Abstract
Parkinson disease (PD) is the second most common neurodegenerative disorder. In most instances, PD is thought to result from a complex interaction between multiple genetic and environmental factors, though rare monogenic forms of the disease do exist. Mutations in 6 genes (SNCA, LRRK2, PRKN, DJ1, PINK1, and ATP13A2) have conclusively been shown to cause familial parkinsonism. In addition, common variation in 3 genes (MAPT, LRRK2, and SNCA) and loss-of-function mutations in GBA have been well-validated as susceptibility factors for PD. The function of these genes and their contribution to PD pathogenesis remain to be fully elucidated. The prevalence, incidence, clinical manifestations, and genetic components of PD are discussed in this review.
Keywords: genetics, neurodegeneration, Parkinson disease
Parkinson Disease
Introduction
Prevalence and Incidence
Parkinson disease ([PD] OMIM #168600) is the second most common neurodegenerative disorder. The incidence is similar worldwide, with the prevalence increasing in proportion to regional increases in population longevity with more than 1% affected over the age of 65 years and more than 4% of the population affected by the age of 85 years.1 In most instances, PD is multifactorial, likely arising from a combination of polygenic inheritance, environmental exposures, and gene–environment interactions. Approximately 20% of patients with PD report a family history of the disease and monogenic forms of PD are relatively rare.2,3
Clinical Manifestations
Clinically, PD has traditionally been defined by the presence of cardinal motor signs: tremor, rigidity, bradykinesia, and postural instability. However, a large body of evidence now indicates that nonmotor features such as autonomic insufficiency, cognitive impairment, depression, olfactory deficits, psychosis, and sleep disturbance are very common during the course of the disease. Motor dysfunction is thought to arise from progressive loss of dopaminergic cells within the substantia nigra pars compacta and becomes evident when approximately 80% of striatal dopamine and 50% of nigral neurons are lost.4 Levodopa is still the most effective treatment for PD, but its use is complicated by the emergence of motor fluctuations and dyskinesias. Other treatment options include dopamine agonists, anticholingergics, amantadine, monoamine oxidase (MAO) inhibitors, and deep brain stimulation.5 However, neuroprotective treatment that delays or arrests neurodegeneration in PD remains an unrealized goal.6
Clinical Diagnosis
The clinical diagnosis of PD is typically based on the presence of cardinal motor features, absence of atypical findings suggestive of an alternate diagnosis, and response to levodopa.5 The diagnostic criteria most frequently used in clinical research are those of the UK Parkinson's Disease Society Brain Bank.7 Differentiating PD from other parkinsonian disorders such as progressive supranuclear palsy (PSP) and multiple system atrophy can be challenging early in the course of the disease. Neuroimaging techniques have been used to facilitate early diagnosis of PD but currently are not widely used in clinical practice.8
Neuropathological Diagnosis
The diagnosis of PD can be confirmed at autopsy. The pathological hallmarks of the disease are dopaminergic cell loss within the substantia nigra pars compacta and the presence of Lewy bodies (LBs) and Lewy neurites (collectively known as Lewy-related pathology) in vulnerable populations of neurons. Lewy bodies are intracytoplasmic inclusions and within the brain stem have a dense eosinophilic core and a clearer surrounding halo on hematoxylin and eosin staining. The principal component of LBs is α-synuclein, a small (140 amino acids) protein that is predominantly expressed in the neocortex, hippocampus, substantia nigra, thalamus, and cerebellum.9 Lewy neurites are nerve cell processes that contain aggregates of α-synuclein and are most numerous in the CA2/3 region of the hippocampus and in the substantia nigra.10 Lewy bodies and Lewy neurites are best visualized immunohistochemically, using an antibody to α-synuclein.10
The neuropathological changes characteristic of PD are thought to evolve sequentially across the brain, and a classification scheme comprising 6 stages has been proposed.11 In the early presymptomatic stages (1 and 2), pathology is limited to the medulla oblongata and pontine tegmentum (including the dorsal motor nucleus of vagus and the locus coeruleus) and olfactory bulb. As patients become symptomatic in stages 3 to 4, the substantia nigra and other nuclei within the midbrain and forebrain, such as the nucleus basalis of Meynert, become involved. Finally, in patients who survive into the late stages of the disease (5-6), pathology advances from mesocortex into neocortex.
Genetics of PD
Introduction
Historically, PD was considered largely sporadic in nature without genetic origin. However, in the past decade, genetic studies of PD families from different geographical regions worldwide have strengthened the hypothesis that PD has a substantial genetic component. The first gene unequivocally tied to PD (SNCA, PARK1 locus) was discovered through analysis of a large multigenerational Italian family (the Contursi Kindred) in which parkinsonism segregated in an autosomal dominant pattern.12,13 Since then a total of 18 PD loci have been nominated through linkage analysis (PARK1-15) or genomewide association studies ([GWASs]; PARK16-18; Tables 1 and 2).3,14-26 Mutations within the genes at 6 of these loci (SNCA, LRRK2, PRKN, DJ1, PINK1, and ATP13A2) have conclusively been demonstrated to cause familial parkinsonism.29 In addition, common polymorphisms within 2 of these same genes (SNCA and LRRK2) and variation in 2 other genes not assigned to a PARK locus (MAPT and GBA) are now well-validated risk factors for PD.27,30-33 Here we discuss the most important genes relevant to PD and their associated clinical features.
Table 1. Genes/Loci Underlying Monogenic Parkinsonism.
| PARK Locus | Gene | Map Position | Inheritance | Type of Parkinsonism |
|---|---|---|---|---|
| Well-validated loci/genes | ||||
| PARK1/PARK4 | SNCA | 4q21 | AD | EOPD |
| PARK2 | Parkin | 6q25.2–q27 | AR | Juvenile and EOPD |
| PARK6 | PINK1 | 1p35-p36 | AR | EOPD |
| PARK7 | DJ-1 | 1p36 | AR | EOPD |
| PARK8 | LRRK2 | 12q12 | AD (incomplete penetrance) | LOPD |
| PARK9 | ATP13A2 | 1p36 | AR | Kufor-Rakeb syndrome |
| Putative loci/genes | ||||
| PARK3 | Unknown | 2p13 | AD | LOPD |
| PARK5 | UCHL1 | 4p14 | AD | LOPD |
| PARK10 | Unknown | 1p32 | Not clear | LOPD |
| PARK11 | GIGYF2 | 2q36–q37 | AD (incomplete penetrance) | LOPD |
| PARK12 | Unknown | Xq21-25 | Not clear | Not clear |
| PARK13 | Omi/HTRA2 | 2p12 | Not clear | Not clear |
| PARK14 | PLA2G6 | 22q13.1 | AR | Adult onset dystonia-parkinsonism |
| PARK15 | FBXO7 | 22q12–q13 | AR | Early onset parkinsonian-pyramidal syndrome |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; EOPD, early onset Parkinson disease; LOPD, late onset Parkinson disease.
Table 2. Susceptibility Genes/Loci for Parkinson Disease.
| PARK Locus | Gene | Map Position | Risk variants | Approximate Odds Ratio for PD |
|---|---|---|---|---|
| Well-validated loci/genes | ||||
| PARK1/PARK4 | SNCA | 4q21 | REP1 repeat polymorphism, multiple SNPs in 3′ half of gene | 1.2-1.4 |
| PARK8 | LRRK2 | 12q12 | G2385R, R1628P | 2.0-2.2 |
| Not assigned | MAPT | 17q21.1 | H1 haplotype | 1.4 |
| Not assigned | GBA | 1q21 | >300 mutations including; N370S and L444P | 5.4 |
| Putative loci/genes | ||||
| PARK16 | Unknown | 1q32 | Multiple SNPs from GWASs | 1.3-1.4 |
| PARK17 | GAK | 4p16 | Multiple SNPs from GWASs | 1.5 |
| PARK18 | HLA-DRA | 6p21.3 | Multiple SNPs from GWASs | 1.3 |
Abbreviations: GWASs, genomewide association studies; PD, Parkinson disease; SNP, single nucleotide polymorphism.
Adapted from PDGene Web site (http://www.pdgene.org) and references 18, 27, and 28.
Genes Associated With Autosomal Dominant PD
PARK1/PARK4: SNCA
Inheritance and clinical features
PARK1/PARK4-linked PD displays autosomal dominant inheritance, is often of early onset, and usually progresses rapidly. Affected family members sometimes have atypical clinical features including myoclonus and hypoventilation. Three missense mutations, A53T,12 A30P,34 and E46K,35 duplications36-39 and triplications40,41 of SNCA are known (Figure 1). The A53T substitution was the first mutation identified in a large family with autosomal dominant disease.13 Later, A30P and E46K substitutions were identified in a German and Spanish family, respectively, with clinical features described as dementia with LBs.34,35 SNCA missense mutations and multiplications are both extremely rare causes of familial parkinsonism.36-41
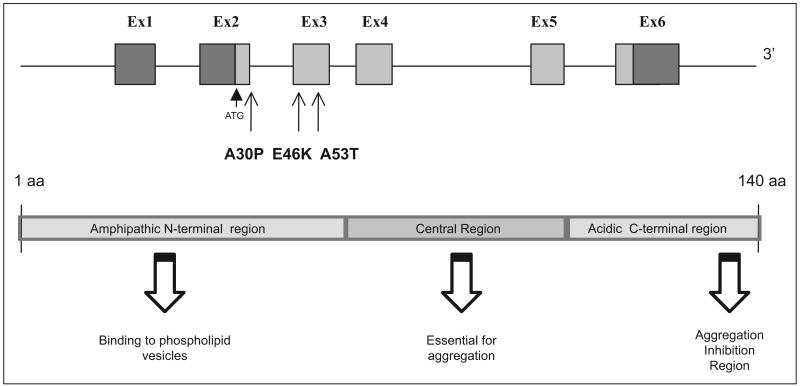
Figure 1.
SNCA (PARK1/PARK4) gene and protein structure. All exons and functional domains are shown. All 3 point mutations identified to date are also shown. ATG indicates the beginning of the coding region (gray); Ex, exon; aa, amino acid.
Gene location and structure. SNCA
Is located on chromosome 4q22.1, has 6 exons, and encodes a 140-amino acid protein. The N-terminus consists of an amphipathic α-helical domain that associates with membrane microdomains, known as lipid rafts.42 The central region contains a fibrillization region and the C-terminus contains an aggregation inhibition region (Figure 1).43
Gene function and expression. SNCA
Is expressed throughout the mammalian brain and enriched in presynaptic nerve terminals.9 The protein can adopt partially folded structures but in its native form is unfolded and can assume both monomeric and oligomeric alpha helix and β-sheet conformations, as well as morphologically diverse aggregates, ranging from those that are amorphous to amyloid-like fibrils.44 These fibrillar moieties are a component of LBs in both familial and idiopathic PD,45 but it is unclear whether the fibrils themselves, or the oligomeric fibrilization intermediates (protofibrils), are toxic to the cell. Interestingly, SNCA genomic multiplications in familial PD are associated with an increase in protein expression,41 and brain samples of triplication mutant carriers show protofibril formation is enhanced with an increase in SNCA expression.46 In vitro, A30P, A53T, and E46K mutant proteins show an increased propensity for self-aggregation and oligomerization into protofibrils, compared with wild-type protein47,48 that may be related to the membrane permeabilization activity of these protofibrils that form pore-like and tubular structures.49 It appears that only A53T and E46K promote formation of the fibrils,50,51 whereas A30P has been reported to disrupt the interaction between α-synuclein and the lipid raft and possibly redistributing the protein away from the synapse.42
A mouse spontaneous deletion strain is viable, fertile, and phenotypically normal,52 whereas overexpression of wild-type SNCA in a mouse model has many features of PD, such as loss of dopaminergic terminals in the striatum, mislocalization and accumulation of insoluble α-synuclein, and motor abnormalities.53-55 Both A30P and A53T mutant mouse models display neuronal cell loss and motor changes.56
Genetic variation
As described in previous sections, multiplications and 3 missense mutations in SNCA have been reported to cause PD, and there is some evidence of genotype–phenotype correlation. For example, American and European families with SNCA triplication show different clinical features than families with a duplication where the phenotype closely resembles idiopathic PD, with late age-of-onset, slow progression, and no atypical features, suggesting that SNCA gene dosage might play a role in disease progression.36,37 SNCA duplications are rarely associated with dementia.38,39 SNCA triplications and the E46K mutation are more commonly associated with dementia than the A30P mutation and gene duplications. The A53T mutation has been associated with dementia and the presence of cortical LBs.57,58
Several common polymorphisms in SNCA have consistently been shown to associate with PD. These variants include a complex repeat polymorphism (REP1) located approximately 10 kb upstream from the translation start site, and several single nucleotide polymorphisms (SNPs) at the 3′ end of the gene.30,59,60 SNCA has been among the genes most significantly associated with PD in all 5 of the PD GWASs conducted in the past 2 years.15-18,61 The mechanism by which common SNCA variants modify susceptibility for PD is not yet known. However, there is evidence to suggest that SNCA alleles associated with increased PD risk are also correlated with higher α-synuclein expression in vitro and with elevated peripheral levels of α-synuclein in vivo.30,62
PARK8: LRRK2
Inheritance and clinical features
Autosomal dominant PARK8-linked PD was first identified in a Japanese family known as the Sagamihara kindred.63 Affected individuals have clinically typical late onset PD and dementia is not a common feature.64 Pathologically, the disease appears to be heterogeneous with the reports of LB pathology, tau pathology, neuronal loss without intracellular inclusions,65,66 and motor neuron disease.64
Gene location and structure
The gene for PARK8 was identified as Leucine Repeat Rich Kinase 2 (LRRK2) in 2004 (also called dardarin, from the Basque word for tremor), in families from the Basque region of Spain, Britain, Western Nebraska, and in an American kindred of German descent.64,67 It is located on chromosome 12p12 and encompasses 144 kb, consisting of 51 exons (7449 bp complementary DNA [cDNA]), and encodes a protein consisting of 2517 amino acids. The LRRK2 gene contains several functional domains including ARM (armadillo domain), ANK (ankyrin repeat domain), LRR (leucine-repeat-rich), ROC (Ras of complex proteins), COR (carboxy terminal of ROC), MAPKKK (mitogen-activated protein kinase kinase kinase), and a WD40 domain that is rich in tryptophan and aspartate repeats.
Gene function and expression
The function of LRRK2 is not well known although it has been identified as a tyrosine kinase–like protein.68 The ROC domain is able to bind guanosine triphosphate (GTP) and is essential for the MAPKKK domain to exert kinase activity but does not have GTPase activity.69 Some of the LRRK2 mutations appear to exert increased kinase activity.70,71 Other functional domains are believed to be important in protein–protein interactions.64 LRRK2 also interacts with other familial PD proteins. For example, LRRK2 appears to interact with parkin through the ROC domain; however, the interaction with parkin does not seem to enhance polyubiquitylation of LRRK2.72 LRRK2 expression has been described in the central nervous system (cerebral cortex, medulla, cerebellum, spinal cord, putamen, and substantia nigra), heart, kidney, lung, liver, and peripheral leukocytes.64,67 LRRK2 protein is found in the cytosol and mitochondrial outer membrane,71 plasma membrane, lysosomes, endosomes, transport vesicles, Golgi apparatus, a cytoskeleton protein microtubule, synaptic vesicles, and lipid rafts.73,74 Interestingly, α-synuclein is also expressed in the presynaptic membranes and lipid rafts.42
There is currently very limited postmortem data on pathogenic LRRK2 mutations, but it appears that typical LB pathology is seen in most LRRK2-related patients. One clinicopathological study reported substantia nigra cell loss, LB formation, and small numbers of cortical LBs.75 In the same study, the 18F-dopa positron emission tomography (PET) in a proband, but not unaffected family members, showed a pattern of nigrostriatal dysfunction typical of idiopathic PD.75 The mechanism that links LRRK2 protein to SNCA protein accumulation remains unknown, but evidence suggests that there may be a direct interaction between LRRK2 and the SNCA protein.76
Genetic variation
Over 40 missense or nonsense mutations have been reported in LRRK2, but the pathogenicity of most of these rare variants has not yet been determined (Figure 2).63,64,67,68 There is currently convincing evidence to suggest that 6 mutations are disease-causing (R1441C, R1441G, R1441H, Y1699C, G2019S, and I2020T). By far the most prevalent mutation is G2019S, which occurs in 1% to 2% of PD patients of European origin, 15% to 20% of Ashkenazi Jewish patients, and approximately 40% of North African Arabs with PD.77-80 G2019S exhibits reduced penetrance, though penetrance estimates at 80 years of age vary widely from 24% to nearly 100%.78,81-84 In contrast, the next most frequent mutations, which occur in codon 1441, are highly penetrant.85
Figure 2.
LRRK2 (PARK8) gene and protein structure. Established pathogenic mutations (boldface) and risk variants (gray) are shown. Variants specific to Asian populations are indicated by an asterisk. ARM indciates Armadillo region; ANK, Ankyrin repeat region; LRR, leucine-rich repeat domain; ROC, Ras of complex; COR, C terminal of Ras (GTPase);. MAPKKK, mitogen-activated protein kinase kinase kinase; Ex, exon; aa, amino acid.
Two common LRRK2 polymorphisms (G2385R and R1628P) that occur only in Asian populations are now well-validated risk factors for PD.86,87 A recent meta-analysis of 9 studies of G2385R reported a combined odds ratio in favor of PD of 2.55 (95% CI, 2.10-3.10).88 Whether LRRK2 polymorphisms that convey risk for PD exist in other populations remains to be determined.
Genes Associated With Autosomal Recessive PD
PARK2
Inheritance and clinical features
PARK2-related parkinsonism is autosomal recessive and usually of early onset. Age at onset is typically between childhood and age 40 years.89 Dystonia is frequently present and patients are levodopa responsive. Pathologically, the substantia nigra undergoes severe neuronal loss and gliosis, whereas the locus coeruleus is much less severely involved and usually no LBs are seen90,91 although rare LB-positive cases have been reported.92-94
Gene location and structure
Linkage analysis of families with autosomal recessive juvenile parkinsonism mapped the PARK2 locus to chromosome 6q26, near the sod2 locus.95-97 By screening a BAC library using the D6S305 marker at this region, a cDNA was cloned containing the 1395 bp open reading frame of the novel PARK2 gene.98 The PARK2 gene contains 12 exons and spans approximately 1.38 Mb.98,99
Gene function and expression. PARK2
Encodes parkin, a 465-amino acid protein that belongs to the “ring between ring fingers” (RBR) family of E3 ubiquitin ligases. The RBR domain of these proteins is composed of 2 RING fingers linked by a cysteine-rich “in-between-RING” (IBR) motif. The RBR domain interacts with ubiquitin-conjugating enzymes (E2s) to catalyze attachment of ubiquitin to protein targets, thus tagging these proteins for destruction by the proteosome.100,101 Many ubiquitination substrates have been proposed for parkin including the aminoacyl-tRNA synthetase cofactor, p38, and a rare, 22-kDa glycosylated form of α-synuclein.102-104 Parkin is predominantly a cytosolic protein but also co-localizes to synaptic vesicles, the Golgi complex, endoplasmic reticulum, and the mitochondrial outer membrane.100,104-107
Many PD-linked point mutations alter wild-type parkin cellular localization, solubility, or propensity to aggregate.108-110 Other mutations, including insertions and deletions, result in parkin loss-of-function. Several lines of PARK2 knockout mice have been generated, but surprisingly none display nigral neuronal degeneration or signs of motor dysfunction.111-113 Subtle behavioral abnormalities and reduced numbers of noradrenergic neurons in the locus ceruleus have been reported in some strains but not others.112-114 In 1 knockout mouse line, reduced numbers of mitochondrial oxidative phosphorylation proteins, a decrease in mitochondrial respiratory capacity, and age-dependent increases in oxidative damage were reported.115 Mitochondrial defects have also been reported in parkin knockout Drosophila, suggesting that defects in parkin ubiquination function might be secondary in the course of pathogenic events.116,117 In vitro studies of a PARK2-knockdown SH-SY5Y cell line showed apoptotic cell death and an increase in the auto-oxidized forms of levodopa and dopamine, suggesting that parkin might have important antioxidative properties.118
Although patients with PARK2-related parkinsonism exhibit loss of pigmented nigral dopamine neurons, LBs are usually not observed.92 However, there are exceptions, as nigral LBs have been reported in 2 patients with compound heterozygous mutations (a 52-year-old with R275W and an exon 3 deletion and a 73-year-old with an exon 7 deletion and del1072T) and in a patient homozygous for an exon 3 deletion.92-94
Genetic variation
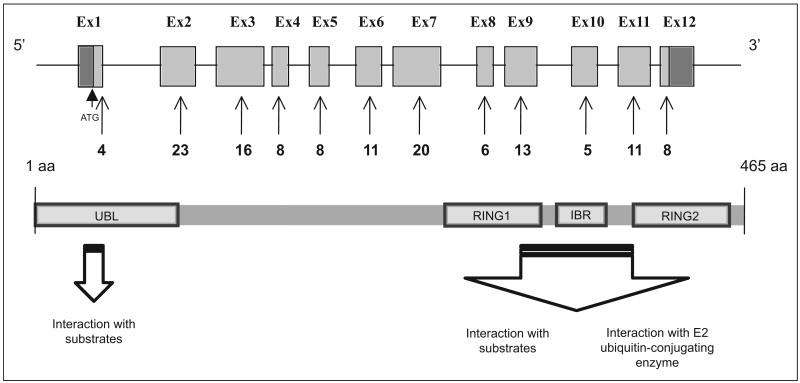
Reported mutations in PARK2 now exceed 100 including missense and nonsense mutations, as well as exonic deletions, rearrangements, and duplications.119-124 These mutations span the entire length of the gene and include the N-terminal UBL domain and the RING-IBR-RING domain (Figure 3). The most common mutations are (1) deletions of exon 4 (n = 28), (2) deletions of exon 3 (n = 27), (3) deletions of exons 3 to 4 (n = 23), (4) a point mutation in exon 7 (924C>T; n = 38), and (5) a single base pair deletion in exon 2 (255/256delA; n = 17). These 5 variants account for 35% of all PARK2 mutations. Hot spots for mutations are concentrated in exons 2 and 7, whereas exon rearrangements are more likely to occur in introns 2 through 4125 (Figure 3).
Figure 3.
PARK2 gene and protein structure. More than 100 mutations have been identified. Number of mutations in each exon is shown as reported in the PD mutation database http://grenada.lumc.nl/LOVD2/TPI/home.php?select_db=PARK2. Hot spots for parkin mutations are concentrated in exons 2 and 7, whereas hot spots for exon rearrangements occur in introns 2 through 4. ATG indicates the beginning of the coding region (gray); UBL, ubiquitin-like domain; Ex, exon; aa, amino acid.
PARK2 mutations account for over 50% of patients with juvenile onset parkinsonism (age at onset ≤20 years), but mutation frequency diminishes substantially with increasing age at onset.89,126,127 In late onset PD (>50 years), the proportion of patients homozygous or compound heterozygous for PARK2 mutations is <1%.123,128,129
Whether simple heterozygotes for PARK2 mutations are at increased risk for PD remains controversial.130 Some studies have reported a significantly higher frequency of PARK2 mutations in patients with PD versus controls.131,132 Also, an analysis of 183 PD families found that the mean age at onset in affected individuals with 1 PARK2 mutation was 11.7 years earlier than affected individuals with no mutations.133 Consistent with these data, Khan and colleagues reported that asymptomatic heterozygous PARK2 carriers show significant striatal dopaminergic dysfunction by 18F-dopa PET, suggesting that such individuals might have haploinsufficiency.134 However, the largest case–control study conducted to date, which included 2091 unselected patients with PD and 1686 controls found no significant difference in mutation frequency between the 2 groups.129 Further larger scale studies will be needed to determine the role of heterozygous PARK2 mutations in the modifying risk of PD.
PARK7: DJ1
Inheritance and clinical features. DJ1
Recessively inherited missense and exonic deletion mutations were first identified in 2 European families with an age of onset of 20 to 40 years.135 PARK7-linked PD appears to be very rare.135-137 Very few DJ1 patients have been reported in the literature and thus there is limited knowledge on the clinical features, neuropathology, and genotype–phenotype correlation for DJ1-related PD. In addition to parkinsonism, clinical characteristics that have been reported in some patients include psychiatric symptoms,138 short statue, and brachydactyly.139
Gene location and structure. DJ1
Has been cloned and is located on chromosome 1p36.23. It has a transcript length of 949 bps with 7 exons.140 It encodes a protein consisting of 189 amino acids.141
Gene function and expression. DJ1
Is a homodimer that belongs to the peptidase C56 family of proteins.142 It is a cytoplasmic protein but can also translocate into the mitochondria143 and appears to act as an antioxidant.140,144-146 Its antioxidant properties may depend on a cysteine residue at position106, which on oxidation forms a disulphide bond.145 DJ1 might act as either a redox-sensor protein that can prevent the aggregation of α-synuclein or an antioxidant.145,147-151 It might also act as a reactive oxygen species scavenger through auto-oxidation.152 These proposed functions for DJ1 could be particularly important in nigral dopamine neurons that are exposed to particularly high levels of oxidative stress.
Expression of DJ1 is ubiquitous and abundant in most mammalian tissues including in the brain where it is found in both neuronal and glial cells.153 Downregulation of endogenous DJ1 protein in neuronal cell lines by small interfering RNA (siRNA) enhances oxidative stress-induced cell death, ER stress, and proteasome inhibition but not by proapoptotic stimulus.152,154 The L166P mutant protein has a reduced antioxidative activity.155 Mutant DJ1 appears to interact with parkin,146 whereby parkin acts as an E3 ligase to remove mutated DJ1. DJ1 null mice are hypersensitive to oxidative stress and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP).156 DJ1 protein expression is increased on oxidative stress induced by paraquat.149
DJ1 does not appear to be an essential component of LBs as only a small proportion of LBs from PD brains display DJ1 immunoreactivity.153 Although DJ1 mutations are rare even in early onset PD, recent studies suggest that DJ1 protein might play an important role in sporadic late-onset PD. Sporadic PD brain has greater oxidative damage to DJ1 and a significant increase in total protein levels, compared with normal controls.157
Genetic variation
In DJ1-related parkinsonism, mutations are found in the homozygous or compound heterozygous state, putatively resulting in a loss of protein function. The L166P mutation causes destabilization through unfolding of the C-terminus, inhibiting dimerization and enhancing degradation by the proteasome.142,158,159 In addition, probably consequent to instability, L166P reduces the neuroprotective function of DJ1.152 Reduced nuclear localization, in favor of the mitochondria, is also seen for L166P, as well as for the M26I and D149A mutations.141,160 The mutations L166P, E64D, M26I, A104T, and D149A have been shown to create structural perturbations of DJ1 protein that lead to global destabilization, unfolding of the protein structure, heterodimer formation, or reduced antioxidant activity.155,161,162
PARK6: PINK1
Inheritance and clinical features
PARK6 was first mapped to chromosome 1p35-p36 in a large consanguineous Italian family with autosomal recessive, early onset PD. Subsequently, phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1) was determined to be the disease-causing gene. PINK1 mutations have been reported to account for approximately 1% to 3% of early onset PD in populations of European ancestry,163-167 8.9% of autosomal recessive PD in a sample of Japanese families,168 and 2.5% of early onset PD in a sample of ethnic Chinese, Malays, and Indians.169,170 Although age at onset for PINK1-related PD is usually in the fourth to fifth decade, clinical features are similar to late onset PD, with slow progression, excellent response to levodopa, and in some instances dementia.164,167,170,171 There is some evidence to suggest that heterozygous carriers of PINK1 mutations might be at increased risk for PD, but definitive large-scale studies have not yet been performed.166,172 In support of this idea, heterozygous PARK6 carriers have been reported to have a 20% to 30% reduction in caudate and putamen 18F-dopa uptake in comparison with controls.173
Gene location and structure. PINK1
Is located on chromosome 1p36.12, has 8 exons, and a cDNA that spans 1.8 kb. It encodes a protein with 581 amino acids. It has a serine/threonine protein kinase domain. However, its function is not known.164 It is a mitochondrial protein located in the matrix and the intermembrane space that is ubiquitously expressed in the brain and systemic organs and contains a mitochondrial-targeting motif and a conserved serine/threonine kinase domain.174
Gene function and expression
Functional studies have shown that PINK1 is localized to mitochondria both in vitro and in vivo.175 Wild-type PINK1 appears to be important in neuroprotection against mitochondrial dysfunction and proteasome-induced apoptosis, whereas the G309D mutation impairs this protective effect, possibly by interfering with adenosine diphosphate (ADP) binding and thus inhibiting kinase activity.164,176 E240K and L489P mutants disrupt PINK1's protectivity by either enhancing the instability of the protein or disrupting the kinase activity of the protein.177 In vitro studies indicate that cells transfected with PINK1 mutants have disrupted mitochondrial membrane potential under stressful conditions.178 Knockout models of the Drosophila PINK1 ortholog have defects in mitochondrial morphology and increased sensitivity to oxidative stress and appear to be rescued by human parkin.179
Genetic variation
The first mutations discovered were the G309D missense and a W437X truncating mutation found in the families of Italian and Spanish descent.164,176 Since then, several point mutations, frameshifts, and truncating mutants have been identified.167,170,180 Interestingly, in contrast to PARK2, the majority of PINK1 mutations reported are either missense or nonsense mutations.165,168,171,176,181 One family with a large homozygous deletion has been reported involving exons 6 to 8.168 Most of the reported point mutations in PINK1 are located in a highly conserved amino acid position in the protein kinase domain.178 However, in a large Sudanese kindred with early onset parkinsonism and dopa-responsive dystonia, the pathogenic mutation (A217D) is located in the highly conserved adenosine triphosphate (ATP) orientation site of the PINK1 kinase domain.182,183
PARK9: ATP13A2
Inheritance and clinical features
Homozygous and compound heterozygous mutations in the P-type ATPase gene (ATP13A2) have been demonstrated in a Jordanian family184,185 and a Chilean family186 with Kufor-Rakeb syndrome, a form of recessively inherited atypical parkinsonism which is clinically characterized by very early age of onset (11-16 years), levodopa-responsive parkinsonism, pyramidal signs, dementia, and supranuclear gaze palsy184,187 Magnetic resonance imaging (MRI) shows significant atrophy of the globus pallidus and the pyramids, and generalized brain atrophy in later stages. Some develop facial, faucial, and finger mini-myoclonus, visual hallucinations, and oculogyric dystonic spasm.187
Gene location and structure
The disease locus designated as PARK9 was mapped to 1p36 with a maximum logarithm of the odds (LOD) score of 3.6, a hot spot for autosomal recessive familial PD. The disease gene was subsequently identified as ATP13A2. The transcript has 29 exons and is 3854 bps in length. The ATP13A2 protein contains 1180 amino acids and has 10 transmembrane domains.186
Gene function and expression
ATP13A2 is a lysosomal membrane protein with an ATPase domain.186 It is a member of the P5 subfamily of ATPases, which transports inorganic cations and other substrates. The exact function of the ATP13A2 protein is still unknown. ATP13A2 is predominantly expressed in brain tissue with the highest levels reported in ventral midbrain (which includes the substantia nigra).186 ATP13A2 mRNA levels are approximately 10-fold higher in the nigral dopamine neurons of sporadic patients than control participant brains
Genetic variation
All known ATP13A2 mutations appear to directly or indirectly affect transmembrane domains.186 In vitro evidence indicates that wild-type ATP13A2 is localized to the lysosome membrane of transiently transfected cells, whereas unstable truncated mutants are retained in the endoplasmic reticulum and degraded by the proteasome.186 A homozygous missense mutation (G504R) has been identified in 1 sporadic case from Brazil with juvenile parkinsonism.188 This patient had symptom onset at age 12, levodopa-responsive severe akinetic-rigid parkinsonism, levodopa-induced motor fluctuations and dyskinesias, severe visual hallucinations, and supranuclear vertical gaze paresis, moderate diffuse atrophy but did not have dementia or pyramidal deficits. In this same study, 2 Italian patients with early onset PD without atypical features each carried a single novel missense mutation (T12M or G533R),188 raising the question of whether heterozygous ATP13A2 mutation carriers are at increased risk of PD.
Genetic Risk Factors for PD
Common variants in 3 genes (LRRK2, MAPT, and SNCA) and loss-of-function mutations in another (GBA) are now well-established risk factors for PD. In addition, 3 new putative susceptibility loci for PD (PARK16-18) have been identified from recent GWASs. Of these 7 genes/loci, LRRK2 and SNCA have been described in previous sections. In this section, we briefly describe the remainder.
GBA
Loss of function mutations in the glucocerebrosidase (GBA) gene, which encodes the enzyme glucocerebrosidase, result in Gaucher disease (GD), an autosomal recessive glycolipid storage disorder with multisystemic manifestations, including involvement of the liver, spleen, bone marrow, lungs, and nervous system.189 Nearly 300 pathogenic mutations have been described, but in populations of European origin, 2 mutations (N370 and L444P) account for approximately two thirds of the disease alleles seen in GD.189,190 A small subset of patients with GD develop parkinsonism with brain stem or diffuse Lewy-related pathology.191 An increased incidence of parkinsonism has also been reported in relatives of patients with GD.192 These observations prompted a number of case–control association analyses seeking to determine whether GBA mutations are a risk factor for PD. In 2009, Sidransky and colleagues published the definitive study on this topic, a pooled analysis of 5691 patients with PD and 4898 controls from 16 centers in 12 countries.27 In the subset of participants in which the entire GBA coding region was screened, loss-of-function mutations were observed in 6.9% of cases and 1.3% of controls (odds ratio, 5.4; 95% CI, 3.9-7.6). Among the subset of individuals of Ashkenazi Jewish ancestry, higher mutation frequencies were seen: 19.3% in cases and 4.1% in controls. The clinical characteristics of patients having PD with and without GBA mutations were very similar. There is also evidence to suggest that GBA mutations are a risk factor for dementia with LBs.193,194
The mechanism by which GBA mutations modify risk for PD is not yet known, but both gain-of- and loss-of-function hypotheses have been proposed. One putative gain-of-function mechanism is based on data suggesting that some GBA mutations result in misfolded protein.195 Misfolded glucocerebrosidase might then contribute to neurodegeneration by inducing lysosomal insufficiency, by impairing autophagic pathways necessary for degrading α-synuclein, or by overwhelming the ubiquitin–proteasome pathway.196 A loss-of-function hypothesis recently proposed centers on observations that α-synuclein binds to lipids in the plasma membrane,197 and that binding of lipid might reduce the formation of fibrillar forms of α-synuclein. Glucocerebrosidase degrades glucocerebroside to ceramide, and thus GBA haploinsuffiency might cause this and other polyunsaturated lipids to accumulate and alter the sphingolipid composition of cell membranes. This in turn could disrupt membrane binding of α-synuclein, thus enhancing its aggregation in the cytoplasm.196
Genetic Variation
Microtubule-Associated Protein Tau
The microtubule-associated protein tau, encoded by the MAPT gene, is primarily expressed in neurons and plays a key role in the organization and integrity of the cytoskeleton.198 Filamentous neuronal tau inclusions define a set of neurodegenerative diseases referred to as the “tauopathies,” which include Alzheimer disease, corticobasal degeneration (CBD), PSP, and frontotemporal dementia with parkinsonism linked to chromosome 17 (FTDP-17).199,200 MAPT was first linked to the pathogenesis of tauopathies by the discovery of mutations resulting in FTDP-17 and subsequently an extended common haplotype (H1) across the gene was shown to associate with disease risk in PSP and CBD.201-203 Because PD shares some clinical features with the tauopathies, a number of studies have been conducted over the past decade on the relationship between common MAPT variants and PD risk. Two large case–control studies published in 2007 provided strong evidence that the MAPT H1 haplotype is associated with PD risk, and a metaanalysis of 18 studies yield an odds ratio for PD (under a dominant model) of 1.40 (95% CI, 1.30-1.50; P = 2 × 10−19).28,204 Subsequently, the MAPT H1 haplotype has been confirmed as a PD risk factor in all 4 of the GWASs conducted in populations of European origin in the past 2 years.16-18,61
The MAPT H1 and H2 haplotypes actually represent 2 distinct families of subhaplotypes, which arose from an inversion of 900 kb on chromosome 17q21 approximately 3 million years ago.205 One such subhaplotype within the H1 family, designated “H1c,” is clearly associated with PSP and CBD.206 However, this same subhaplotype is not associated with PD, so it appears that different MAPT variants convey risk of PD versus the tauopathies.203
In FTDP-17, MAPT mutations are thought to alter tau function in a manner that promotes tau fibrillization and impairs its ability to regulate microtubule dynamics, thus causing neurotoxicity.199,207 However, the mechanism by which common MAPT variation modifies risk of PD, a disease in which tau aggregates are rarely seen, remains to be determined.
PARK16-18
Recent GWASs have nominated 3 new susceptibility loci. Satake and colleagues reported an association between multiple SNPs on chromosome 1q32 and PD in a large, 2-tiered GWASs of participants of Japanese ancestry.15 This region, designated PARK16, contains several candidate genes, including RAB7L1, a small GTP-binding protein that regulates exo- and endocytotic pathways, and NUCKS1, a nuclear DNA-binding protein expressed in brain and other tissues that might regulate chromatin structure and its activity.208-210
Pankratz and colleagues performed a GWASs in 857 familial PD cases and 867 controls.17 Although none of their results met genomewide significance, the region most significantly associated with PD under an additive model occurred on chromosome 4 (rs11248060, odds ratio 1.69, P = 3.4 × 10−6). This region was later replicated in a second, larger GWASs and designated PARK17.18 The PARK17 locus contains several genes, include GAK (cyclin G-associated kinase), a promising candidate gene that functions as a cell cycle regulator and is differentially expressed within the substantia nigra of PD brains in comparison to controls.17,211
Finally, the most recently conducted GWASs by Hamza and colleagues nominated the HLA region, designated PARK18, as a putative susceptibility locus for PD.18 This fits well with a growing body of literature, indicating that chronic inflammation and humoral immunity play a role in the pathogenesis PD.212,213
Because of concerns for type I error that are inherent in all GWASs, PARK16-18 must be replicated in multiple independent samples before being considered bona fide PD susceptibility loci.
Summary and Conclusions
A total of 6 genes (SNCA, LRRK2, PRKN, DJ1, PINK1, and ATP13A2,) have conclusively been linked to monogenic forms of parkinsonism, and all but ATP13A2 result in disease that closely resembles the clinical features of idiopathic PD. Common variation in 3 genes (MAPT, LRRK2, and SNCA) and loss-of-function mutations in GBA are now well-established risk factors for PD. Recent PD GWASs have nominated 3 new putative susceptibility loci (PARK16-18), though these findings require rigorous replication before firm conclusions can be drawn.
Additional susceptibility loci will likely be uncovered in the near future, as the wealth of recent data from GWASs is further analyzed. Such efforts will include meta-analysis, consideration of gene × gene and gene × environment interaction, and analysis of copy number variation. Traditional linkage analysis will also continue to be an invaluable tool for gene discovery, and efforts to examine understudied populations across the world could prove particularly fruitful.
Although important progress has been made, the mechanisms by which variation in PD-linked genes leads to neurodegeneration remains poorly understood. However, data accumulated thus far has implicated mitochondrial dysfunction, oxidative damage, aberrant protein aggregation, and deficits in ubiquitin-mediated protein degradation as playing key roles in the etiopathogenesis of PD.
Acknowledgments
Funding: The author(s) disclosed receipt of the following financial support for the research and/or authorship of this article: the Department of Veterans Affairs (1I01BX000531, Office and Research and Development, Biomedical Laboratory Research Program) and the National Institutes of Health (P50 NS062684, R01 NS065070 and T32 AG000258).
Footnotes
Declaration of Conflicting Interests: The author(s) declared no conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1.de Rijk MC, Launer LJ, Berger K, et al. Prevalence of Parkinson's disease in Europe: a collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology. 2000;54(11 suppl 5):S21–S23. [PubMed] [Google Scholar]
- 2.Sellbach AN, Boyle RS, Silburn PA, Mellick GD. Parkinson's disease and family history. Parkinsonism Relat Disord. 2006;12(7):399–409. doi: 10.1016/j.parkreldis.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Farrer MJ. Genetics of Parkinson disease: paradigm shifts and future prospects. Nat Rev Genet. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 4.Fearnley JM, Lees AJ. Ageing and Parkinson's disease: substantia nigra regional selectivity. Brain. 1991;114(pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 5.Samii A, Nutt JG, Ransom BR. Parkinson's disease. Lancet. 2004;363(9423):1783–1793. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 6.Jankovic J. An update on the treatment of Parkinson's disease. Mt Sinai J Med. 2006;73(4):682–689. [PubMed] [Google Scholar]
- 7.Gibb WR, Lees AJ. A comparison of clinical and pathological features of young- and old-onset Parkinson's disease. Neurology. 1988;38(9):1402–1406. doi: 10.1212/wnl.38.9.1402. [DOI] [PubMed] [Google Scholar]
- 8.Brooks DJ. Imaging approaches to Parkinson disease. J Nucl Med. 2010;51(4):596–609. doi: 10.2967/jnumed.108.059998. [DOI] [PubMed] [Google Scholar]
- 9.George JM. The synucleins. Genome Biol. 2002;3(1) doi: 10.1186/gb-2001-3-1-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Love S. Neuropathological investigation of dementia: a guide for neurologists. J Neurol Neurosurg Psychiatry. 2005;76(suppl 5):v8–v14. doi: 10.1136/jnnp.2005.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson's disease-related pathology. Cell Tissue Res. 2004;318(1):121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 12.Polymeropoulos MH, Higgins JJ, Golbe LI, et al. Mapping of a gene for Parkinson's disease to chromosome 4q21-q23. Science. 1996;274(5290):1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 13.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 14.Belin AC, Westerlund M. Parkinson's disease: a genetic perspective. FEBS J. 2008;275(7):1377–1383. doi: 10.1111/j.1742-4658.2008.06301.x. [DOI] [PubMed] [Google Scholar]
- 15.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 16.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankratz N, Wilk JB, Latourelle JC, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamza TH, Zabetian CP, Tenesa A, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet. 2010;42(9):781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paisan-Ruiz C, Bhatia KP, Li A, et al. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65(1):19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Fonzo A, Dekker MC, Montagna P, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72(3):240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 21.Hicks AA, Petursson H, Jonsson T, et al. A susceptibility gene for late-onset idiopathic Parkinson's disease. Ann Neurol. 2002;52(5):549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- 22.Lautier C, Goldwurm S, Durr A, et al. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am J Hum Genet. 2008;82(4):822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leroy E, Boyer R, Auburger G, et al. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 24.Gasser T, Muller-Myhsok B, Wszolek ZK, et al. A susceptibility locus for Parkinson's disease maps to chromosome 2p13. Nat Genet. 1998;18(3):262–265. doi: 10.1038/ng0398-262. [DOI] [PubMed] [Google Scholar]
- 25.Strauss KM, Martins LM, Plun-Favreau H, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet. 2005;14(15):2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 26.Pankratz N, Nichols WC, Uniacke SK, et al. Genome-wide linkage analysis and evidence of gene-by-gene interactions in a sample of 362 multiplex Parkinson disease families. Hum Mol Genet. 2003;12(20):2599–2608. doi: 10.1093/hmg/ddg270. [DOI] [PubMed] [Google Scholar]
- 27.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N Engl J Med. 2009;361(17):1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goris A, Williams-Gray CH, Clark GR, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson's disease. Ann Neurol. 2007;62(2):145–153. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 29.Lesage S, Brice A. Parkinson's disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18(R1):R48–R59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- 30.Mata I, Samii A, Factor S, et al. Variation in the Alpha-Synuclein Gene, Independent of REPI, Modifies Risk for Parkinson's Disease. Neurology. 2009;72(11):A393–A394. [Google Scholar]
- 31.Paisan-Ruiz C. LRRK2 gene variation and its contribution to Parkinson disease. Hum Mutat. 2009;30(8):1153–1160. doi: 10.1002/humu.21038. [DOI] [PubMed] [Google Scholar]
- 32.Mata I, Checkoway H, Hutter C, et al. Common variation in the LRRK2 gene is a risk factor for Parkinson's disease. Neurology. 2009;72(11):A394–A394. doi: 10.1002/mds.25226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabetian C, Hutter CM, Factor S, et al. Association analysis of the MAPT H1 haplotype and subhaplotypes in Parkinson's disease. Neurology. 2007;68(12):A303–A303. doi: 10.1002/ana.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat Genet. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 35.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 36.Chartier-Harlin MC, Kachergus J, Roumier C, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 37.Ibanez P, Bonnet AM, Debarges B, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet. 2004;364(9440):1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 38.Nishioka K, Hayashi S, Farrer MJ, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson's disease. Ann Neurol. 2006;59(2):298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs J, Nilsson C, Kachergus J, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68(12):916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 40.Singleton AB, Farrer M, Johnson J, et al. Alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 41.Farrer M, Kachergus J, Forno L, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55(2):174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 42.Fortin DL, Troyer MD, Nakamura K, Kubo S, Anthony MD, Edwards RH. Lipid rafts mediate the synaptic localization of alpha-synuclein. J Neurosci. 2004;24(30):6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bisaglia M, Mammi S, Bubacco L. Structural insights on physiological functions and pathological effects of alpha-synuclein. FASEB J. 2009;23(2):329–340. doi: 10.1096/fj.08-119784. [DOI] [PubMed] [Google Scholar]
- 44.Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21(2):211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 45.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 46.Miller DW, Hague SM, Clarimon J, et al. Alpha-synuclein in blood and brain from familial Parkinson disease with SNCA locus triplication. Neurology. 2004;62(10):1835–1838. doi: 10.1212/01.wnl.0000127517.33208.f4. [DOI] [PubMed] [Google Scholar]
- 47.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4(11):1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 48.Pandey N, Schmidt RE, Galvin JE. The alpha-synuclein mutation E46K promotes aggregation in cultured cells. Exp Neurol. 2006;197(2):515–520. doi: 10.1016/j.expneurol.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 49.Lashuel HA, Petre BM, Wall J, et al. Alpha-synuclein, especially the Parkinson's disease-associated mutants, forms pore-like annular and tubular protofibrils. J Mol Biol. 2002;322(5):1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- 50.Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry. 2000;39(10):2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 51.Greenbaum EA, Graves CL, Mishizen-Eberz AJ, et al. The E46K mutation in alpha-synuclein increases amyloid fibril formation. J Biol Chem. 2005;280(9):7800–7807. doi: 10.1074/jbc.M411638200. [DOI] [PubMed] [Google Scholar]
- 52.Specht CG, Schoepfer R. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2001;2:11. doi: 10.1186/1471-2202-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rockenstein E, Mallory M, Hashimoto M, et al. Differential neuropathological alterations in transgenic mice expressing alpha-synuclein from the platelet-derived growth factor and Thy-1 promoters. J Neurosci Res. 2002;68(5):568–578. doi: 10.1002/jnr.10231. [DOI] [PubMed] [Google Scholar]
- 54.Masliah E, Rockenstein E, Veinbergs I, et al. Dopaminergic loss and inclusion body formation in alpha-synuclein mice: implications for neurodegenerative disorders. Science. 2000;287(5456):1265–1269. doi: 10.1126/science.287.5456.1265. [DOI] [PubMed] [Google Scholar]
- 55.Fleming SM, Salcedo J, Fernagut PO, et al. Early and progressive sensorimotor anomalies in mice overexpressing wild-type human alpha-synuclein. J Neurosci. 2004;24(42):9434–9440. doi: 10.1523/JNEUROSCI.3080-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melrose HL, Lincoln SJ, Tyndall GM, Farrer MJ. Parkinson's disease: a rethink of rodent models. Exp Brain Res. 2006;173(2):196–204. doi: 10.1007/s00221-006-0461-3. [DOI] [PubMed] [Google Scholar]
- 57.Golbe LI, Di Iorio G, Bonavita V, Miller DC, Duvoisin RC. A large kindred with autosomal dominant Parkinson's disease. Ann Neurol. 1990;27(3):276–282. doi: 10.1002/ana.410270309. [DOI] [PubMed] [Google Scholar]
- 58.Golbe LI. The genetics of Parkinson's disease: a reconsideration. Neurology. 1990;40(10 suppl 3):suppl 7–14. discussion 14-16. [PubMed] [Google Scholar]
- 59.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296(6):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 60.Kay DM, Factor SA, Samii A, et al. Genetic association between alpha-synuclein and idiopathic Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(7):1222–1230. doi: 10.1002/ajmg.b.30758. [DOI] [PubMed] [Google Scholar]
- 61.Edwards TL, Scott WK, Almonte C, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74(2):97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10(26):3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 63.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51(3):296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 64.Zimprich A, Muller-Myhsok B, Farrer M, et al. The PARK8 locus in autosomal dominant parkinsonism: confirmation of linkage and further delineation of the disease-containing interval. Am J Hum Genet. 2004;74(1):11–19. doi: 10.1086/380647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wszolek ZK, Pfeiffer RF, Tsuboi Y, et al. Autosomal dominant parkinsonism associated with variable synuclein and tau pathology. Neurology. 2004;62(9):1619–1622. doi: 10.1212/01.wnl.0000125015.06989.db. [DOI] [PubMed] [Google Scholar]
- 66.Nicholl DJ, Vaughan JR, Khan NL, et al. Two large British kindreds with familial Parkinson's disease: a clinicopathological and genetic study. Brain. 2002;125(pt 1):44–57. doi: 10.1093/brain/awf013. [DOI] [PubMed] [Google Scholar]
- 67.Paisan-Ruiz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 68.Mata IF, Ross OA, Kachergus J, et al. LRRK2 mutations are a common cause of Parkinson's disease in Spain. Eur J Neurol. 2006;13(4):391–394. doi: 10.1111/j.1468-1331.2006.01256.x. [DOI] [PubMed] [Google Scholar]
- 69.Ito G, Okai T, Fujino G, et al. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry. 2007;46(5):1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 70.Gloeckner CJ, Kinkl N, Schumacher A, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15(2):223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 71.West AB, Moore DJ, Biskup S, et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102(46):16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith WW, Pei Z, Jiang H, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102(51):18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biskup S, Moore DJ, Celsi F, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60(5):557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 74.Hatano T, Kubo S, Imai S, et al. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet. 2007;16(6):678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- 75.Khan NL, Jain S, Lynch JM, et al. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson's disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128(pt 12):2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 76.Lin X, Parisiadou L, Gu XL, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson's-disease-related mutant alpha-synuclein. Neuron. 2009;64(6):807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Orr-Urtreger A, Shifrin C, Rozovski U, et al. The LRRK2 G2019S mutation in Ashkenazi Jews with Parkinson disease: is there a gender effect? Neurology. 2007;69(16):1595–1602. doi: 10.1212/01.wnl.0000277637.33328.d8. [DOI] [PubMed] [Google Scholar]
- 78.Ozelius LJ, Senthil G, Saunders-Pullman R, et al. LRRK2 G2019S as a cause of Parkinson's disease in Ashkenazi Jews. N Engl J Med. 2006;354(4):424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 79.Lesage S, Durr A, Tazir M, et al. LRRK2 G2019S as a cause of Parkinson's disease in North African Arabs. N Engl J Med. 2006;354(4):422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 80.Zabetian CP, Hutter CM, Yearout D, et al. LRRK2 G2019S in families with Parkinson disease who originated from Europe and the Middle East: evidence of two distinct founding events beginning two millennia ago. Am J Hum Genet. 2006;79(4):752–758. doi: 10.1086/508025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kachergus J, Mata IF, Hulihan M, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: evidence of a common founder across European populations. Am J Hum Genet. 2005;76(4):672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Healy DG, Falchi M, O'Sullivan SS, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson's disease: a case-control study. Lancet Neurol. 2008;7(7):583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Clark LN, Wang Y, Karlins E, et al. Frequency of LRRK2 mutations in early- and late-onset Parkinson disease. Neurology. 2006;67(10):1786–1791. doi: 10.1212/01.wnl.0000244345.49809.36. [DOI] [PubMed] [Google Scholar]
- 84.Goldwurm S, Zini M, Mariani L, et al. Evaluation of LRRK2 G2019S penetrance: relevance for genetic counseling in Parkinson disease. Neurology. 2007;68(14):1141–1143. doi: 10.1212/01.wnl.0000254483.19854.ef. [DOI] [PubMed] [Google Scholar]
- 85.Haugarvoll K, Rademakers R, Kachergus JM, et al. Lrrk2 R1441C parkinsonism is clinically similar to sporadic Parkinson disease. Neurology. 2008;70(16 pt 2):1456–1460. doi: 10.1212/01.wnl.0000304044.22253.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Di Fonzo A, Wu-Chou YH, Lu CS, et al. A common missense variant in the LRRK2 gene, Gly2385Arg, associated with Parkinson's disease risk in Taiwan. Neurogenetics. 2006;7(3):133–138. doi: 10.1007/s10048-006-0041-5. [DOI] [PubMed] [Google Scholar]
- 87.Ross OA, Wu YR, Lee MC, et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson's disease. Ann Neurol. 2008;64(1):88–92. doi: 10.1002/ana.21405. [DOI] [PubMed] [Google Scholar]
- 88.Zabetian CP, Yamamoto M, Lopez AN, et al. LRRK2 mutations and risk variants in Japanese patients with Parkinson's disease. Mov Disord. 2009;24(7):1034–1041. doi: 10.1002/mds.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lucking CB, Durr A, Bonifati V, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 90.Takahashi H, Ohama E, Suzuki S, et al. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44(3 pt 1):437–441. doi: 10.1212/wnl.44.3_part_1.437. [DOI] [PubMed] [Google Scholar]
- 91.Mori H, Kondo T, Yokochi M, et al. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51(3):890–892. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- 92.Farrer M, Chan P, Chen R, et al. Lewy bodies and parkinsonism in families with parkin mutations. Ann Neurol. 2001;50(3):293–300. doi: 10.1002/ana.1132. [DOI] [PubMed] [Google Scholar]
- 93.Pramstaller PP, Schlossmacher MG, Jacques TS, et al. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann Neurol. 2005;58(3):411–422. doi: 10.1002/ana.20587. [DOI] [PubMed] [Google Scholar]
- 94.Sasaki S, Shirata A, Yamane K, Iwata M. Parkin-positive autosomal recessive juvenile Parkinsonism with alpha-synuclein-positive inclusions. Neurology. 2004;63(4):678–682. doi: 10.1212/01.wnl.0000134657.25904.0b. [DOI] [PubMed] [Google Scholar]
- 95.Matsumine H, Saito M, Shimoda-Matsubayashi S, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am J Hum Genet. 1997;60(3):588–596. [PMC free article] [PubMed] [Google Scholar]
- 96.Jones AC, Yamamura Y, Almasy L, et al. Autosomal recessive juvenile parkinsonism maps to 6q25.2-q27 in four ethnic groups: detailed genetic mapping of the linked region. Am J Hum Genet. 1998;63(1):80–87. doi: 10.1086/301937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tassin J, Durr A, de Broucker T, et al. Chromosome 6-linked autosomal recessive early-onset Parkinsonism: linkage in European and Algerian families, extension of the clinical spectrum, and evidence of a small homozygous deletion in one family. The French Parkinson's Disease Genetics Study Group, and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Am J Hum Genet. 1998;63(1):88–94. doi: 10.1086/301934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 99.Asakawa S, Tsunematsu K, Takayanagi A, et al. The genomic structure and promoter region of the human parkin gene. Biochem Biophys Res Commun. 2001;286(5):863–868. doi: 10.1006/bbrc.2001.5490. [DOI] [PubMed] [Google Scholar]
- 100.Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25(3):302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 101.Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci U S A. 1999;96(20):11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shimura H, Schlossmacher MG, Hattori N, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 103.Corti O, Hampe C, Koutnikova H, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12(12):1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- 104.von Coelln R, Dawson VL, Dawson TM. Parkin-associated Parkinson's disease. Cell Tissue Res. 2004;318(1):175–184. doi: 10.1007/s00441-004-0924-4. [DOI] [PubMed] [Google Scholar]
- 105.Kubo SI, Kitami T, Noda S, et al. Parkin is associated with cellular vesicles. J Neurochem. 2001;78(1):42–54. doi: 10.1046/j.1471-4159.2001.00364.x. [DOI] [PubMed] [Google Scholar]
- 106.Mouatt-Prigent A, Muriel MP, Gu WJ, et al. Ultrastructural localization of parkin in the rat brainstem, thalamus and basal ganglia. J Neural Transm. 2004;111(10-11):1209–1218. doi: 10.1007/s00702-004-0144-9. [DOI] [PubMed] [Google Scholar]
- 107.Darios F, Corti O, Lucking CB, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12(5):517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 108.Cookson MR, Lockhart PJ, McLendon C, O'Farrell C, Schlossmacher M, Farrer MJ. RING finger 1 mutations in Parkin produce altered localization of the protein. Hum Mol Genet. 2003;12(22):2957–2965. doi: 10.1093/hmg/ddg328. [DOI] [PubMed] [Google Scholar]
- 109.Gu WJ, Corti O, Araujo F, et al. The C289G and C418R missense mutations cause rapid sequestration of human Parkin into insoluble aggregates. Neurobiol Dis. 2003;14(3):357–364. doi: 10.1016/j.nbd.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 110.Wang C, Tan JM, Ho MW, et al. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson's disease-linked point mutations. J Neurochem. 2005;93(2):422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- 111.Goldberg MS, Fleming SM, Palacino JJ, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278(44):43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- 112.Itier JM, Ibanez P, Mena MA, et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum Mol Genet. 2003;12(18):2277–2291. doi: 10.1093/hmg/ddg239. [DOI] [PubMed] [Google Scholar]
- 113.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci U S A. 2005;102(6):2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Von Coelln R, Thomas B, Savitt JM, et al. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101(29):10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Palacino JJ, Sagi D, Goldberg MS, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279(18):18614–18622. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 116.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100(7):4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pesah Y, Pham T, Burgess H, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131(9):2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 118.Machida Y, Chiba T, Takayanagi A, et al. Common anti-apoptotic roles of parkin and alpha-synuclein in human dopaminergic cells. Biochem Biophys Res Commun. 2005;332(1):233–240. doi: 10.1016/j.bbrc.2005.04.124. [DOI] [PubMed] [Google Scholar]
- 119.Hattori N, Matsumine H, Asakawa S, et al. Point mutations (Thr240Arg and Gln311Stop) [correction of Thr240Arg and Ala311Stop] in the Parkin gene. Biochem Biophys Res Commun. 1998;249(3):754–758. doi: 10.1006/bbrc.1998.9134. [DOI] [PubMed] [Google Scholar]
- 120.Abbas N, Lucking CB, Ricard S, et al. A wide variety of mutations in the parkin gene are responsible for autosomal recessive parkinsonism in Europe. French Parkinson's Disease Genetics Study Group and the European Consortium on Genetic Susceptibility in Parkinson's Disease. Hum Mol Genet. 1999;8(4):567–574. doi: 10.1093/hmg/8.4.567. [DOI] [PubMed] [Google Scholar]
- 121.Klein C, Pramstaller PP, Kis B, et al. Parkin deletions in a family with adult-onset, tremor-dominant parkinsonism: expanding the phenotype. Ann Neurol. 2000;48(1):65–71. [PubMed] [Google Scholar]
- 122.Kann M, Jacobs H, Mohrmann K, et al. Role of parkin mutations in 111 community-based patients with early-onset parkinsonism. Ann Neurol. 2002;51(5):621–625. doi: 10.1002/ana.10179. [DOI] [PubMed] [Google Scholar]
- 123.Klein C, Hedrich K, Wellenbrock C, et al. Frequency of parkin mutations in late-onset Parkinson's disease. Ann Neurol. 2003;54(3):415–416. doi: 10.1002/ana.10737. author reply 416-417. [DOI] [PubMed] [Google Scholar]
- 124.Hedrich K, Marder K, Harris J, et al. Evaluation of 50 probands with early-onset Parkinson's disease for Parkin mutations. Neurology. 2002;58(8):1239–1246. doi: 10.1212/wnl.58.8.1239. [DOI] [PubMed] [Google Scholar]
- 125.Hedrich K, Eskelson C, Wilmot B, et al. Distribution, type, and origin of Parkin mutations: review and case studies. Mov Disord. 2004;19(10):1146–1157. doi: 10.1002/mds.20234. [DOI] [PubMed] [Google Scholar]
- 126.Periquet M, Latouche M, Lohmann E, et al. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain. 2003;126(pt 6):1271–1278. doi: 10.1093/brain/awg136. [DOI] [PubMed] [Google Scholar]
- 127.Mata IF, Lockhart PJ, Farrer MJ. Parkin genetics: one model for Parkinson's disease. Hum Mol Genet. 2004;13(spec no 1):R127–R133. doi: 10.1093/hmg/ddh089. [DOI] [PubMed] [Google Scholar]
- 128.Oliveri RL, Zappia M, Annesi G, et al. The parkin gene is not a major susceptibility locus for typical late-onset Parkinson's disease. Neurol Sci. 2001;22(1):73–74. doi: 10.1007/s100720170053. [DOI] [PubMed] [Google Scholar]
- 129.Kay DM, Stevens CF, Hamza TH, et al. A comprehensive analysis of deletions, multiplications, and copy number variations in PARK2. Neurology. 2010;75:1189–1194. doi: 10.1212/WNL.0b013e3181f4d832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foroud T, Uniacke SK, Liu L, et al. Heterozygosity for a mutation in the parkin gene leads to later onset Parkinson disease. Neurology. 2003;60(5):796–801. doi: 10.1212/01.wnl.0000049470.00180.07. [DOI] [PubMed] [Google Scholar]
- 131.Pankratz N, Kissell DK, Pauciulo MW, et al. Parkin dosage mutations have greater pathogenicity in familial PD than simple sequence mutations. Neurology. 2009;73(4):279–286. doi: 10.1212/WNL.0b013e3181af7a33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lesage S, Lohmann E, Tison F, Durif F, Durr A, Brice A. Rare heterozygous parkin variants in French early-onset Parkinson disease patients and controls. J Med Genet. 2008;45(1):43–46. doi: 10.1136/jmg.2007.051854. [DOI] [PubMed] [Google Scholar]
- 133.Sun M, Latourelle JC, Wooten GF, et al. Influence of heterozygosity for parkin mutation on onset age in familial Parkinson disease: the GenePD study. Arch Neurol. 2006;63(6):826–832. doi: 10.1001/archneur.63.6.826. [DOI] [PubMed] [Google Scholar]
- 134.Khan NL, Scherfler C, Graham E, et al. Dopaminergic dysfunction in unrelated, asymptomatic carriers of a single parkin mutation. Neurology. 2005;64(1):134–136. doi: 10.1212/01.WNL.0000148725.48740.6D. [DOI] [PubMed] [Google Scholar]
- 135.Bonifati V, Rizzu P, Squitieri F, et al. DJ-1(PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003;24(3):159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- 136.Hague S, Rogaeva E, Hernandez D, et al. Early-onset Parkinson's disease caused by a compound heterozygous DJ-1 mutation. Ann Neurol. 2003;54(2):271–274. doi: 10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 137.Hering R, Strauss KM, Tao X, et al. Novel homozygous p.E64D mutation in DJ1 in early onset Parkinson disease (PARK7) Hum Mutat. 2004;24(4):321–329. doi: 10.1002/humu.20089. [DOI] [PubMed] [Google Scholar]
- 138.Dekker MC, van Swieten JC, Houwing-Duistermaat JJ, et al. A clinical-genetic study of Parkinson's disease in a genetically isolated community. J Neurol. 2003;250(9):1056–1062. doi: 10.1007/s00415-003-0151-z. [DOI] [PubMed] [Google Scholar]
- 139.Dekker MC, Galjaard RJ, Snijders PJ, Heutink P, Oostra BA, van Duijn CM. Brachydactyly and short stature in a kindred with early-onset parkinsonism. Am J Med Genet A. 2004;130A(1):102–104. doi: 10.1002/ajmg.a.30021. [DOI] [PubMed] [Google Scholar]
- 140.Nagakubo D, Taira T, Kitaura H, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231(2):509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 141.Bonifati V, Rizzu P, van Baren MJ, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 142.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson's disease, confers reduced protein stability and impairs homooligomerization. J Neurochem. 2003;87(6):1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 143.Zhang L, Shimoji M, Thomas B, et al. Mitochondrial localization of the Parkinson's disease related protein DJ-1: implications for pathogenesis. Hum Mol Genet. 2005;14(14):2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 144.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann Neurol. 2003;54(3):283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 145.Canet-Aviles RM, Wilson MA, Miller DW, et al. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteinesulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci U S A. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Moore DJ, Zhang L, Troncoso J, et al. Association of DJ-1 and parkin mediated by pathogenic DJ-1 mutations and oxidative stress. Hum Mol Genet. 2005;14(1):71–84. doi: 10.1093/hmg/ddi007. [DOI] [PubMed] [Google Scholar]
- 147.Batelli S, Albani D, Rametta R, et al. DJ-1 modulates alpha-synuclein aggregation state in a cellular model of oxidative stress: relevance for Parkinson's disease and involvement of HSP70. PLoS One. 2008;3(4):e1884. doi: 10.1371/journal.pone.0001884. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 148.Mitsumoto A, Nakagawa Y. DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radic Res. 2001;35(6):885–893. doi: 10.1080/10715760100301381. [DOI] [PubMed] [Google Scholar]
- 149.Mitsumoto A, Nakagawa Y, Takeuchi A, Okawa K, Iwamatsu A, Takanezawa Y. Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radic Res. 2001;35(3):301–310. doi: 10.1080/10715760100300831. [DOI] [PubMed] [Google Scholar]
- 150.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem. 2005;280(52):43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 151.Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J Mol Biol. 2006;356(4):1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 152.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5(2):213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bandopadhyay R, Kingsbury AE, Cookson MR, et al. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson's disease. Brain. 2004;127(pt 2):420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- 154.Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312(4):1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 155.Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem Biophys Res Commun. 2004;320(2):389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 156.Kim RH, Smith PD, Aleyasin H, et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci U S A. 2005;102(14):5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Choi J, Sullards MC, Olzmann JA, et al. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006;281(16):10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Miller DW, Ahmad R, Hague S, et al. L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278(38):36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
- 159.Olzmann JA, Brown K, Wilkinson KD, et al. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J Biol Chem. 2004;279(9):8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 160.Xu J, Zhong N, Wang H, et al. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum Mol Genet. 2005;14(9):1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 161.Anderson PC, Daggett V. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson's disease. Biochemistry. 2008;47(36):9380–9393. doi: 10.1021/bi800677k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Malgieri G, Eliezer D. Structural effects of Parkinson's disease linked DJ-1 mutations. Protein Sci. 2008;17(5):855–868. doi: 10.1110/ps.073411608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Healy DG, Abou-Sleiman PM, Gibson JM, et al. PINK1 (PARK6) associated Parkinson disease in Ireland. Neurology. 2004;63(8):1486–1488. doi: 10.1212/01.wnl.0000142089.38301.8e. [DOI] [PubMed] [Google Scholar]
- 164.Valente EM, Bentivoglio AR, Dixon PH, et al. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35-p36. Am J Hum Genet. 2001;68(4):895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rohe CF, Montagna P, Breedveld G, Cortelli P, Oostra BA, Bonifati V. Homozygous PINK1 C-terminus mutation causing early-onset parkinsonism. Ann Neurol. 2004;56(3):427–431. doi: 10.1002/ana.20247. [DOI] [PubMed] [Google Scholar]
- 166.Valente EM, Salvi S, Ialongo T, et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56(3):336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 167.Bonifati V, Rohe CF, Breedveld GJ, et al. Early-onset parkinsonism associated with PINK1 mutations: frequency, genotypes, and phenotypes. Neurology. 2005;65(1):87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- 168.Li Y, Tomiyama H, Sato K, et al. Clinicogenetic study of PINK1 mutations in autosomal recessive early-onset parkinsonism. Neurology. 2005;64(11):1955–1957. doi: 10.1212/01.WNL.0000164009.36740.4E. [DOI] [PubMed] [Google Scholar]
- 169.Tan EK, Yew K, Chua E, et al. Analysis of PINK1 in Asian patients with familial parkinsonism. Clin Genet. 2005;68(5):468–470. doi: 10.1111/j.1399-0004.2005.00500.x. [DOI] [PubMed] [Google Scholar]
- 170.Tan EK, Yew K, Chua E, et al. PINK1 mutations in sporadic early-onset Parkinson's disease. Mov Disord. 2006;21(6):789–793. doi: 10.1002/mds.20810. [DOI] [PubMed] [Google Scholar]
- 171.Hatano Y, Sato K, Elibol B, et al. PARK6-linked autosomal recessive early-onset parkinsonism in Asian populations. Neurology. 2004;63(8):1482–1485. doi: 10.1212/01.wnl.0000142258.29304.fe. [DOI] [PubMed] [Google Scholar]
- 172.Brooks J, Ding J, Simon-Sanchez J, Paisan-Ruiz C, Singleton AB, Scholz SW. Parkin and PINK1 mutations in early-onset Parkinson's disease: comprehensive screening in publicly available cases and control. J Med Genet. 2009;46(6):375–381. doi: 10.1136/jmg.2008.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Khan NL, Valente EM, Bentivoglio AR, et al. Clinical and subclinical dopaminergic dysfunction in PARK6-linked parkinsonism: an 18F-dopa PET study. Ann Neurol. 2002;52(6):849–853. doi: 10.1002/ana.10417. [DOI] [PubMed] [Google Scholar]
- 174.Silvestri L, Caputo V, Bellacchio E, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14(22):3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 175.Gandhi S, Muqit MM, Stanyer L, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129(pt 7):1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 176.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 177.Petit A, Kawarai T, Paitel E, et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280(40):34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 178.Abou-Sleiman PM, Muqit MM, McDonald NQ, et al. A heterozygous effect for PINK1 mutations in Parkinson's disease? Ann Neurol. 2006;60(4):414–419. doi: 10.1002/ana.20960. [DOI] [PubMed] [Google Scholar]
- 179.Clark IE, Dodson MW, Jiang C, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 180.Ibanez P, Lesage S, Lohmann E, et al. Mutational analysis of the PINK1 gene in early-onset parkinsonism in Europe and North Africa. Brain. 2006;129(pt 3):686–694. doi: 10.1093/brain/awl005. [DOI] [PubMed] [Google Scholar]
- 181.Hatano Y, Li Y, Sato K, et al. Novel PINK1 mutations in early-onset parkinsonism. Ann Neurol. 2004;56(3):424–427. doi: 10.1002/ana.20251. [DOI] [PubMed] [Google Scholar]
- 182.Leutenegger AL, Salih MA, Ibanez P, et al. Juvenile-onset Parkinsonism as a result of the first mutation in the adenosine triphosphate orientation domain of PINK1. Arch Neurol. 2006;63(9):1257–1261. doi: 10.1001/archneur.63.9.1257. [DOI] [PubMed] [Google Scholar]
- 183.Tan EK, Skipper LM. Pathogenic mutations in Parkinson disease. Hum Mutat. 2007;28(7):641–653. doi: 10.1002/humu.20507. [DOI] [PubMed] [Google Scholar]
- 184.Najim al-Din AS, Wriekat A, Mubaidin A, Dasouki M, Hiari M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: Kufor-Rakeb syndrome. Acta Neurol Scand. 1994;89(5):347–352. doi: 10.1111/j.1600-0404.1994.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 185.Myhre R, Steinkjer S, Stormyr A, et al. Significance of the parkin and PINK1 gene in Jordanian families with incidences of young-onset and juvenile parkinsonism. BMC Neurol. 2008;8(1):47. doi: 10.1186/1471-2377-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ramirez A, Heimbach A, Grundemann J, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38(10):1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 187.Williams DR, Hadeed A, al-Din AS, Wreikat AL, Lees AJ. Kufor Rakeb disease: autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov Disord. 2005;20(10):1264–1271. doi: 10.1002/mds.20511. [DOI] [PubMed] [Google Scholar]
- 188.Di Fonzo A, Chien HF, Socal M, et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68(19):1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 189.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29(5):567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 190.Kaplan P, Andersson HC, Kacena KA, Yee JD. The clinical and demographic characteristics of nonneuronopathic Gaucher disease in 887 children at diagnosis. Arch Pediatr Adolesc Med. 2006;160(6):603–608. doi: 10.1001/archpedi.160.6.603. [DOI] [PubMed] [Google Scholar]
- 191.Wong K, Sidransky E, Verma A, et al. Neuropathology provides clues to the pathophysiology of Gaucher disease. Mol Genet Metab. 2004;82(3):192–207. doi: 10.1016/j.ymgme.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 192.Halperin A, Elstein D, Zimran A. Increased incidence of Parkinson disease among relatives of patients with Gaucher disease. Blood Cells Mol Dis. 2006;36(3):426–428. doi: 10.1016/j.bcmd.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 193.Mata IF, Samii A, Schneer SH, et al. Glucocerebrosidase gene mutations: a risk factor for Lewy body disorders. Arch Neurol. 2008;65(3):379–382. doi: 10.1001/archneurol.2007.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Clark LN, Kartsaklis LA, Wolf Gilbert R, et al. Association of glucocerebrosidase mutations with dementia with lewy bodies. Arch Neurol. 2009;66(5):578–583. doi: 10.1001/archneurol.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sawkar AR, Adamski-Werner SL, Cheng WC, et al. Gaucher disease-associated glucocerebrosidases show mutation-dependent chemical chaperoning profiles. Chem Biol. 2005;12(11):1235–1244. doi: 10.1016/j.chembiol.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 196.Velayati A, Yu WH, Sidransky E. The role of glucocerebrosidase mutations in Parkinson disease and Lewy body disorders. Curr Neurol Neurosci Rep. 2010;10(3):190–198. doi: 10.1007/s11910-010-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. Alpha-Synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275(44):34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 198.Paglini G, Peris L, Mascotti F, Quiroga S, Caceres A. Tau protein function in axonal formation. Neurochem Res. 2000;25(1):37–42. doi: 10.1023/a:1007531230651. [DOI] [PubMed] [Google Scholar]
- 199.Lee VM, Goedert M, Trojanowski JQ. Neurodegenerative tauopathies. Annu Rev Neurosci. 2001;24:1121–1159. doi: 10.1146/annurev.neuro.24.1.1121. [DOI] [PubMed] [Google Scholar]
- 200.Pittman AM, Fung HC, de Silva R. Untangling the tau gene association with neurodegenerative disorders. Hum Mol Genet. 2006;15(suppl 2):R188–R195. doi: 10.1093/hmg/ddl190. [DOI] [PubMed] [Google Scholar]
- 201.Baker M, Litvan I, Houlden H, et al. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8(4):711–715. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 202.Houlden H, Baker M, Morris HR, et al. Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology. 2001;56(12):1702–1706. doi: 10.1212/wnl.56.12.1702. [DOI] [PubMed] [Google Scholar]
- 203.Poorkaj P, Bird TD, Wijsman E, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43(6):815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 204.Zabetian CP, Hutter CM, Factor SA, et al. Association analysis of MAPT H1 haplotype and subhaplotypes in Parkinson's disease. Ann Neurol. 2007;62(2):137–144. doi: 10.1002/ana.21157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 206.Pittman AM, Myers AJ, Abou-Sleiman P, et al. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet. 2005;42(11):837–846. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.LeBoeuf AC, Levy SF, Gaylord M, et al. FTDP-17 mutations in Tau alter the regulation of microtubule dynamics: an “alternative core” model for normal and pathological Tau action. J Biol Chem. 2008;283(52):36406–36415. doi: 10.1074/jbc.M803519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shimizu F, Katagiri T, Suzuki M, et al. Cloning and chromosome assignment to 1q32 of a human cDNA (RAB7L1) encoding a small GTP-binding protein, a member of the RAS superfamily. Cytogenet Cell Genet. 1997;77(3-4):261–263. doi: 10.1159/000134591. [DOI] [PubMed] [Google Scholar]
- 209.Ostvold AC, Norum JH, Mathiesen S, Wanvik B, Sefland I, Grundt K. Molecular cloning of a mammalian nuclear phosphoprotein NUCKS, which serves as a substrate for Cdk1 in vivo. Eur J Biochem. 2001;268(8):2430–2440. doi: 10.1046/j.1432-1327.2001.02120.x. [DOI] [PubMed] [Google Scholar]
- 210.Ziolkowski P, Gamian E, Osiecka B, Zougman A, Wisniewski JR. Immunohistochemical and proteomic evaluation of nuclear ubiquitous casein and cyclin-dependent kinases substrate in invasive ductal carcinoma of the breast. J Biomed Biotechnol. 2009;2009:919645. doi: 10.1155/2009/919645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Grunblatt E, Mandel S, Jacob-Hirsch J, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111(12):1543–1573. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 212.Orr CF, Rowe DB, Mizuno Y, Mori H, Halliday GM. A possible role for humoral immunity in the pathogenesis of Parkinson's disease. Brain. 2005;128(pt 11):2665–2674. doi: 10.1093/brain/awh625. [DOI] [PubMed] [Google Scholar]
- 213.McGeer PL, McGeer EG. Glial reactions in Parkinson's disease. Mov Disord. 2008;23(4):474–483. doi: 10.1002/mds.21751. [DOI] [PubMed] [Google Scholar]