Abstract
Background
Autosomal recessive congenital ichthyosis (ARCI) is a rare hereditary disorder of cornification. Mutations in the transglutaminase-1 (TGM1) gene, which encodes for the epidermal enzyme transglutaminase-1 (TGase-1), are one of the causes of ARCI.
Methods
The TGM1 mutation spectrum was characterised and genotype–phenotype correlations investigated in 104 patients with ARCI ascertained through the National Registry for Ichthyosis and Related Disorders in the USA.
Methods
Germline mutations in TGM1 were identified in 55% (57/104) of patients with ARCI. Arginine residues in TGase-1 were mutated in 39% (22/57) of patients overall and 54% (20/37) of those with missense mutations. In total, 55% (12/22) of missense mutations were within CpG dinucleotides and 92% (11/12) of these mutations were C→T or G→A transitions. The genotype–phenotype investigation found that ARCI with TGM1 mutations was significantly associated with presence of collodion membrane at birth (p = 0.006), ectropion (p = 0.001), plate-like scales (p = 0.005) and alopecia (p = 0.001). Patients who had at least one mutation predicted to truncate TGase-1 were more likely to have more severe hypohidrosis (p = 0.001) and overheating (p = 0.0007) at onset of symptoms than were those with exclusively TGM1 missense mutations. A logistic model was developed, which predicted that individuals with collodion membrane, alopecia and/or eye problems are about four times more likely to have TGM1 mutations than patients without these findings.
Conclusion
This is the largest investigation of patients with ARCI to date. It expands the TGM1 mutation spectrum and confirms that despite genetic and phenotypic heterogeneity in ARCI, TGM1 is the main causative gene for this disorder. The high frequency of mutated arginine codons in TGM1 may be due to the deamination of CpG dinucleotides.
Autosomal recessive congenital ichthyosis (ARCI; OMIM 242100, 242300) classifies a clinically diverse group of autosomal recessive hereditary disorders of cornification characterised by epidermal scaling. ARCI is rare, with estimated incidence rates of 1:200 000–1:300 000 in the USA1 and as high as 1:91 000 in Norway, owing to a founder effect.2 Patients with ARCI are often born encased in a very tight, translucent, thick sheath called a collodion membrane. Although the collodion membrane usually desquamates within the first few weeks of life, ectropion, eclabium and digital and joint contractures are some of its more long-lasting sequelae. After the shedding of the membrane, a variable amount of epidermal scaling soon appears. Patients may also have alopecia, hypohidrosis and palmar–plantar hyperkeratosis.
ARCI has traditionally been divided into two major clinical subtypes: lamellar ichthyosis (LI; OMIM 242300 and non-bullous congenital ichthyosiform erythroderma (NBCIE; OMIM 242100.3 LI, characterised by large, dark, plate-like cutaneous scales with minimal erythema, represents one end of this spectrum and NBCIE, characterised by erythroderma with overlying fine, white scales, represents the other end. However, patients may display cutaneous finding of both subtypes, making it difficult to classify them either as NBCIE or LI, as a patient may lie in the middle of the wide ARCI spectrum.
Russell et al mapped the LI locus to the long arm of chromosome 14 (14q11), showed linkage to the TGM1 gene and identified germline mutations in TGM1 in patients with classic LI.4, 5 Since then, > 70 unique TGM1 germline mutations have been reported.5–33 Approximately 35–40% of patients with ARCI have germline mutations in TGM1.34 A founder mutation, IVS5-2A→G (also reported as A2526G or +3366A→G), has been identified in 80% of TGM1 mutated alleles in patients with LI or NBCIE in the Norwegian population.2 This mutation is also prevalent in families in North America with ARCI, accounting for 9.6% of TGM1 mutated alleles.29
TGM1 consists of 15 exons and encodes a 90 kDa, 817-amino-acid protein called transglutaminase-1 (TGase-1).35 TGase-1 is one of eight catalytic transglutaminases identified in humans and one of three transglutaminases found in the epidermis.36 TGase-1 is a Ca2+-dependent, membrane-bound enzyme that functions in the formation of the cornified cell envelope (CCE). The CCE, an insoluble 10–20-nm structure composed of proteins cross-linked by Nε-(γ-glutamyl)lysine bonds, is laid down on the inside of the plasma membrane in keratinocytes in the stratum corneum. The CCE acts as a mechanical barrier and protects against water loss and infectious agents. TGase-1 is responsible for catalysing the critical Nε-(γ-glutamyl) lysine cross-linking of ARP preserver proteins such as involucrin and loricrin during the formation of the CCE.37, 38 Decreased to absent enzyme activity in TGase-1 has been reported in patients with germline mutations in TGM1.6–9, 14, 17, 25, 32, 39, 40 Knockout TGM1 mice present at birth with ARCI-like features, including erythematous, tight, translucent skin, reminiscent of a collodion membrane. Newborn TGM1−/−knockout mice do not feed and consequently develop severe dehydration, dying within 4–5 hours of birth.41
ARCI is a genetically heterogenous disease and to date, germline mutations in five other genes besides TGM1 have been identified in patients with ARCI. ABCA12 encodes an ATP binding epidermal transporter involved in lipid trafficking.42, 43 ABCA12 mutations causing premature termination of the protein lead to harlequin ichthyosis,44, 45 whereas missense mutations have been reported in patients with LI.46 Germline mutations in ALOX12B and ALOXE3, which encode for epidermal lipoxygenases,47 and in Ichthyin48 and CYP4F22,49 have also been associated with ARCI.
Because of the genetic heterogeneity and the rarity of the disorder, few genotype–phenotype association studies have been conducted. Some studies suggest that patients with ARCI TGM1 mutations are associated with LI, although only some patients with NBCIE have TGM1 mutations.15, 29, 50, 51 In this study, we identified novel TGM1 mutations and genotype–phenotype correlations in 104 patients with ARCI identified through the National Registry for Ichthyosis and Related Disorders in the US. This is the largest and most comprehensive study of ARCI patients with TGM1 mutations to date.
METHODS
Patient ascertainment and study design
From 1995 to 2004, subjects with inherited scaling skin disorders were ascertained for enrolment in the National Registry for Ichthyosis and Related Disorders through announcements by the Foundation for Ichthyosis and Related Skin Types (FIRST), in dermatology journals, at national dermatology meetings and through recruitment letters mailed to members of the American Academy of Dermatology. Enrolment in the Registry included a telephone interview, medical record review and blood samples and/or buccal swabs.
Patients were enrolled in the Registry through their local dermatologist. All subjects were enrolled in a protocol approved by the University of Washington’s institutional review board. Each patient or their guardian provided written informed consent/assent before participation.
Every participant had a dermatological evaluation and a standard clinical-information form was completed by their dermatologist. Patients or their guardians were mailed a 10-page questionnaire and a 45–60 minute telephone interview, based on the standardised questionnaire, was administered by the Registry coordinator, who was blinded to patients’ TGM1 mutation status. The telephone interview elicited information regarding various aspects of dermatological history, such as presence or absence of scales, erythema, sweating, temperature regulation, skin infections, ectropion, eclabium, alopecia, palm or sole involvement and specified areas of skin involvement. Each of these topics had accompanying questions regarding severity at its onset, currently or at its worst, using a scale of mild, mild/moderate, moderate, moderate/severe and severe. Patients were also asked about their family, personal (including eye problems), birth and treatment medical histories and their quality of life. Eye problems included ectropion, corneal erosions, scarring, impaired vision, glaucoma, cataracts and history of eye surgery.
In total, 610 patients were enrolled in the study, of whom 206 were diagnosed with ARCI. None of the patients had a diagnosis of bathing-suit ichthyosis. All 206 subjects with ARCI were invited to submit buccal swabs or blood for sequencing of TGM1 and 104 (62 female, 42 male; 94 white, 10 Hispanic; average age at the time of interview 24.4 years, range 2 months to 72 years) provided buccal samples for analysis.
Mutation analysis
DNA was extracted from blood and/or buccal swabs according to standard procedures.52 DNA from the submitted specimens was amplified by PCR for analysis of exons 2–15 of the TGM1 gene and their flanking splice-sites. Bidirectional sequence was obtained for analysis and compared with the published gene sequence. DNA from at least 160 unrelated control individuals was examined for each disease-associated sequence variant.
Statistical analysis
Data collected from patients with ARCI addressed the following specific features: presence of collodion membrane, plate-like scales, erythema, lack of sweating, temperature regulation, skin infections, ectropion, eclabium, alopecia and palm or sole involvement. Analyses comparing two dichotomous parameters were performed using the Fisher exact test. The Mehta modification to this test was used to compare an unordered categorical parameter to a dichotomous parameter.53 An exact Cochran–Armitage test for trend was used to determine the association between a dichotomous parameter and ordered categorical parameters.54 Associations between two ordered categorical parameters, such as the number of mutations present and the level of severity of a clinical feature, were determined using an exact Jonckheere–Terpstra test for trend.55 All p values were two-sided and were not adjusted for multiple comparisons. In view of the exploratory nature of the study and the large number of comparisons performed, only p <0.005 was considered significant.
We performed a logistic regression analysis to determine whether the TGM1 mutation status of patients with ARCI could be predicted by the presence of a combination of the following clinical variables: collodion membrane, ectropion, other eye problems, alopecia, plate-like scales, use of systemic retinoids and use of topical retinoids. Using the backward selection method, a model was developed.
Literature review
We performed an electronic literature search from 1994 to 2008 designed to capture all reported cases of ARCI with TGM1 germline mutations. Cases were included in our review even if the method of mutation detection was not included. In total, 36 articles were included.2, 5–33, 40, 50, 51, 56–58
RESULTS
TGM1 mutation analysis
Using direct sequencing, we identified TGM1 mutations in 57 patients (55% TGM1 mutation detection rate), giving a total of 103 mutated alleles.
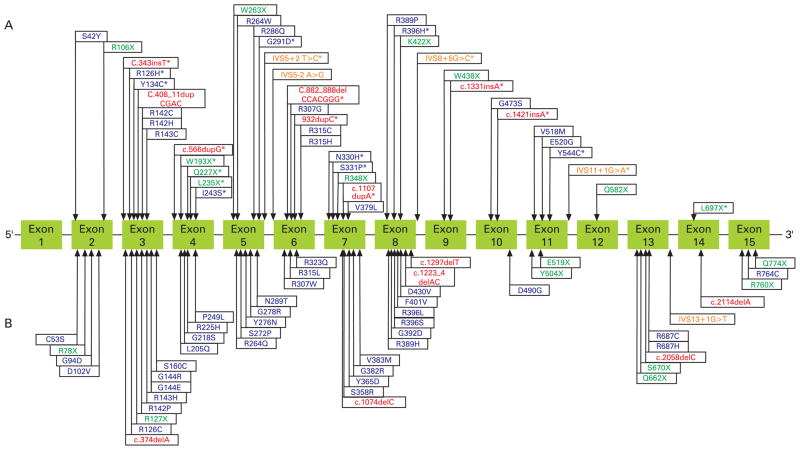
Direct sequence analysis of the 14 coding exons and splice-site junctions of TGM1 showed 44 different TGM1 germline mutations including 22 missense, 10 nonsense, 8 frameshift and 4 splice-site mutations (Figure 1A). Of these 44 mutations, 22 were novel, including 8 missense (R126H, Y134C, I243S, G291D, N330H, S331P, R396H, Y544C), 7 frameshifts (c.343insT, c.566dupG, c.882_888delCCACGGG, c.932dupC, c.1107dupA, c.1331insA, c.1421insA), 4 nonsense (W193X, Q227X, L235X, L697X) and 3 putative splice-site mutations (IVS5+2T→C (also known as c.876+2T→C), IVS8+5G→C, IVS11+1G→A) (fig 1A). The identified TGM1 mutations affected all exons, with the exception of exons 1, 13 and 15 (fig 1A). The most 5′ mutation (S42Y) occurred in exon 2 and the most 3′ mutation (L697X) occurred in exon 15.
Figure 1.
TGM1 mutations reported in this investigation and previous studies. (A) The 44 germline TGM1 mutations identified. Asterisks denote the 22 novel mutations identified; mutations identified in both this and previous studies are not marked with asterisks. (B) The 50 previously reported germline TGM1 mutations that were not identified in the present study. Blue, missense; green, nonsense; red, frameshifts, orange, putative splice site.
In total, 65% (37/57) of patients (48 mutated alleles) had at least one missense mutation. Of the 37 patients with missense mutations, 27 (73%) had at least one mutation in the catalytic core, 9 had mutations in the β-sandwich domain only and 1 had a mutation in the anchoring domain. The catalytic core was the most commonly mutated protein domain, accounting for 67% (32/48) of all missense mutated alleles, the β-sandwich was second with 31% (15/48), followed by the anchoring domain with one mutation. No missense mutations were found in either of the β-barrel domains. Of patients who had at least one missense mutation, 54% (20/37) had at least one missense mutation at an arginine residue. Of the six codons that encode arginine, four contained CpG dinucleotides (CGG, CGC, CGT, CGA). We found that 54% (12/22) of TGM1 missense mutations in patients with ARCI were within CpG dinucleotides and 92% (11/12) of these mutations were G→A or C→T transitions. Overall, arginine residues were mutated in 39% (22/57) of all patients with ARCI with TGM1 mutations. Of the 57 patients with TGM1 mutations, 19% (11/57) had at least one nonsense mutation and 12% (7/57) had at least one frameshift mutation.
Overall, the most common TGM1 mutation was the splice-site mutation IVS5-2A→G (also referred to as c.877-2A→G and A2526G), which was present in 39% (22/57) of patients (7 homozygous and 22 compound heterozygous). TGM1 mutations in exon 3 were found in 28% (16/57) of patients (18 mutated alleles). The missense mutation R142H was the most common mutation in exon 3 and the second most common mutation in our cohort overall (5 patients with 7 mutated alleles).
Sequence alignments
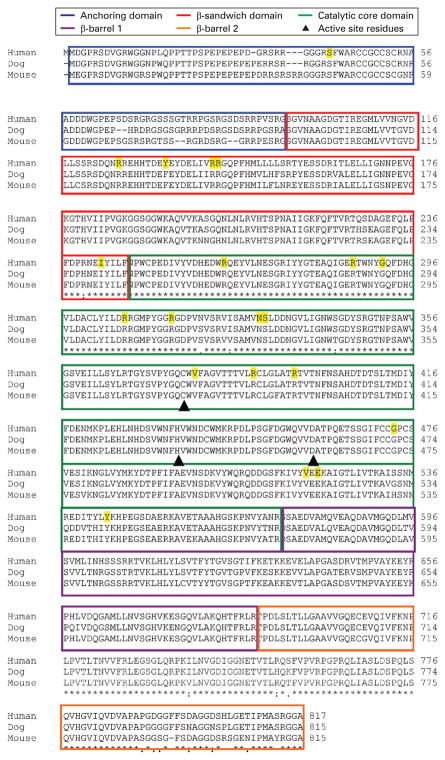
To evaluate the sequence conservation in TGase-1, we ran a BLAST analysis of the amino acid sequences of human, dog, mouse TGase-1 and found that TGase-1 is highly conserved among species (fig 2).59 We found that human and mouse TGase-1 had 88% identity and 91% similarity and human and dog TGase-1 had 89% identity and 92% physical and chemical similarity. Our study showed that missense mutations in TGM1 occurred at evolutionarily conserved residues across species. We identified 22 missense mutations in TGM1 in 20 different residues in TGase-1. Most (95%; 19/20) missense mutations identified in patients with ARCI in this study occurred at residues identical to those of dog and mouse TGase-1(fig 2).
Figure 2.
TGase-1 alignments showing amino acid sequence comparisons among human, dog and mouse. Boxes indicate amino acid residues within the TGase-1 domains. Yellow characters indicate the amino acid positions where missense mutations were identified. Triangles represent active site residues (Cys377, His436, Asp459). The 22 TGM1 missense mutations identified in our cohort are marked.
Genotype–phenotype analyses
Patients with autosomal recessive congenital ichthyosis with TGM1 mutations versus patients without TGM1 mutations
Patients with ARCI with TGM1 mutations (n = 57) were significantly associated with increased probability of collodion membrane at birth (p = 0.006), alopecia (p = 0.001), plate-like scales (p = 0.005), ectropion (p = 0.001) and systemic retinoid use (p = 0.002) compared with patients with ARCI without TGM1 mutations (n = 47) (table 1). We found that subjects without TGM1 mutations reported only slightly higher frequency of erythema (77%) compared with ARCI subjects with a TGM1 mutation (67%), but they were not significantly different (p = 0.029) (table 1).
Table 1.
Analyses of clinical features comparing patients with and without at least one TGM1 mutation
| Clinical features or treatment | Patients, % (n/total n) |
p Value | |
|---|---|---|---|
| Without TGM1 mutations (n = 47) | With at least one TGM1 mutation (n = 57) | ||
| Collodion membrane | 44 (18/41) | 73 (38/52) | 0.006* |
| Alopecia | 21 (10/47) | 53 (30/57) | 0.001* |
| Plate-like scale | 64 (30/47) | 88 (50/57) | 0.005* |
| Ectropion | 60 (28/47) | 88 (50/57) | 0.001* |
| Severity at onset‡ | |||
| 1 or 2 (n = 28) | 50 (14) | 50 (14) | 0.019‡ |
| 3 (n = 23) | 39 (9) | 61 (14) | |
| 4 or 5 (n = 23) | 17 (4) | 83 (19) | |
| Severity at worst‡ | |||
| 1 or 2 (n = 26) | 50 (13) | 50 (13) | 0.034‡ |
| 3 (n = 20) | 40 (8) | 60 (12) | |
| 4 or 5 (n = 30) | 23 (7) | 77 (23) | |
| Eye problems | 40 (19/47) | 65 (37/57) | 0.018* |
| Erythema | 77 (36/47) | 67 (38/57) | 0.29* |
| Fine scales | 96 (45/47) | 89 (51/57) | 0.29* |
| Eclabium | 15 (7/47) | 23 (13/57) | 0.33* |
| Temperature regulation | 96 (45/47) | 91 (52/57) | 0.45* |
| Hearing problems | 51 (24/47) | 58 (33/57) | 0.55* |
| Hypohidrosis | 89 (42/47) | 84 (48/57) | 0.57* |
| Soles | 94 (44/47) | 89 (51/57) | 0.51* |
| Palms | 89 (42/47) | 84 (48/57) | 0.57* |
| Skin infections | 51 (24/47) | 51 (29/57) | 1.0* |
| Treatment | |||
| Systemic retinoids | 13 (6/46) | 42 (24/57) | 0.002* |
| Topical retinoids | 25 (11/44) | 51 (27/53) | 0.012* |
| Systemic steroids | 7 (3/44) | 0 (0/53) | 0.090* |
Fisher’s exact test.
Cochran–Armitage exact trend test.
Severity scale.
Severity: 1 or 2, mild or mild/moderate; 3, moderate; 4 or 5, moderate/severe or severe.
Bold p values indicate significance (p<0.005).
There was a trend towards differences between patients with ARCI with and without TGM1 mutations in severity of ectropion at its onset (p = 0.019) and its worst (p = 0.034) (table 1). More patients with TGM1 mutations reported moderate/severe or severe ectropion at its onset and at its worst than did patients without TGM1 mutations. Of 23 patients reporting moderate/severe or severe ectropion at onset, 83% had TGM1 mutations compared with 17% without TGM1 mutations (table 1).
Patients with autosomal recessive congenital ichthyosis who had at least one mutation predicted to truncate transglutaminase versus those who had only missense TGM1 mutations
Patients with ARCI who had only mutations predicted to truncate TGase-1 were more likely to have sweating abnormalities than were patients who had at least one TGM1 missense mutation (p = 0.041) (table 2). More specifically, when asked about hypohidrosis, all 20 patients with mutations predicted to truncate TGase-1 reported hypohidrosis, compared with 76% of patients who had at least one missense mutation (p = 0.020). Similarly, we found a trend for increasing severity of overheating among patients who had only truncating mutations compared with patients who had at least one TGM1 missense mutation (p = 0.022).
Table 2.
Analyses of clinical features comparing patients who had only mutations predicted to truncated TGase-1 versus patients who had at least one TGM1 missense mutation
| Clinical features | Patients, % (n/total n) |
p Value | |
|---|---|---|---|
| With mutations predicted only to truncate TGase-1 (n = 20) | With at least one TGM1 missense mutation (n = 37) | ||
| Sweating abnormality | 100 (20/20) | 78 (29/37) | 0.041* |
| Hypohidrosis | 100 (20/20) | 76 (28/37) | 0.020* |
| Overheating | |||
| Severity at worst‡ | |||
| 1 or 2 (n = 11) | 9 (1) | 91 (10) | 0.022† |
| 3 (n = 16) | 37 (6) | 63 (10) | |
| 4 or 5 (n = 24) | 50 (12) | 50 (12) | |
Fisher’s exact test.
Cochran–Armitage exact trend test.
Severity: 1 or 2, mild or mild/moderate; 3, moderate; 4 or 5, moderate/severe or severe.
Patients with autosomal recessive congenital ichthyosis who had only mutations predicted to truncate transglutaminase-1 versus patients who had at least one TGM1 missense mutation
Similar to the findings above, patients with ARCI who had at least one mutation predicted to truncate TGase-1 were significantly more likely to have moderate/severe or severe hypohidrosis at its onset (p = 0.005) or at its worst (p = 0.001) compared with patients who had only TGM1 missense mutations (table 3). Of the patients reporting moderate/severe or severe hypohidrosis at its worst, 83% had at least one mutation predicted to truncate TGase-1 and the remaining 17% had only missense mutations. There was also a significant association between severity of overheating at its worst and the presence of at least one truncating TGase-1 mutation (p = 0.0007) (table 3). Of patients who reported moderate/severe or severe overheating at worst, 92% had mutations predicted to truncate TGase-1, and the remaining 8% had only missense mutations. Finally, we found that patients who had at least one truncating mutation were less likely to report eclabium than were patients with only missense mutations (p = 0.007) (table 3), but this association did not reach statistical significance.
Table 3.
Analyses of clinical features comparing patients who had at least one mutation predicted to truncate TGase-1 versus patients who had only TGM1 missense mutations
| Clinical features | Patients, % (n/total n) |
p Value | |
|---|---|---|---|
| With at least one mutation predicted to truncate TGase-1 (n = 37) | With TGM1 missense mutations only (n = 20) | ||
| Hypohidrosis | 92 (34/37) | 70 (14/20) | 0.054* |
| Severity at onset | |||
| 1 or 2 (n = 2) | 0 | 100 (2) | 0.005† |
| 3 (n = 8) | 50 (4) | 50 (4) | |
| 4 or 5 (n = 34) | 82 (28) | 18 (6) | |
| Severity currently | |||
| 1 or 2 (n = 2) | 0 | 100 (2) | 0.027† |
| 3 (n = 12) | 67 (8) | 33 (4) | |
| 4 or 5 (n = 30) | 80 (24) | 20 (6) | |
| Severity at worst | |||
| 1 or 2 (n = 2) | 0 | 100 (2) | 0.001† |
| 3 (n = 8) | 37 (3) | 63 (5) | |
| 4 or 5 (n = 35) | 83 (28) | 17 (6) | |
| Overheating | 97 (36/37) | 80 (16/20) | 0.047* |
| Severity at onset‡ | |||
| 1 or 2 (n = 12) | 42 (5) | 58 (7) | 0.039† |
| 3 (n = 28) | 79 (22) | 21 (6) | |
| 4 or 5 (n = 9) | 78 (7) | 22 (2) | |
| Severity at worst‡ | |||
| 1 or 2 (n = 11) | 36 (4) | 64 (7) | 0.0007† |
| 3 (n = 16) | 63 (10) | 37 (6) | |
| 4 or 5 (n = 24) | 92 (22) | 8 (2) | |
Fisher’s exact test.
Cochran–Armitage exact trend test.
Severity: 1 or 2, mild or mild/moderate; 3, moderate; 4 or 5, moderate/severe or severe.
Bolded p values indicate significance (p<0.005).
Modelling results
Using a backward selection method and adjusting for the set of parameters which remain in the final model, we found that patients with a collodion membrane were 4.24 times (95% CI 1.58 to 11.39; p = 0.004) more likely to have a TGM1 mutation, those with alopecia were 4.13 times (95% CI 1.49 to 11.5; p = 0.007) more likely to have a TGM1 mutation and patients with eye problems were 3.60 times (95% CI 1.37 to 9.46; p = 0.009) more likely to have a TGM1 mutation. Eye problems included ectropion, corneal erosions, scarring, impaired vision, glaucoma, cataracts and history of eye surgery. The specificity and sensitivity of the backward selection model for predicting who has a mutation, based on presentation of clinical features, were 76% and 69%, respectively.
Review of published reports of TGM1 germline mutations
We reviewed cases with published TGM1 germline mutations and general clinical characteristics associated with ARCI. The number of affected individuals in a family ranged from one to seven. Excluding the present study, 72 unique TGM1 germline mutations have been previously reported, comprising 49 missense, 13 nonsense, 7 insertion/deletion and 2 putative splice-site mutations (fig 1A,B).2, 5–16, 19–27, 29–33, 40, 50, 56–58 These previously published TGM1 mutations affected all translated exons (215). Of the 72 TGM1 germline mutations reported to date, only 22 mutations were also identified in the present study (fig 1A).5, 10–17, 19, 20, 23, 29, 33 In combination with previous reports,5–33 our 22 novel TGM1 mutations expand the number of unique published TGM1 mutations to 94, comprising 57 missense, 18 nonsense, 14 insertion/deletion and 5 splice-site mutations.
Excluding the present study, the hotspot mutation IVS5–2A→G was the most common single mutation previously reported to date, being found in 64 individuals from 58 ARCI families.2, 11, 12, 14–17, 23, 26, 29, 57 Exon 3 was the most common coding sequence site for mutations, with 30 affected individuals from 23 unrelated ARCI families having TGM1 mutations.5, 8, 11, 15, 17, 20, 23, 25, 32, 57 The most 5′ and 3′ TGM1 mutations occurred in exon 2 (c.125C→A (S42Y))16 and the most 3′ mutation reported occurred in exon 15 (c.2320C→T (Q774X))13
DISCUSSION
We identified three main genotype–phenotype correlations in the largest series to date of 104 patients with ARCI ascertained through the National Registry for Ichthyosis and Related Disorders in the USA. We found that TGM1 mutations are a major cause of ARCI in the USA, with 55% of patients with ARCI tested having germline TGM1 mutations. We characterised 22 novel mutations and in combination with previous reports this study expands the number of identified TGM1 germline mutations to 94.2, 5–33, 40, 50, 51, 56–58 Our study showed that missense mutations in TGM1 occur in highly conserved amino acids, suggesting the functional importance of this enzyme in evolution. Our investigation revealed that 40% of patients affected by TGM1 mutations had mutations in TGase-1 arginine residues. In combination with the previous reports, this indicates that 41% of all TGM1 mutations occur at arginine residues.5–7, 11, 13, 15–17, 19, 20, 22–24, 28–30 Arginine is the most commonly mutated amino acid, accounting for 15% of all mutations in genetic diseases.60 Arginine residues are very highly mutable due to the deamination of 5′-CpG dinucleotides of arginine codons.61, 62 Four of the six codons that encode arginine contain CpG dinucleotides, thus partly explaining the tendency of the arginine codon to mutate.61, 63 The deamination of CpG dinucleotides occurs due to methylation-mediated deamination of 5-methylcystosine in CpG dinucleotides.61, 62 In phenylketonuria, arginine mutations also have the highest frequency of all PAH gene mutations.63 Furthermore, it has been shown that specific arginine residues in keratin K-14 (K14-R125) and K-10 (K10-R156) are mutated at high frequency in patients with severe epidermal bullosa simplex and epidermolytic hyperkeratosis because both codons contain CpG dinucleotides.64
In contrast to most previous studies,2, 15, 20, 51, 57 our large sample size and detailed clinical data allowed us to conduct a comprehensive genotype–phenotype investigation and identify four main genotype–phenotype associations. We found that patients with ARCI and TGM1 mutations were significantly more likely to report the presence of (1) a collodion membrane at birth, (2) ectropion, (3) plate-like scales and (4) alopecia compared with patients without TGM1 mutations. In general, our investigation suggests that patients with TGM1 mutations were more severely affected than patients without TGM1 mutations. These findings are consistent with the literature and confirm the previously observation that patients with TGM1 mutations more commonly have collodion membrane at birth and ectropion compared with patients without TGM1 mutations.50 The fact that two independent groups identified similar significant genotype–phenotype correlations in different populations is significantly important.
In agreement with Ganemo et al,50 we found that patients with ARCI with TGM1 mutations reported alopecia more often than those without TGM1 mutations. However, we found a strong correlation between patients with TGM1 mutations and alopecia. It is possible that the disparate findings may be related to specific TGM1 mutations within American, Swedish and Estonian populations. TGase-1 is expressed in the cortex and medulla cells as well as the outer and inner root sheath cells of normal hair follicles.65 The abundant expression of TGase-1 suggests involvement of this enzyme in hair-follicle formation.65 Therefore, mutations in TGM1 may explain the alopecia present in our patients with ARCI.
We found that patients with ARCI who only had mutations predicted to truncate TGase-1 were more likely to report sweating abnormalities, hypohidrosis and overheating than were patients having at least one missense mutation. Similarly, patients who had at least one mutation predicted to truncate TGase-1 were more likely to develop hypohidrosis than were patients having only missense mutations. Mutations predicted to truncate TGase-1 (nonsense, frameshift and splice-site mutations) have been shown to result in drastically reduced to absent TGase-1 mRNA transcripts, protein levels or enzyme activity in the epidermis of affected people.16, 39, 66 Mutant mRNA transcribed from genomic DNA of individuals with TGM1 mutations predicted to truncate TGase-1 is likely to be degraded by nonsense-mediated decay (NMD), a surveillance mechanism that eliminates mRNAs with premature termination codons.67 If NMD effectively eliminates the TGM1 mutant mRNAs, mutation carriers with truncating mutations may show more phenotypic severity because of the absence of TGase-1 protein. Hypohidrosis can result in overheating and heat stroke, which can result in a life-threatening situation. The pathophysiology of hypohidrosis in ARCI is not well understood, but obstruction of sweat glands by scale was initially suggested as the causative factor.68 However, in 1998, Yoneda et al showed that normal secretory and intradermal eccrine duct cells contain abundant TGase-1 mRNA, challenging this hypothesis.65 They hypothesised that hypohidrosis in patients with ARCI is due to dysfunction of TGase-1 in sweat glands and that TGase-1 may contribute to the reinforcement of the characteristic spiral architecture of the acrosyringia.65 Several case series support our finding of an association between the presence of mutations predicted to truncate TGase-1 and hypohidrosis and/or heat intolerance.15, 17, 69 Consistent with our findings, Yotsumoto et al reported a Japanese patient with ARCI and homozygosity for TGM1 missense mutations, who had reduced TGase-1 enzyme activity but normal sweating.32
Based on our findings we have developed the first model to predict the likelihood a TGM1 mutation in patients with ARCI with certain clinical features. Our model predicts that individuals born encased in a collodion membrane, with eye problems and/or alopecia are about four times more likely to test positive for a TGM1 mutation by direct DNA sequencing. Our model may be clinically useful to dermatologists and geneticists in determining which patients with ARCI are more likely to have mutations in TGM1. However, this model remains to be validated in a separate population of subjects with ARCI in a future study.
In this study, IVS5-2A→G, the most common TGM1 mutation, accounted for 28% of the TGM1 mutated alleles. This mutation allele frequency is almost three times that previously reported (9.6%).29 IVS5-2A→G affects the canonical splice acceptor site of intron 5, leading to two different consequences on TGM1 mRNA processing.2, 17, 29 It has been shown that 90% of transcripts had intron 5 retained and 10% of transcripts had an inserted G nucleotide.29 This mutation has been reported in ARCI families with diverse ethnic and racial backgrounds.12, 29, 57, 58 Shevchenko et al previously showed that the IVS5-2A→G splice-site mutation is common among North American patients, owing to a founder effect.29 A common haplotype around TGM1 was found among North American and Norwegian patients with ARCI with the IVS5-2A→G, supporting a possible common origin for this mutation.29 In the Norwegian population, where the incidence of ARCI is twice that of the North American population, IVS5-2A→G is also the most common ARCI-causing TGM1 mutation, again owing to a founder effect.2 In the middle of the 14th century, a bottleneck effect resulting from the bubonic plague reduced the Norwegian population by half,29 which could account for the higher incidence of this mutation among Norwegians. Using the Luria–Delbruck method, it was estimated that the IVS5-2A→G mutation originated in Germany and was introduced in the Norwegian population around 1000–1100 AD.29 It was also hypothesised that German families from Westphalia immigrating to the USA introduced this mutation to the North American population.29
This study is the largest genetic investigation of TGM1 mutations in patients with ARCI to date, providing us with statistical power to detect genotype–phenotype associations. Owing to the rarity of ARCI, previous studies on ARCI have been limited to case reports or small case series. The large sample size allowed us to detect significant differences between clinical phenotype and type or location of TGM1 mutation.50 Our study was registry-based, which avoids the ascertainment and assessment biases associated with hospital and clinical series. Other strengths of our study are the comprehensive clinical and genetic data that were available from our large cohort of subjects with a rare condition.
Our study has some limitations. Despite the large size of our cohort of patients with ARCI, the sample size did not have sufficient power to detect effects by stratification on analyses. Because some participants were referred by their dermatologist, our study population could be biased towards patients who sought medical care or had symptoms. Recall bias is a limitation in studies requiring patients or their parents to remember specific dates and symptoms at birth, infancy or in the past. As this was a cross-sectional study, young TGM1 mutation carriers may develop additional symptoms in the future. Prospective follow-up of patients with ARCI with TGM1 mutations will be required to evaluate this issue further and determine its effect on the results.
In conclusion, our study expands the TGM1 mutation spectrum and shows that, despite genetic and phenotypic heterogeneity in ARCI, TGM1 is the main susceptibility gene for ARCI. Our comprehensive investigation identified various genotype–phenotype associations and validated some of the findings of a previous ARCI study.50 The high frequency of mutated TGase-1 arginine residues in patients with ARCI identified in our study may be due to the deamination of 5′-CpG dinucleotides. Based on our results, we have developed the first model to predict the likelihood that patients with ARCI with specific clinical features have a TGM1 mutation. These findings could provide guidance for the clinical evaluation and practical utility in genetic testing of patients with ARCI.
Acknowledgments
We thank the ARCI families for their participation in the National Registry of Ichthyosis and Related Disorders. We also thank the members of the American Academy of Dermatology for their help in the recruitment of ARCI families and the members of the Foundation of Ichthyosis and Related Skin Types (FIRST). SF is a fellow of the National Institutes of Health (NIH) Clinical Research Training Program, a public–private partnership supported jointly by the NIH and Pfizer, Inc (via a grant to the Foundation for NIH from Pfizer, Inc). SJB is president of GeneDx (Gaithersburg, MD), and SF, M-HW, MH, SMS, DJL and JRT work for the US government. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organisations imply endorsement by the US government. This research was supported in part by the Intramural Research Program of the NIH and FIRST. The project described was also supported by a grant (no 025014–01) from the National Center for Research Resources (NCRR), a component of the NIH, and the NIH Roadmap for Medical Research, and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Re-engineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp.
Footnotes
Competing interests: None.
References
- 1.Bale SJ, Doyle SZ. The genetics of ichthyosis: a primer for epidemiologists. J Invest Dermatol. 1994;102:49S–50S. doi: 10.1111/1523-1747.ep12388591. [DOI] [PubMed] [Google Scholar]
- 2.Pigg M, Gedde-Dahl T, Jr, Cox D, Hausser I, Anton-Lamprecht I, Dahl N. Strong founder effect for a transglutaminase 1 gene mutation in lamellar ichthyosis and congenital ichthyosiform erythroderma from Norway. Eur J Hum Genet. 1998;6:589–96. doi: 10.1038/sj.ejhg.5200224. [DOI] [PubMed] [Google Scholar]
- 3.Williams ML, Elias PM. Heterogeneity in autosomal recessive ichthyosis. Clinical and biochemical differentiation of lamellar ichthyosis and nonbullous congenital ichthyosiform erythroderma. Arch Dermatol. 1985;121:477–88. doi: 10.1001/archderm.121.4.477. [DOI] [PubMed] [Google Scholar]
- 4.Russell LJ, DiGiovanna JJ, Hashem N, Compton JG, Bale SJ. Linkage of autosomal recessive lamellar ichthyosis to chromosome 14q. Am J Hum Genet. 1994;55:1146–52. [PMC free article] [PubMed] [Google Scholar]
- 5.Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ. Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet. 1995;9:279–83. doi: 10.1038/ng0395-279. [DOI] [PubMed] [Google Scholar]
- 6.Akiyama M, Takizawa Y, Kokaji T, Shimizu H. Novel mutations of TGM1 in a child with congenital ichthyosiform erythroderma. Br J Dermatol. 2001;144:401–7. doi: 10.1046/j.1365-2133.2001.04037.x. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama M, Takizawa Y, Suzuki Y, Ishiko A, Matsuo I, Shimizu H. Compound heterozygous TGM1 mutations including a novel missense mutation L204Q in a mild form of lamellar ichthyosis. J Invest Dermatol. 2001;116:992–5. doi: 10.1046/j.0022-202x.2001.01367.x. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama M, Takizawa Y, Suzuki Y, Shimizu H. A novel homozygous mutation 371delA in TGM1 leads to a classic lamellar ichthyosis phenotype. Br J Dermatol. 2003;148:149–53. doi: 10.1046/j.1365-2133.2003.05041.x. [DOI] [PubMed] [Google Scholar]
- 9.Becker K, Csikos M, Sardy M, Szalai ZS, Horvath A, Karpati S. Identification of two novel nonsense mutations in the transglutaminase 1 gene in a Hungarian patient with congenital ichthyosiform erythroderma. Exp Dermatol. 2003;12:324–9. doi: 10.1034/j.1600-0625.2003.120313.x. [DOI] [PubMed] [Google Scholar]
- 10.Bichakjian CK, Nair RP, Wu WW, Goldberg S, Elder JT. Prenatal exclusion of lamellar ichthyosis based on identification of two new mutations in the transglutaminase 1 gene. J Invest Dermatol. 1998;110:179–82. doi: 10.1046/j.1523-1747.1998.00104.x. [DOI] [PubMed] [Google Scholar]
- 11.Cserhalmi-Friedman PB, Milstone LM, Christiano AM. Diagnosis of autosomal recessive lamellar ichthyosis with mutations in the TGM1 gene. Br J Dermatol. 2001;144:726–30. doi: 10.1046/j.1365-2133.2001.04126.x. [DOI] [PubMed] [Google Scholar]
- 12.Esposito G, Auricchio L, Rescigno G, Paparo F, Rinaldi M, Salvatore F. Transglutaminase 1 gene mutations in Italian patients with autosomal recessive lamellar ichthyosis. J Invest Dermatol. 2001;116:809–12. doi: 10.1046/j.1523-1747.2001.01314.x. [DOI] [PubMed] [Google Scholar]
- 13.Esposito G, Tadini G, Paparo F, Viola A, Ieno L, Pennacchia W, Messina F, Giordano L, Piccirillo A, Auricchio L. Transglutaminase 1 deficiency and corneocyte collapse: an indication for targeted molecular screening in autosomal recessive congenital ichthyosis. Br J Dermatol. 2007;157:808–10. doi: 10.1111/j.1365-2133.2007.08070.x. [DOI] [PubMed] [Google Scholar]
- 14.Hennies HC, Raghunath M, Wiebe V, Vogel M, Velten F, Traupe H, Reis A. Genetic and immunohistochemical detection of mutations inactivating the keratinocyte transglutaminase in patients with lamellar ichthyosis. Hum Genet. 1998;102:314–18. doi: 10.1007/s004390050697. [DOI] [PubMed] [Google Scholar]
- 15.Hennies HC, Kuster W, Wiebe V, Krebsova A, Reis A. Genotype/phenotype correlation in autosomal recessive lamellar ichthyosis. Am J Hum Genet. 1998;62:1052–61. doi: 10.1086/301818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D. Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science. 1995;267:525–8. doi: 10.1126/science.7824952. [DOI] [PubMed] [Google Scholar]
- 17.Huber M, Yee VC, Burri N, Vikerfors E, Lavrijsen AP, Paller AS, Hohl D. Consequences of seven novel mutations on the expression and structure of keratinocyte transglutaminase. J Biol Chem. 1997;272:21018–26. doi: 10.1074/jbc.272.34.21018. [DOI] [PubMed] [Google Scholar]
- 18.Jessen BA, Phillips MA, Hovnanian A, Rice RH. Role of Sp1 response element in transcription of the human transglutaminase 1 gene. J Invest Dermatol. 2000;115:113–17. doi: 10.1046/j.1523-1747.2000.00027.x. [DOI] [PubMed] [Google Scholar]
- 19.Kon A, Takeda H, Sasaki H, Yoneda K, Nomura K, Ahvazi B, Steinert PM, Hanada K, Hashimoto I. Novel transglutaminase 1 gene mutations (R348X/Y365D) in a Japanese family with lamellar ichthyosis. J Invest Dermatol. 2003;120:170–2. doi: 10.1046/j.1523-1747.2003.19522.x. [DOI] [PubMed] [Google Scholar]
- 20.Laiho E, Ignatius J, Mikkola H, Yee VC, Teller DC, Niemi KM, Saarialho-Kere U, Kere J, Palotie A. Transglutaminase 1 mutations in autosomal recessive congenital ichthyosis: private and recurrent mutations in an isolated population. Am J Hum Genet. 1997;61:529–38. doi: 10.1086/515498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lugassy J, Hennies HC, Indelman M, Khamaysi Z, Bergman R, Sprecher E. Rapid detection of homozygous mutations in congenital recessive ichthyosis. Arch Dermatol Res. 2008;300:81–5. doi: 10.1007/s00403-007-0815-0. [DOI] [PubMed] [Google Scholar]
- 22.Mizrachi-Koren M, Shemer S, Morgan M, Indelman M, Khamaysi Z, Petronius D, Bitterman-Deutsch O, Hennies HC, Bergman R, Sprecher E. Homozygosity mapping as a screening tool for the molecular diagnosis of hereditary skin diseases in consanguineous populations. J Am Acad Dermatol. 2006;55:393–401. doi: 10.1016/j.jaad.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Oji V, Hautier JM, Ahvazi B, Hausser I, Aufenvenne K, Walker T, Seller N, Steijlen PM, Küster W, Hovnanian A, Hennies HC, Traupe H. Bathing suit ichthyosis is caused by transglutaminase-1 deficiency: evidence for a temperature-sensitive phenotype. Hum Mol Genet. 2006;15:3083–97. doi: 10.1093/hmg/ddl249. [DOI] [PubMed] [Google Scholar]
- 24.Parmentier L, Blanchet-Bardon C, Nguyen S, Prud’homme JF, Dubertret L, Weissenbach J. Autosomal recessive lamellar ichthyosis: identification of a new mutation in transglutaminase 1 and evidence for genetic heterogeneity. Hum Mol Genet. 1995;4:1391–5. doi: 10.1093/hmg/4.8.1391. [DOI] [PubMed] [Google Scholar]
- 25.Petit E, Huber M, Rochat A, Bodemer C, Teillac-Hamel D, Müh JP, Revuz J, Barrandon Y, Lathrop M, de Prost Y, Hohl D, Hovnanian A. Three novel point mutations in the keratinocyte transglutaminase (TGK) gene in lamellar ichthyosis: significance for mutant transcript level, TGK immunodetection and activity. Eur J Hum Genet. 1997;5:218–28. [PubMed] [Google Scholar]
- 26.Pigg M, Gedde-Dahl T, Jr, Cox DW, Haugen G, Dahl N. Haplotype association and mutation analysis of the transglutaminase 1 gene for prenatal exclusion of lamellar ichthyosis. Prenat Diagn. 2000;20:132–7. doi: 10.1002/(sici)1097-0223(200002)20:2<132::aid-pd765>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 27.Raghunath M, Hennies HC, Ahvazi B, Vogel M, Reis A, Steinert PM, Traupe H. Self-healing collodion baby: a dynamic phenotype explained by a particular transglutaminase-1 mutation. J Invest Dermatol. 2003;120:224–8. doi: 10.1046/j.1523-1747.2003.12032.x. [DOI] [PubMed] [Google Scholar]
- 28.Schorderet DF, Huber M, Laurini RN, Von Moos G, Gianadda B, Délèze G, Hohl D. Prenatal diagnosis of lamellar ichthyosis by direct mutational analysis of the keratinocyte transglutaminase gene. Prenat Diagn. 1997;17:483–6. doi: 10.1002/(sici)1097-0223(199705)17:5<483::aid-pd80>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Shevchenko YO, Compton JG, Toro JR, DiGiovanna JJ, Bale SJ. Splice-site mutation in TGM1 in congenital recessive ichthyosis in American families: molecular, genetic, genealogic, and clinical studies. Hum Genet. 2000;106:492–9. doi: 10.1007/s004390000284. [DOI] [PubMed] [Google Scholar]
- 30.Tok J, Garzon MC, Cserhalmi-Friedman P, Lam HM, Spitz JL, Christiano AM. Identification of mutations in the transglutaminase 1 gene in lamellar ichthyosis. Exp Dermatol. 1999;8:128–33. doi: 10.1111/j.1600-0625.1999.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 31.Yang JM, Ahn KS, Cho MO, Yoneda K, Lee CH, Lee JH, Lee ES, Candi E, Melino G, Ahvazi B, Steinert PM. Novel mutations of the transglutaminase 1 gene in lamellar ichthyosis. J Invest Dermatol. 2001;117:214–18. doi: 10.1046/j.0022-202x.2001.01429.x. [DOI] [PubMed] [Google Scholar]
- 32.Yotsumoto S, Akiyama M, Yoneda K, Fukushige T, Kobayashi K, Saheki T, Kanzaki T. Analyses of the transglutaminase 1 gene mutation and ultrastructural characteristics in a Japanese patient with lamellar ichthyosis. J Dermatol Sci. 2000;24:119–25. doi: 10.1016/s0923-1811(00)00087-6. [DOI] [PubMed] [Google Scholar]
- 33.Rice RH, Crumrine D, Uchida Y, Gruber R, Elias PM. Structural changes in epidermal scale and appendages as indicators of defective TGM1 activity. Arch Dermatol Res. 2005;297:127–33. doi: 10.1007/s00403-005-0591-7. [DOI] [PubMed] [Google Scholar]
- 34.Oji V, Traupe H. Ichthyoses: differential diagnosis and molecular genetics. Eur J Dermatol. 2006;16:349–59. [PubMed] [Google Scholar]
- 35.Boeshans KM, Mueser TC, Ahvazi B. A three-dimensional model of the human transglutaminase 1: insights into the understanding of lamellar ichthyosis. J Mol Model. 2007;13:233–46. doi: 10.1007/s00894-006-0144-9. [DOI] [PubMed] [Google Scholar]
- 36.Kim IG, McBride OW, Wang M, Kim SY, Idler WW, Steinert PM. Structure and organization of the human transglutaminase-1 gene. J Biol Chem. 1992;267:7710–17. [PubMed] [Google Scholar]
- 37.Robinson NA, Lapic S, Welter JF, Eckert RL. S100A11, S100A10, annexin I, desmosomal proteins, small proline-rich proteins, plasminogen activator inhibitor-2, and involucrin are components of the cornified envelope of cultured human epidermal keratinocytes. J Biol Chem. 1997;272:12035–46. doi: 10.1074/jbc.272.18.12035. [DOI] [PubMed] [Google Scholar]
- 38.Steinert PM, Marekov LN. The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J Biol Chem. 1995;270:17702–11. doi: 10.1074/jbc.270.30.17702. [DOI] [PubMed] [Google Scholar]
- 39.Candi E, Melino G, Lahm A, Ceci R, Rossi A, Kim IG, Ciani B, Steinert PM. Transglutaminase 1 mutations in lamellar ichthyosis. Loss of activity due to failure of activation by proteolytic processing. J Biol Chem. 1998;273:13693–702. doi: 10.1074/jbc.273.22.13693. [DOI] [PubMed] [Google Scholar]
- 40.Muramatsu S, Suga Y, Kon J, Matsuba S, Hashimoto Y, Ogawa H. A Japanese patient with a mild form of lamellar ichthyosis harbouring two missense mutations in the core domain of the transglutaminase 1 gene. Br J Dermatol. 2004;150:390–2. doi: 10.1111/j.1365-2133.2003.05803.x. [DOI] [PubMed] [Google Scholar]
- 41.Matsuki M, Yamashita F, Ishida-Yamamoto A, Yamada K, Kinoshita C, Fushiki S, Ueda E, Morishima Y, Tabata K, Yasuno H, Hashida M, Iizuka H, Ikawa M, Okabe M, Kondoh G, Kinoshita T, Takeda J, Yamanishi K. Defective stratum corneum and early neonatal death in mice lacking the gene for transglutaminase 1 (keratinocyte transglutaminase) Proc Natl Acad Sci U S A. 1998;95:1044–9. doi: 10.1073/pnas.95.3.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akiyama M. Harlequin ichthyosis and other autosomal recessive congenital ichthyoses: the underlying genetic defects and pathomechanisms. J Dermatol Sci. 2006;42:83–9. doi: 10.1016/j.jdermsci.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Annilo T, Shulenin S, Chen ZQ, Arnould I, Prades C, Lemoine C, Maintoux-Larois C, Devaud C, Dean M, Denèfle P, Rosier M. Identification and characterization of a novel ABCA subfamily member, ABCA12, located in the lamellar ichthyosis region on 2q34. Cytogenet Genome Res. 2002;98:169–76. doi: 10.1159/000069811. [DOI] [PubMed] [Google Scholar]
- 44.Akiyama M, Sugiyama-Nakagiri Y, Sakai K, McMillan JR, Goto M, Arita K, Tsuji-Abe Y, Tabata N, Matsuoka K, Sasaki R, Sawamura D, Shimizu H. Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J Clin Invest. 2005;115:1777–84. doi: 10.1172/JCI24834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelsell DP, Norgett EE, Unsworth H, Teh MT, Cullup T, Mein CA, Dopping-Hepenstal PJ, Dale BA, Tadini G, Fleckman P, Stephens KG, Sybert VP, Mallory SB, North BV, Witt DR, Sprecher E, Taylor AE, Ilchyshyn A, Kennedy CT, Goodyear H, Moss C, Paige D, Harper JI, Young BD, Leigh IM, Eady RA, O’Toole EA. Mutations in ABCA12 underlie the severe congenital skin disease harlequin ichthyosis. Am J Hum Genet. 2005;76:794–803. doi: 10.1086/429844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefèvre C, Audebert S, Jobard F, Bouadjar B, Lakhdar H, Boughdene-Stambouli O, Blanchet-Bardon C, Heilig R, Foglio M, Weissenbach J, Lathrop M, Prud’homme JF, Fischer J. Mutations in the transporter ABCA12 are associated with lamellar ichthyosis type 2. Hum Mol Genet. 2003;12:2369–78. doi: 10.1093/hmg/ddg235. [DOI] [PubMed] [Google Scholar]
- 47.Jobard F, Lefèvre C, Karaduman A, Blanchet-Bardon C, Emre S, Weissenbach J, Ozgüc M, Lathrop M, Prud’homme JF, Fischer J. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet. 2002;11:107–13. doi: 10.1093/hmg/11.1.107. [DOI] [PubMed] [Google Scholar]
- 48.Lefèvre C, Bouadjar B, Karaduman A, Jobard F, Saker S, Ozguc M, Lathrop M, Prud’homme JF, Fischer J. Mutations in ichthyin a new gene on chromosome 5q33 in a new form of autosomal recessive congenital ichthyosis. Hum Mol Genet. 2004;13:2473–82. doi: 10.1093/hmg/ddh263. [DOI] [PubMed] [Google Scholar]
- 49.Lefèvre C, Bouadjar B, Ferrand V, Tadini G, Mégarbané A, Lathrop M, Prud’homme JF, Fischer J. Mutations in a new cytochrome P450 gene in lamellar ichthyosis type 3. Hum Mol Genet. 2006;15:767–76. doi: 10.1093/hmg/ddi491. [DOI] [PubMed] [Google Scholar]
- 50.Ganemo A, Pigg M, Virtanen M, Kukk T, Raudsepp H, Rossman-Ringdahl I, Westermark P, Niemi KM, Dahl N, Vahlquist A. Autosomal recessive congenital ichthyosis in Sweden and Estonia: clinical, genetic and ultrastructural findings in eighty-three patients. Acta Derm Venereol. 2003;83:24–30. doi: 10.1080/00015550310002666. [DOI] [PubMed] [Google Scholar]
- 51.Laiho E, Niemi KM, Ignatius J, Kere J, Palotie A, Saarialho-Kere U. Clinical and morphological correlations for transglutaminase 1 gene mutations in autosomal recessive congenital ichthyosis. Eur J Hum Genet. 1999;7:625–32. doi: 10.1038/sj.ejhg.5200353. [DOI] [PubMed] [Google Scholar]
- 52.Richards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, Parad RB, Witt D, Klinger KW. Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Mol Genet. 1993;2:159–63. doi: 10.1093/hmg/2.2.159. [DOI] [PubMed] [Google Scholar]
- 53.Mehta CR, Patel NR. A network algorithm for performing Fisher’s exact test in r × c contingency tables. J Am Stat Assoc. 1983;78:427–34. [Google Scholar]
- 54.Agresti A. Categorical data analysis. New York: Johns Wiley and Sons, Inc; 1990. [Google Scholar]
- 55.Hollander M, Wolfe DA. Nonparametric statistical analysis. 2. New York: John Wiley and Sons Inc; 1999. [Google Scholar]
- 56.Arita K, Jacyk WK, Wessagowit V, van Rensburg EJ, Chaplin T, Mein CA, Akiyama M, Shimizu H, Happle R, McGrath JA. The South African “bathing suit ichthyosis” is a form of lamellar ichthyosis caused by a homozygous missense mutation, p. R315L, in transglutaminase 1. J Invest Dermatol. 2007;127:490–3. doi: 10.1038/sj.jid.5700550. [DOI] [PubMed] [Google Scholar]
- 57.Shawky RM, Sayed NS, Elhawary NA. Mutations in transglutaminase 1 gene in autosomal recessive congenital ichthyosis in Egyptian families. Dis Markers. 2004;20:325–32. doi: 10.1155/2004/965968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huber M, Rettler I, Bernasconi K, Wyss M, Hohl D. Lamellar ichthyosis is genetically heterogeneous–cases with normal keratinocyte transglutaminase. J Invest Dermatol. 1995;105:653–4. doi: 10.1111/1523-1747.ep12324122. [DOI] [PubMed] [Google Scholar]
- 59.Atschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vitkup D, Sander C, Church GM. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–5. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- 62.Krawczak M, Ball EV, Cooper DN. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am J Hum Genet. 1998;63:474–88. doi: 10.1086/301965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lüleyap HU, Alptekin D, Pazarbasi A, Kasap M, Kasap H, Demirhindi H, Mungan N, Ozer G, Froster UG. The importance of arginine mutation for the evolutionary structure and function of phenylalanine hydroxylase gene. Mutat Res. 2006;601:39–45. doi: 10.1016/j.mrfmmm.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Letai A, Coulombe PA, McCormick MB, Yu QC, Hutton E, Fuchs E. Disease severity correlates with position of keratin point mutations in patients with epidermolysis bullosa simplex. Proc Natl Acad Sci U S A. 1993;90:3197–201. doi: 10.1073/pnas.90.8.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoneda K, Akiyama M, Morita K, Shimizu H, Imamura S, Kim SY. Expression of transglutaminase 1 in human hair follicles, sebaceous glands and sweat glands. Br J Dermatol. 1998;138:37–44. doi: 10.1046/j.1365-2133.1998.02024.x. [DOI] [PubMed] [Google Scholar]
- 66.McIntosh I, Hamosh A, Dietz HC. Nonsense mutations and diminished mRNA levels. Nat Genet. 1993;4:219. doi: 10.1038/ng0793-219. [DOI] [PubMed] [Google Scholar]
- 67.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 68.Phillips MA, Baden HP. Ichthyosiform dermatoses. In: Fitzpatrick TB, Eisen AZ, Wolff K, Freedberg IM, Austen KF, editors. Dermatology in general medicine. 4. New York: McGraw-Hill, Inc; 1993. pp. 531–44. [Google Scholar]
- 69.Rossmann-Ringdahl I, Anton-Lamprecht I, Swanbeck G. A mother and two children with nonbullous congenital ichthyosiform erythroderma. Arch Dermatol. 1986;122:559–64. [PubMed] [Google Scholar]