA1) (33) and three primary explants of keratinocytes from independent oral SCC tumors (T1-T3; Figure 4F), using an established oral cancer stem cell marker CD44 (56). We have demonstrated that CD44hi cells were more clonogenic than CD44lo cells in oral SCC cell lines (5PT and CA1; Supplemental Figures S1F and S1G). Interestingly, we have found that CD44hi and CD44lo cancer cells retain similar expression levels of FOXM1 in both day 3 (Figure 4F) and day 12 after FACS-isolation (data not shown), suggesting that FOXM1 expression is uniformly expressed in tumor cells regardless of their lineage hierarchy within the tumor subpopulations. The homogenous FOXM1 expression found in tumor is also consistent with our previous report that FOXM1 protein was abundantly expressed throughout the tumor mass of oral SCCs in vivo (5). In this study, we have shown that contrarily to its heterogeneous expression pattern found in normal oral mucosal tissue which retains an intact program of epithelial differentiation, FOXM1 is uniformly expressed in the whole population of tumor-derived keratinocytes. This suggests that the perturbation of epithelial differentiation during malignancy may further perpetuate FOXM1 expression in tumor cells. This is in agreement with our current hypothesis that a tumor may arise from a common pre-malignant progenitor which has acquired high levels of FOXM1 and later giving rise to progeny malignant cells (regardless of their ‘stemness’) with a hallmark of elevated FOXM1 expression. Furthermore, malignant cells are characterised by multiple genetic hits, many of which (e.g. RAS, loss of p53 function, c-MYC, p16/Rb pathway inactivation, EGFR) are known to lead to constitutive upregulation of FOXM1 regardless of ‘cancer stem cell’ status.
In summary, this study demonstrated the first evidence that FOXM1B has a role in the proliferation of normal epithelial stem/progenitor cell. The aberrant upregulation of FOXM1B in these cells may perturb the balance between stem/progenitor expansion and commitment to terminal differentiation. These findings are consistent with the general consensus regarding epithelial carcinogenesis whereby a premalignant progenitor, possibly one with aberrant activation of FOXM1B, undergoes excessive rounds of clonal expansion within the epithelium. We have now demonstrated that in order to achieve a pro-oncogenic ‘hit’, FOXM1B upregulation ought to be acquired during a ‘permissive window’ during stem/progenitor cell expansion prior to their commitment to differentiation (Fig. 5). Although unable to revert the differentiated phenotype of committed keratinocytes, FOXM1B induced the expansion of stem/progenitor compartment, which may explain why oncogenic stimulation upstream of FOXM1, such as RAS, is most efficient when targeted to the immature stem/progenitor epithelial compartment (30, 31, 57). Although large scale whole-genome sequencing analysis across multiple human cancer types (58) did not find any DNA mutations within the FOXM1 gene (59), we have previously shown that environmental factors such as nicotine (5), and ultraviolet light (6) could directly stabilize and activate endogenous FOXM1B in primary human oral and epidermal cells respectively, while others have demonstrated that various oncogenic pathways lead to the activation of FOXM1B (1, 2). This study provides a novel role for FOXM1B in the initiation of human epithelial neoplasia, while our previous finding that ectopic FOXM1B expression contributed to genomic instability (5, 6) may also explain why this gene is implicated in both malignant progression and metastatic invasion (1, 2).
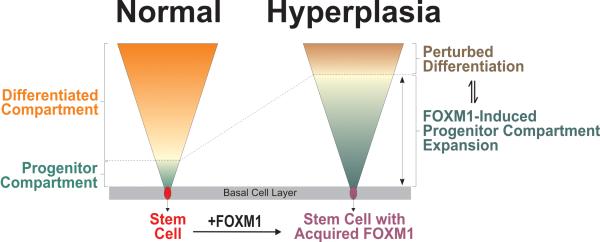
Figure 5.
A schematic model showing the mechanism for FOXM1-induced expansion of epithelial progenitor compartment. We have previously shown that environmental factors such as nicotine (5) and ultraviolet light (6) can directly activate endogenous FOXM1 (1st oncogenic hit) in primary human oral and skin keratinocytes, respectively. Based on multiple lines of evidence found in this study, we showed that aberrant upregulation of FOXM1 in epithelial stem cells produce progenitor cells with enhanced proliferative capacity which impacts on terminal differentiation, resulting in a hyperplastic phenotype displaying perturbed differentiation characteristics. The deregulated differentiation pathway found in hyperplastic tissues may predispose the ‘expanded’ progenitor compartment to subsequent ‘2nd’ oncogenic hit necessary for malignant conversion. We hypothesized that FOXM1 mediates a window of cancer susceptibility in the progenitor cells whereby anticancer therapeutic intervention at this stage may prevent cancer initiation.
Supplementary Material
Acknowledgements
We are indebted to Professor Farida Fortune (Institute of Dentistry, Barts and the London School of Medicine and Dentistry) for her encouragement and support. We thank Professor Ian Mackenzie (Centre for Cutaneous Research, Blizard Institute of Cell and Molecular Science) for his critical advice and precious gifts of various HNSCC cell lines used in this study. The authors would like to thank Professor Irene Leigh (University of Dundee) for her advice on the proliferation-specific regulation of keratins.
Financial Support This study was co-funded by the Wellcome Trust (M.T.T, E.G), Medical Research Council (M.T.T, E.G), the Institute of Dentistry, Barts and The London School of Medicine and Dentistry, and the Norwegian Research Council (grant no. 178601) (D.E.C).
Footnotes
Conflicts of interests No potential conflicts of interests were disclosed.
References
- 1.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 2007;7(11):847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 2.Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388(12):1257–74. doi: 10.1515/BC.2007.159. [DOI] [PubMed] [Google Scholar]
- 3.Laoukili J, Kooistra MR, Bras A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 4.Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res. 2002;62(16):4773–80. [PubMed] [Google Scholar]
- 5.Gemenetzidis E, Bose A, Riaz AM, et al. FOXM1 upregulation is an early event in human squamous cell carcinoma and it is enhanced by nicotine during malignant transformation. PLoS ONE. 2009;4(3):e4849. doi: 10.1371/journal.pone.0004849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teh MT, Gemenetzidis E, Chaplin T, Young BD, Philpott MP. Upregulation of FOXM1 induces genomic instability in human epidermal keratinocytes. Mol Cancer. 2010;9(1):45. doi: 10.1186/1476-4598-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma RY, Tong TH, Cheung AM, Tsang AC, Leung WY, Yao KM. Raf/MEK/MAPK signaling stimulates the nuclear translocation and transactivating activity of FOXM1c. J Cell Sci. 2005;118(Pt 4):795–806. doi: 10.1242/jcs.01657. [DOI] [PubMed] [Google Scholar]
- 8.Kalin TV, Wang IC, Ackerson TJ, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66(3):1712–20. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalinichenko VV, Major ML, Wang X, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18(7):830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gusarova GA, Wang IC, Major ML, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117(1):99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang IC, Meliton L, Ren X, et al. Deletion of Forkhead Box M1 transcription factor from respiratory epithelial cells inhibits pulmonary tumorigenesis. PLoS ONE. 2009;4(8):e6609. doi: 10.1371/journal.pone.0006609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 13.Jaks V, Barker N, Kasper M, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40(11):1291–9. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 14.Snippert HJ, Haegebarth A, Kasper M, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 327(5971):1385–9. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 15.Ito M, Liu Y, Yang Z, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–4. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 16.Jones PH, Harper S, Watt FM. Stem cell patterning and fate in human epidermis. Cell. 1995;80(1):83–93. doi: 10.1016/0092-8674(95)90453-0. [DOI] [PubMed] [Google Scholar]
- 17.Li A, Simmons PJ, Kaur P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc Natl Acad Sci U S A. 1998;95(7):3902–7. doi: 10.1073/pnas.95.7.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci U S A. 1987;84(8):2302–6. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurelli R, Zambruno G, Guerra L, et al. Inactivation of p16INK4a (inhibitor of cyclin-dependent kinase 4A) immortalizes primary human keratinocytes by maintaining cells in the stem cell compartment. FASEB J. 2006;20(9):1516–8. doi: 10.1096/fj.05-4480fje. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrini G, Dellambra E, Golisano O, et al. p63 identifies keratinocyte stem cells. Proc Natl Acad Sci U S A. 2001;98(6):3156–61. doi: 10.1073/pnas.061032098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73(4):713–24. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 22.Wan H, Yuan M, Simpson C, et al. Stem/progenitor cell-like properties of desmoglein 3dim cells in primary and immortalized keratinocyte lines. Stem Cells. 2007;25(5):1286–97. doi: 10.1634/stemcells.2006-0304. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Endo K, Kinoshita S. Identification of human oral keratinocyte stem/progenitor cells by neurotrophin receptor p75 and the role of neurotrophin/p75 signaling. Stem Cells. 2007;25(3):628–38. doi: 10.1634/stemcells.2006-0494. [DOI] [PubMed] [Google Scholar]
- 24.Zhu AJ, Haase I, Watt FM. Signaling via beta1 integrins and mitogen-activated protein kinase determines human epidermal stem cell fate in vitro. Proc Natl Acad Sci U S A. 1999;96(12):6728–33. doi: 10.1073/pnas.96.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu AJ, Watt FM. beta-catenin signalling modulates proliferative potential of human epidermal keratinocytes independently of intercellular adhesion. Development. 1999;126(10):2285–98. doi: 10.1242/dev.126.10.2285. [DOI] [PubMed] [Google Scholar]
- 26.Gandarillas A, Watt FM. c-Myc promotes differentiation of human epidermal stem cells. Genes Dev. 1997;11(21):2869–82. doi: 10.1101/gad.11.21.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold I, Watt FM. c-Myc activation in transgenic mouse epidermis results in mobilization of stem cells and differentiation of their progeny. Curr Biol. 2001;11(8):558–68. doi: 10.1016/s0960-9822(01)00154-3. [DOI] [PubMed] [Google Scholar]
- 28.Barrandon Y, Morgan JR, Mulligan RC, Green H. Restoration of growth potential in paraclones of human keratinocytes by a viral oncogene. Proc Natl Acad Sci U S A. 1989;86(11):4102–6. doi: 10.1073/pnas.86.11.4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholes AG, Woolgar JA, Boyle MA, et al. Synchronous oral carcinomas: independent or common clonal origin? Cancer Res. 1998;58(9):2003–6. [PubMed] [Google Scholar]
- 30.Bailleul B, Surani MA, White S, et al. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell. 1990;62(4):697–708. doi: 10.1016/0092-8674(90)90115-u. [DOI] [PubMed] [Google Scholar]
- 31.Greenhalgh DA, Rothnagel JA, Quintanilla MI, et al. Induction of epidermal hyperplasia, hyperkeratosis, and papillomas in transgenic mice by a targeted v-Ha-ras oncogene. Mol Carcinog. 1993;7(2):99–110. doi: 10.1002/mc.2940070208. [DOI] [PubMed] [Google Scholar]
- 32.Ye H, Kelly TF, Samadani U, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626–41. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harper LJ, Piper K, Common J, Fortune F, Mackenzie IC. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J Oral Pathol Med. 2007;36(10):594–603. doi: 10.1111/j.1600-0714.2007.00617.x. [DOI] [PubMed] [Google Scholar]
- 34.Bickenbach JR, Mackenzie IC. Identification and localization of label-retaining cells in hamster epithelia. J Invest Dermatol. 1984;82(6):618–22. doi: 10.1111/1523-1747.ep12261460. [DOI] [PubMed] [Google Scholar]
- 35.Morris DJ, Reis A. A YAC contig spanning the nevoid basal cell carcinoma syndrome, Fanconi anaemia group C, and xeroderma pigmentosum group A loci on chromosome 9q. Genomics. 1994;23(1):23–9. doi: 10.1006/geno.1994.1454. [DOI] [PubMed] [Google Scholar]
- 36.Okumura T, Shimada Y, Imamura M, Yasumoto S. Neurotrophin receptor p75(NTR) characterizes human esophageal keratinocyte stem cells in vitro. Oncogene. 2003;22(26):4017–26. doi: 10.1038/sj.onc.1206525. [DOI] [PubMed] [Google Scholar]
- 37.Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A. 2006;103(32):11958–63. doi: 10.1073/pnas.0601886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanco-Bose WE, Murphy MJ, Ehninger A, et al. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48(4):1302–11. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
- 39.Ueno H, Nakajo N, Watanabe M, Isoda M, Sagata N. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development. 2008;135(11):2023–30. doi: 10.1242/dev.019893. [DOI] [PubMed] [Google Scholar]
- 40.Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nat Rev Cancer. 2005;5(2):127–35. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 41.Owens DM, Watt FM. Contribution of stem cells and differentiated cells to epidermal tumors. Nat Rev Cancer. 2003;3(6):444–51. doi: 10.1038/nrc1096. [DOI] [PubMed] [Google Scholar]
- 42.Paladini RD, Coulombe PA. Directed expression of keratin 16 to the progenitor basal cells of transgenic mouse skin delays skin maturation. J Cell Biol. 1998;142(4):1035–51. doi: 10.1083/jcb.142.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ta BM, Gallagher GT, Chakravarty R, Rice RH. Keratinocyte transglutaminase in human skin and oral mucosa: cytoplasmic localization and uncoupling of differentiation markers. J Cell Sci. 1990;95(Pt 4):631–8. doi: 10.1242/jcs.95.4.631. [DOI] [PubMed] [Google Scholar]
- 44.Lindberg K, Rheinwald JG. Suprabasal 40 kd keratin (K19) expression as an immunohistologic marker of premalignancy in oral epithelium. Am J Pathol. 1989;134(1):89–98. [PMC free article] [PubMed] [Google Scholar]
- 45.Youssef KK, Van Keymeulen A, Lapouge G, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12(3):299–305. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 46.Waseem A, Dogan B, Tidman N, et al. Keratin 15 expression in stratified epithelia: downregulation in activated keratinocytes. J Invest Dermatol. 1999;112(3):362–9. doi: 10.1046/j.1523-1747.1999.00535.x. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd C, Yu QC, Cheng J, et al. The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J Cell Biol. 1995;129(5):1329–44. doi: 10.1083/jcb.129.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casanova ML, Bravo A, Jorcano JL. Simple Epithelial Keratins: Expression, Function and Disease. In: Paramio J, editor. Intermediate Filaments. Springer; US: 2006. pp. 110–9. [Google Scholar]
- 49.Liu Y, Lyle S, Yang Z, Cotsarelis G. Keratin 15 promoter targets putative epithelial stem cells in the hair follicle bulge. J Invest Dermatol. 2003;121(5):963–8. doi: 10.1046/j.1523-1747.2003.12600.x. [DOI] [PubMed] [Google Scholar]
- 50.Michel M, Torok N, Godbout MJ, et al. Keratin 19 as a biochemical marker of skin stem cells in vivo and in vitro: keratin 19 expressing cells are differentially localized in function of anatomic sites, and their number varies with donor age and culture stage. J Cell Sci. 1996;109(Pt 5):1017–28. doi: 10.1242/jcs.109.5.1017. [DOI] [PubMed] [Google Scholar]
- 51.Vaughan MB, Ramirez RD, Andrews CM, Wright WE, Shay JW. H-ras expression in immortalized keratinocytes produces an invasive epithelium in cultured skin equivalents. PLoS ONE. 2009;4(11):e7908. doi: 10.1371/journal.pone.0007908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quann EJ, Khwaja F, Zavitz KH, Djakiew D. The aryl propionic acid R-flurbiprofen selectively induces p75NTR-dependent decreased survival of prostate tumor cells. Cancer Res. 2007;67(7):3254–62. doi: 10.1158/0008-5472.CAN-06-3657. [DOI] [PubMed] [Google Scholar]
- 53.Khwaja F, Allen J, Lynch J, Andrews P, Djakiew D. Ibuprofen inhibits survival of bladder cancer cells by induced expression of the p75NTR tumor suppressor protein. Cancer Res. 2004;64(17):6207–13. doi: 10.1158/0008-5472.CAN-03-3814. [DOI] [PubMed] [Google Scholar]
- 54.Jin H, Pan Y, Zhao L, et al. p75 neurotrophin receptor suppresses the proliferation of human gastric cancer cells. Neoplasia. 2007;9(6):471–8. doi: 10.1593/neo.07175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nalbandian A, Pang AL, Rennert OM, Chan WY, Ravindranath N, Djakiew D. A novel function of differentiation revealed by cDNA microarray profiling of p75NTR-regulated gene expression. Differentiation. 2005;73(8):385–96. doi: 10.1111/j.1432-0436.2005.00040.x. [DOI] [PubMed] [Google Scholar]
- 56.Prince ME, Sivanandan R, Kaczorowski A, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104(3):973–8. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown K, Strathdee D, Bryson S, Lambie W, Balmain A. The malignant capacity of skin tumors induced by expression of a mutant H-ras transgene depends on the cell type targeted. Curr Biol. 1998;8(9):516–24. doi: 10.1016/s0960-9822(98)70203-9. [DOI] [PubMed] [Google Scholar]
- 58.Bignell GR, Greenman CD, Davies H, et al. Signatures of mutation and selection in the cancer genome. Nature. 2010;463(7283):893–8. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forbes SA, Tang G, Bindal N, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;(38):D652–7. doi: 10.1093/nar/gkp995. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.