INTRODUCTION
Diffusely infiltrating astrocytomas are the most common primary brain tumors in adults, being classified into four grades according to the World Health Organization (WHO).1 Glioblastoma (GBM), grade IV astrocytoma, is the most frequent,2 and presents median survival rarely exceeding 12 months in spite of currently available treatment approaches.3 GBM may manifest rapidly de novo (primary GBM), or may develop slowly from grade II or grade III astrocytomas (secondary GBM), suggesting that they are distinct disease entities that evolve through different genetic pathways.
In recent genome‐wide analyses, high rates of spontaneous mutations in the gene encoding cytosolic NADP‐dependent isocitrate dehydrogenase 1 (IDH1) have been reported in diffuse gliomas including WHO grades II and III astroglial and oligodendroglial lineages.4–8 Mutations of IDH1 are rare in primary GBM (<10%) and frequent in secondary GBM (>80%).4–7,9–11 Thus, IDH1 mutations are strong predictors of more favorable prognosis and a highly selective molecular marker of secondary GBM that complements clinical criteria for distinguishing them from primary GBM. Intriguingly, mutations of IDH1 predominantly occurred in younger patients and were preferentially found in tumors harboring TP53 mutations.12 These results corroborate the fact that primary and secondary GBMs originate from different progenitor cells.
IDH1, located on 2q33.3, encodes the cytosolic NADP+ specific isocitrate dehydrogenase, which catalyzes the oxidative decarboxylation of isocitrate to α‐ketoglutarate.13 IDH1 is configured as a homodimer with two enzymatically active sites, and most of its activity is detected in the cytosol and in peroxisomes. The other four members of the IDH family are exclusively localized in mitochondria.14 Glioma‐specific mutations in IDH1 always affect the amino acid arginine 132 located in an evolutionarily highly conserved region at the binding site for isocitrate.11 Mutations in IDH1 are of somatic origin and heterozygous, and inactivate enzyme activity.7
We studied the frequency of IDH1 mutations in a series of 161 GBM patients from the Brazilian population according to patient age, gender, GBM type and survival time.
Tumor samples were collected during surgical procedures by the Neurosurgery Groups of different institutions from the state of São Paulo: 93 from Hospital das Clínicas, School of Medicine of University of São Paulo; 38 from Barretos Cancer Hospital; 13 from Paulista School of Medicine, Federal University of São Paulo; 10 from Albert Einstein Jewish Hospital; and 7 from Nove de Julho Hospital. Informed consent was obtained from each patient, and the study was approved by the local ethics committee. The samples included frozen tissues, collected upon surgical removal and immediately snap‐frozen in liquid nitrogen15 and paraffin‐embedded blocks. The mean age of 161 GBM patients was 56 years, with 59 females and 102 males. A total of 155 cases were primary GBMs (mean age 55 years), and 6 cases were diagnosed as secondary GBM (mean age 31 years) with histological evidence of a previous less malignant astrocytoma.
DNA was extracted from the frozen tissues by a standard phenol/chloroform method or by Trizol (Invitrogen Inc, Carlsbad, CA, USA), following the manufacturer's instructions, and by QiaAmp DNA Micro kit (Qiagen, Hilden, Germany) from paraffin‐embedded sections.
Polymerase chain reaction (PCR) followed by DNA sequencing was applied to detect IDH1 mutation. Primers sequences synthesized by IDT (Integrated DNA Technologies, Inc, Coralville, IA, USA) for PCR amplification of exon 4 were (5′‐3′): CCATCACTGCAGTTGTAGGTT and CATACAAGTTGGAAATTTCTGG. PCR products were generated in a 25 µL reaction mixture including 100 ng of DNA, 50 mM KCl, 50 µM of each dNTP, 10 mM Tris‐HCl (pH 9.0), 1.5 mM MgCl2, 10 pmol of each primer and 1 unit of Taq DNA polymerase (GE Healthcare, Piscataway, NJ, USA). The PCR was performed with an initial denaturating step at 94°C for 5 min, followed by 35 cycles consisting of 94°C for 30 s, 54°C for 30 s and at 72°C for 30 s. After the final cycle, an extension period of 10 min at 72°C was performed. The PCR products (436 bp) of amplification were checked, purified with a GFX column (GE Healthcare) and sequenced on an ABI Prism 3130 DNA automated sequencer using the Big Dye™ Terminator Cycle Sequencing Ready Reaction Kit version 3.1 (Applied Biosystems, Foster City, CA, USA). Primers used for the sequencing were the same as those used for PCR. Results were analyzed and compared with the public sequence of IDH1 cDNA (GenBank), accession no. NM_005896.
The statistical analyses and their associations with patient characteristics were performed by chi‐square test (χ2). Overall survival (OS) was calculated as the interval between the surgery and day of death, in months. The log‐rank test was used for univariate analysis to estimate differences in survival time for IDH1 mutation status, according to the Kaplan–Meier method. Calculations were performed using STATA, version 7 (STATA Corp., College Station, TX, USA) and SPSS 15.0 software (SPSS, Chicago, IL, USA), with statistical significance of p<0.05.
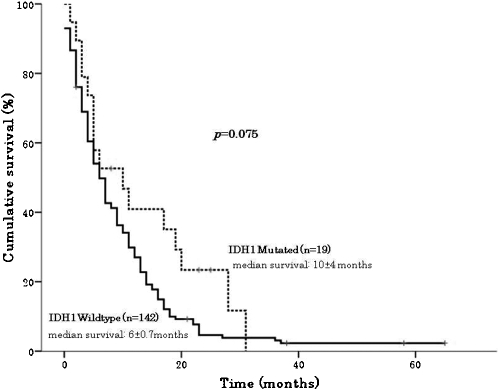
We found IDH1 mutations in 11.8% (19 out of 161) of samples tested, with a higher mutation rate in GBMs diagnosed as secondary, 66.7% (4 out of 6), than in cases of primary GBMs, 9.7% (15 out of 155), p<0.001. All mutations were heterozygous, located at codon 132, resulting in amino acid change from arginine to histidine. We found a higher frequency of IDH1 mutation in females (18.6%) than in males (7.8%) (p = 0.041). GBM patients carrying IDH1 mutations were significantly younger (diagnosed before age 50 years), mean age of 44 years, than patients with wild‐type IDH1 (diagnosed at age 50 years or older), mean age of 56 years, p = 0.011 (Table 1). The mean survival time of all GBM patients with and without IDH1 mutations was 12 months (19 cases) and 9 months (142 cases), respectively (p = 0.075, log‐rank test), as shown in Figure 1.
Table 1.
IDH1 mutation status of glioblastomas according to age, gender and tumor subtype in Brazilian patients.
| IDH1 mutation | |||||
| Characteristics | GBM patients (%) | Positive* | Negative | % | p value |
| 161 | 19 | 142 | 11.8 | ||
| Gender | |||||
| Female | 59 (36.7) | 11 | 48 | 18.6 | 0.041 |
| Male | 102 (63.3) | 8 | 94 | 7.8 | |
| Age at diagnosis (years) | |||||
| <50 | 52 (32.3) | 11 | 41 | 21.1 | 0.011 |
| ≥50 | 109 (67.7) | 8 | 101 | 7.3 | |
| (Age, mean±SE) | 54.6±13.4 | 43.9±19.1 | 56.1±11.9 | ||
| Tumor subtype | |||||
| Primary | 155 (96.3) | 15 | 140 | 9.7 | <0.001 |
| Secondary | 6 (3.7) | 4 | 2 | 66.7 | |
IDH1 mutation at R132H in heterozygous form.
Figure 1.
Survival of glioblastoma patients according to their IDH1 mutation status (positive vs. negative). Glioblastoma patients carrying an IDH1 mutation had longer overall survival (log‐rank test; Mantel–Cox test; p = 0.075).
DISCUSSION
A higher rate of IDH1 mutation in secondary compared with primary GBM cases (66.7% vs. 9.7%) was observed in our study. Additionally, patients carrying IDH1 mutations were younger (44 years) than those patients without the mutation (56 years), and IDH1 status has been shown to have an association trend with an increase in the overall survival of GBM patients, as described by others.4–7,9–12 The lack of statistical impact of the overall survival time might be attributed to a low number of cases with IDH1 mutation reflecting a low incidence of secondary GBM in our series. We have also previously reported a low prevalence of TP53 mutations, usually detected among secondary GBM cases, because of a higher frequency of primary GBM in our series.16 Both results concerning TP53 and IDH1 mutation status point out the molecular differences between primary and secondary GBM. These results reinforce the concept that, despite the histological similarities, primary and secondary GBMs are genetically and clinically distinct entities.6,12
In summary, this study established the frequency of IDH1 mutation in a Brazilian series of GBM, confirmed IDH1 mutation as a genetic marker for secondary GBM, and therefore as complementary information to help predict the outcome of patients with GBM.
ACKNOWLEDGMENTS
This study was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, process #04/12133‐6), the Ludwig Institute for Cancer Research, Conselho Nacional de Pesquisa (CNPq) and Albert Einstein Jewish Hospital. We sincerely thank the neurosurgeons from the participating institutions for the therapeutic and diagnostic procedures of all patients included in this study. We also thank the Psychiatry Institute for the logistical help in the surgical therapy at USP‐SP, and Diagnostika Laboratory for preparation of paraffin‐embedded material from Albert Einstein Jewish Hospital and Nove de Julho Hospital.
REFERENCES
- 1. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. 10.1007/s00401‐007‐0243‐4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. von Deimling A, Louis DN, Schramm J, Wiestler OD. Astrocytic gliomas: characterization on a molecular genetic basis. Recent Results Cancer Res. 1994:33–42. doi: 10.1007/978-3-642-85039-4_5. [DOI] [PubMed] [Google Scholar]
- 3. Korshunov A, Sycheva R, Golanov A. The prognostic relevance of molecular alterations in glioblastomas for patients age <50 years. Cancer. 2005;104:825–32. doi: 10.1002/cncr.21221. 10.1002/cncr.21221 [DOI] [PubMed] [Google Scholar]
- 4. Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. 10.1007/s00401‐008‐0455‐2 [DOI] [PubMed] [Google Scholar]
- 5. Ichimura K, Pearson DM, Kocialkowski S, Bäcklund LM, Chan R, Jones DT, et al. IDH1 mutations are present in the majority of common adult gliomas but are rare in primary glioblastomas. Neurol Oncol. 2009;11:341–7. doi: 10.1215/15228517-2009-025. 10.1215/15228517‐2009‐025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–53. doi: 10.2353/ajpath.2009.080958. 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gravendeel LA, Kloosterhof NK, Bralten LB, van Marion R, Dubbink HJ, Dinjens W, et al. Segregation of non‐p.R132H mutations in IDH1 in distinct molecular subtypes of glioma. Hum Mutat. 2010;31:E1186–99. doi: 10.1002/humu.21201. 10.1002/humu.21201 [DOI] [PubMed] [Google Scholar]
- 9. Bleeker FE, Lamba S, Leenstra S, Troost D, Hulsebos T, Vandertop WP, et al. IDH1 mutations at residue p.R132 (IDH1(R132) occur frequently in high‐grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. 10.1002/humu.20937 [DOI] [PubMed] [Google Scholar]
- 10. Kang MR, Kim MS, Oh JE, Kim YR, Song SY, Seo SI, et al. Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int J Cancer. 2009;125:353–5. doi: 10.1002/ijc.24379. 10.1002/ijc.24379 [DOI] [PubMed] [Google Scholar]
- 11. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res. 2009;15:6002–7. doi: 10.1158/1078-0432.CCR-09-0715. 10.1158/1078‐0432.CCR‐09‐0715 [DOI] [PubMed] [Google Scholar]
- 13. Narahara K, Kimura S, Kikkawa K, Takahashi Y, Wakita Y, Kasai R, et al. Probable assignment of soluble isocitrate dehydrogenase (IDH1) to 2q33.3. Hum Genet. 1985;71:37–40. doi: 10.1007/BF00295665. 10.1007/BF00295665 [DOI] [PubMed] [Google Scholar]
- 14. Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)‐dependent isocitrate dehydrogenase. J Biol Chem. 1999;274:30527–33. doi: 10.1074/jbc.274.43.30527. 10.1074/jbc.274.43.30527 [DOI] [PubMed] [Google Scholar]
- 15. Oba‐Shinjo SM, Bengtson MH, Winnischofe SMB, Colin C, Vedoy CG, Mendonça Z, et al. Identification of novel differentially expressed genes in human astrocytomas by cDNA representational difference analysis. Mol Brain Res. 2005;140:25–33. doi: 10.1016/j.molbrainres.2005.06.015. 10.1016/j.molbrainres.2005.06.015 [DOI] [PubMed] [Google Scholar]
- 16. Uno M, Oba‐Shinjo SM, de Aguiar PH, Leite CC, Rosemberg S, Miura FK, et al. Detection of somatic TP53 splice site mutations in diffuse astrocytomas. Cancer Lett. 2005;224:321–7. doi: 10.1016/j.canlet.2004.10.022. 10.1016/j.canlet.2004.10.022 [DOI] [PubMed] [Google Scholar]