Summary
The outermost layer of the Bacillus anthracis spore consists of an exosporium comprised of two distinct layers, an outer hair-like nap layer and an internal basal layer. The hair-like nap is primarily comprised of the glycosylated collagen-like protein BclA. BclA is found in a trimeric form in close association with many other exosporium proteins in high MW complexes. We previously had characterized an N-terminal sequence of BclA that is sufficient for incorporation into the exosporium. Here we utilized site-directed mutagenesis to identify BclA residues critical to two steps in this process, positioning of the protein at the site of the developing exosporium basal layer and stable incorporation which includes a proteolytic cleavage of BclA after residue 19. The BxpB (ExsFA) protein is known to be important for proper incorporation of BclA onto the exposporium. BxpB and BclA were found to be expressed at the same time in sporulating cells of B. anthracis and immediately co-localize to high molecular weight complexes. The BxpB protein was found to be in close proximity to the BclA-NTD. BxpB and BclA are co-dependent for exosporium incorporation, with the BclA NTD being sufficient to deliver BxpB to the exosporium.
Introduction
The genus Bacillus is comprised of Gram-positive endospore-forming species that can shift to an alternative developmental pathway, sporulation, when growth conditions become unfavorable (Henriques and Moran, 2007). B. anthracis, B. cereus, and B. thuringiensis belong to a subset of Bacillus that has evolved to become occasional pathogens of mammals and insects. Spores from B. anthracis in particular are the infectious particles which initiate infection upon entry into the host organism. As such, a better understanding of the interaction between these spores and their hosts is of paramount importance.
Spores of the Bacillus cereus family (B. anthracis, B. cereus, and B. thuringiensis) diverge from the spore architecture of the well-characterized B. subtilis in that they contain an additional outer structure known as the exosporium. The exosporium is comprised of two distinct sublayers, the basal layer and the outer hair-like nap layer (Hachisuka et al., 1966; Kramer and Roth, 1968; Driks, 2002). Separating the exosporium from the rest of the spore structures is an area deemed the interspace region of the spore (Giorno et al., 2009).
Many exosporium proteins of B. anthracis and B. cereus have been identified (Charlton et al., 1999; Lai et al., 2003; Todd et al., 2003; Redmond et al., 2004). The basal layer associated proteins include ExsY, CotY, ExsFA/BxpB [hereafter designated BxpB], and ExsFB, (Steichen et al., 2003; Redmond et al., 2004; Swiecki et al., 2006, Steichen et al., 2005; Sylvestre et al., 2005; Boydston et al., 2006). An exsY mutant resulted in a highly destabilized exosporium with fragments released from the spores (Boydston et al., 2006; Johnson et al., 2006). CotY, a homolog of ExsY, plays a role in exosporium formation in B. cereus (Johnson et al., 2006). Other proteins, including ExsY, CotY, BxpA, ExsK, and BclB have also been found in high molecular weight complexes often associated with the exosporium of Bacillus spp. (Steichen et al., 2003; Redmond et al., 2004; Swiecki et al., 2006, Steichen et al., 2005; Boydston et al., 2006; Severson et al., 2009).
Filaments of the outer “hair-like” nap layer are composed primarily of the collagen-like protein BclA (Sylvestre et al., 2002; Steichen et al., 2003; Sylvestre et al., 2003). BclA forms a trimeric complex and is glycosylated at its internal collagen-like repeat sequences (Sylvestre et al., 2003; Daubenspeck et al., 2004). A second collagen-like protein, BclB, is also stably incorporated into the exosporium of B. anthracis (Waller et al., 2005; Thompson et al., 2007). Both BclA and BclB are glycosylated with O-methyl rhamnose, rhamnose, and galactosamine moieties (Daubenspeck et al., 2004; Waller et al., 2005). BclA and BxpB both play a partial role in the incorporation of ExsK into the outer spore layers (Severson et al., 2009).
The components of the exosporium are produced in the mother cell and are subsequently assembled on the developing spore. Most of the exosporium determinants have promoter elements specific for the σK-containing late stage sporulation RNA polymerase (Hilbert and Piggot, 2004; Henriques and Moran, 2007). The assembly of these late stage proteins is dependent on a number of earlier produced proteins that permit the proper attachment of the exosporium to the spore coat layers. One of these, CotE, is crucial for attachment of the exosporium. A lack of CotE resulted in production of spores with released and partially attached exosporium “strips” (Giorno et al., 2007).
BclA is the outermost known component of the exosporium, and as such plays a role in the early interaction between spores and macrophages/dendritic cells in the host (Bozue et al., 2007; Oliva et al., 2008). In addition, loss of BclA leads to an increase in overall spore hydrophilic properties and changes the spore’s interaction with host tissue matrix substances (Brahmbhatt et al., 2007). Late in sporulation, BclA incorporation begins at the central pole of the spore and gradually proceeds across the spore until it is fully encompassed (Ohye and Murrell 1973; Thompson and Stewart, 2008; Giorno et al., 2009). When BclA is positioned at the exosporium, a progressive cleavage and release of its N-terminal 19 amino acids occurs resulting in its stable incorporation into the exosporium (Sylvestre et al., 2002; Thompson and Stewart, 2008). The localization of BclA to the exosporium layer is dependent on a motif(s) between amino acids 25–35 (LVGPTLPPIPP, Thompson and Stewart, 2008; Tan et al., 2010). Removal of this region results in loss of BclA localization around the developing spore and a diminished level of BclA incorporated into the exosporium. The specific role of amino acids 20–25 is still debated, but removal of these 6 residues results in localization of BclA to the exosporium but without its stable attachment (Thompson and Stewart, 2008; Tan et al., 2010).
BxpB has been shown to be important in interactions between the basal layer and the hair-like nap protein BclA (Steichen et al., 2005; Sylvestre et al., 2005). Absence of BxpB results in a dramatic decrease in the levels of BclA and either partial or total loss of the hair-like nap of the exosporium (Sylvestre et al., 2005; Steichen et al., 2005; Giorno et al., 2009). The ExsFB protein has been shown to play a minor role in hair-like nap development, and an ExsFB-GFP fusion has a unique ovoid pattern of localization in the developing spore (Sylvestre et al., 2005; Giorno et al., 2009). These studies demonstrate that both BxpB and possibly ExsFB influence the assembly of BclA into the hair-like nap of the exosporium of B. anthracis.
Herein we describe the interaction between BclA and BxpB in detail, determine if this interaction is direct or indirect, and suggest a mechanism by which ExsFB may be involved in BclA assembly.
Results
BxpB and BclA are co-expressed and co-localize to the developing exosporium
We previously demonstrated the assembly of BclA from its expression, its localization to the developing exosporium layer by means of a “localization motif,” and its eventual incorporation into the exosporium and associated cleavage at the 19th residue (Thompson and Stewart, 2008). Since BxpB appears to be required for proper assembly of BclA, it is likely that BxpB assembly precedes, or coincides with, that of BclA (Sylvestre et al., 2005; Steichen et al., 2005). To compare the expression kinetics of BxpB and BclA, we created C-terminal fusions of BxpB to the monomeric fluorescence protein mCherry and BclA to eGFP and then followed progression of these fusion proteins in synchronous sporulation cultures (Shaner et al., 2005). Fusions were driven by their endogenous promoter, RBS, and start codons and introduced into B. anthracis via the compatible Gram-positive shuttle vectors pMK4 (Sullivan et al. 1984) and pHPS (this study).
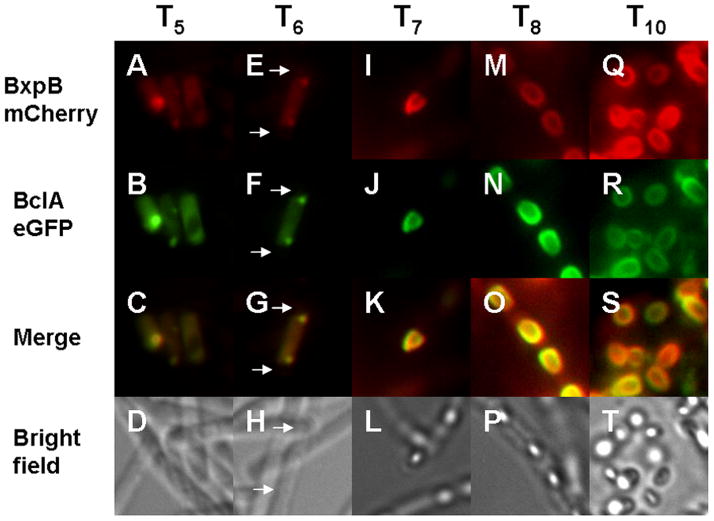
The BxpB fusion is initially expressed at low levels in the mother cell cytoplasm beginning between T4 and T5 of sporulation (Fig. 1A). This coincided with the appearance of the BclA-eGFP fusion in the mother cell cytoplasm (Fig. 1A-C). At T6, both BxpB and BclA fusions co-localized to the centrally-located pole of the developing spore (Fig. 1E-G). This is the point from which the exosporium layer initiates its assembly (Ohye and Murrell, 1973; Giorno et al. 2007; Thompson and Stewart, 2008). The BxpB fluorescence seen at the central pole of the spore was weak relative to the levels observed with the BclA fusion (Fig. 1E-G). From T7-T8, both BxpB and BclA concentrated at the spore surface and spread across the spore surface to eventually envelop the spore (Fig. 1I-P). After T8, spores were released from their mother cells and free spores were surrounded with both BxpB and BclA fusion protein fluorescence (Fig. 1Q-T). The localization and assembly patterns of the BxpB fusion were identical when imaged independently of the BclA fusion (data not shown). Additionally, expression and assembly of BclA fusions were identical to that reported previously (Thompson and Stewart, 2008). The merging of BclA and BxpB fluorescent micrographs fully demonstrated the concurrent expression, localization, and assembly of these two proteins (Fig. 1C, G, K, O, and S). Because BclA and BxpB are assembled in close spatial and temporal proximity to each other, they could assemble either as a complex or by distinct mechanisms that occur concurrently.
Fig. 1. Time course micrographs of synchronous cultures expressing BxpB mCherry fusions and BclA eGFP fusions.
Time points are designated above the corresponding micrographs. Arrows denote the site of developing spores. Note the almost exact overlap of BxpB and BclA fluorescence over time as they localize to the developing exosporium (mother cell pole of the spore, T6) and continue to co-envelope the spore from T7-T8. Free spores express high levels of both BclA-eGFP and BxpB-mCherry fluorescence (T10). BxpB-mCherry fluorescence (panels A, E, I, M, Q); BclA-eGFP fluorescence (B, F, J, N, R); merged fluorescent images (C, G, K, O, and S); and bright field images of the sporulating cells and free spores (D, H, L, P, T).
BxpB and the BclA N-terminus fusions are sufficiently close together to allow FRET
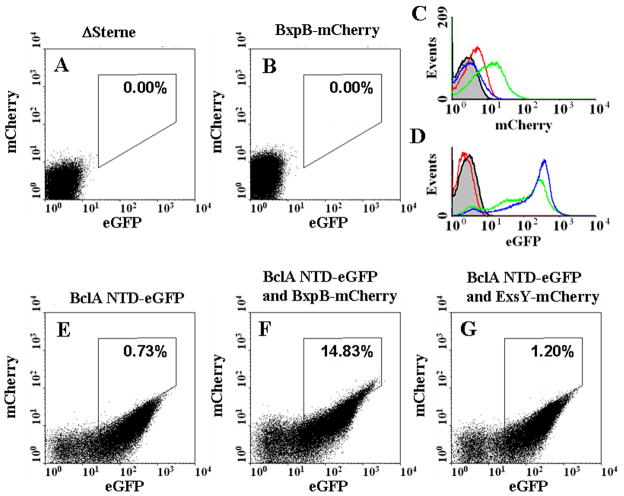
To determine if BclA and BxpB are assembled in close proximity to each other, we performed flow cytometric analysis of spores containing A) no fusions, B) a BxpB-mCherry fusion, C) the BclA N-terminal 35 amino acids fused to eGFP, and D) dual expression of both fusions. The mCherry and eGFP reporters are efficient Forster Resonance Energy Transfer (FRET) partners and their spectral overlap make observations of FRET easy to demonstrate (Albertazzi et al., 2009). As shown in Fig. 2B-C, the fluorescence of mCherry was weak upon excitement with the suboptimal 488 nm laser of the flow cytometer (only 8% of maximum emission possible). When the BxpB mCherry fusion was analyzed in the presence of a BclA NTD-eGFP fusion, the fluorescence of BxpB mCherry fluorophore increased 2.3-fold (Fig. 2C and F). The increased BxpB-mCherry signal obtained in the presence of the BclA NTD-eGFP fusion resulted in a decreased eGFP signal (2.1-fold lower), further demonstrating the passage of photons from the eGFP to the mCherry fluorophores (Fig. 2D). Appearance of FRET was also observed by shifting of events into the FRET channel, denoted by the boxes in Fig. 2A-G. The percentage of FRET positive spores is illustrated in the upper right hand corner of each dot plot. In comparison, a lack of intense FRET activity was seen when spores containing both a fusion to the known exosporium protein ExsY (C-terminal mCherry) and the BclA NTD 35 amino acid eGFP fusion were co-expressed (Fig. 2G). FRET was not observed to any significant extent despite the high expression of the ExsY-mCherry fusion (data not shown). Additionally, no FRET was observed between full-length BclA-eGFP fusions and the BxpB-mCherry fusions which place the eGFP reporter at the C-terminus of the ~70 kDa filamentous BclA protein and away from the mCherry reporter of BxpB (data not shown). Taken together, this demonstrated that mCherry of BxpB and the eGFP on the BclA N-terminal 35 amino acids are within FRET distance (<10 nm) of each other. The percentage (14.8%) of FRET positive spores would likely have been even greater if not for the nonfluorescently tagged native BclA and BxpB proteins competing with the fusions for binding partners.
Fig. 2. Flow Cytometry of spores expressing BxpB and BclA fusions.
Comparisons between overall eGFP fluorescence (X axis) and overall mCherry fluorescence (Y axis) are shown. (A, B, E, F and G) A) ΔSterne containing no fusion reporter; B) BxpB-mCherry fusion spores alone; E) BclA NTD-eGFP fusion alone, F) BclA NTD-eGFP and BxpB-mCherry fusions co-expressed, and G) BclA NTD-eGFP and an ExsY-mCherry fusion co-expressed. The box in each panel denotes the percentage of FRET positive cells. Note the FRET positive cells observed in panel F but not present in panel G. (C and D) C) Total mCherry fluorescence of spores of ΔSterne (gray area), BclA NTD-eGFP (bleed through, red line), BxpB-mCherry alone (blue), and a dual BclA NTD-eGFP and BxpB-mCherry demonstrating enhanced fluorescence (green line). D) Identical samples in the eGFP channel, with BxpB-mCherry as the bleed through channel.
BclA and BxpB co-assemble into complexes
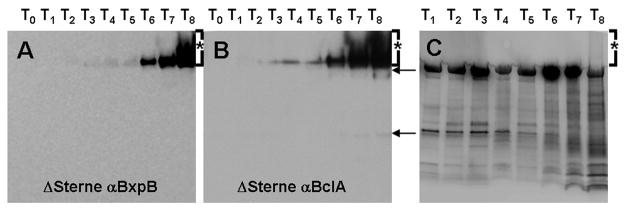
Coincident patterns of expression, localization, and assembly and the appearance of FRET between the N-terminal 35 amino acids of BclA and BxpB suggested a close association between the two proteins. We further examined their patterns of assembly via western blot analysis of time-matched wild-type sporulating cells. Initial, albeit weak expression of both BclA and BxpB appeared in sporulating cell extracts at T3 (Fig. 3A and B). Interestingly, BclA was never observed as a ~70 kDa unglycosylated monomer (the position this protein migrates on SDS PAGE gels). at early time points, suggesting the trimerization of BclA and/or possibly glycosylation of BclA are immediate upon expression of the protein in the sporulating cell (Fig. 3B, T3-T5). Additionally, BxpB did not appear as a low MW species, suggesting it too was immediately placed into a large MW complex upon its expression (Fig. 3A). A small amount of monomeric BxpB was previously reported to be present in extracts of mature spores (Sylvestre et al., 2005; Steichen et al., 2005). BxpB appeared in greater quantities as a >250 kDa complex from T6-T8. These same complexes were identified when the blot was stripped and re-probed for BclA (T6-T8, Fig. 3A-B). T6-T8 corresponded to the time points at which BclA and BxpB fusions co-localized to the central pole of the spore and assembled around the developing spore (Fig. 1). At T8, appearance of a high MW heterogeneous species containing both BxpB and BclA with increased levels of glycosylation was apparent (Fig. 3A-C). This glycosylated complex can be detected as a high molecular weight smear at the same time point in a corresponding glycoprotein-stained SDS-PAGE gel containing identical samples taken from the same sporulation time points (Fig. 3C, asterisk). Glycosylation of BxpB and BclA complexes prior to T8 could not be determined due to the interfering presence of a yet unknown glycosylated protein complex of a similar MW found between T1-T8 (Fig. 3C).
Fig. 3. Western blots and glycoprotein stained synchronous sporulating culture samples.
Samples from synchronous cultures were taken, lysed, and resolved on 4–20% Tris-glycine gels. Identical immunoblots blotted with A) polyclonal anti-BxpB antibodies and B) polyclonal anti-BclA antibodies. Time points correspond to those of Fig. 1. The positions of a minor BclA complex (upper arrow) and monomeric BclA (lower arrow) are indicated. C) Glycoprotein stain of identical samples as A and B. Although a unknown glycoprotein complex is found at the same molecular weight as the initial BclA and BxpB complexes found in A and B, at T8 a smear of glycosylation is present ( >250 kDa) that corresponds to the increase in size of both BclA and BxpB complexes at that time point (asterisk).
BclA and BxpB are co-dependent for their assembly into the exosporium
It was reported that bxpB mutants do not contain substantial amounts of BclA and are deficient in the hair-like nap layer (Sylvestre et al., 2005; Steichen et al., 2005). To examine this phenomenon in detail, a BclA-eGFP fusion was expressed in a bxpB-null mutant background. At T8, BclA-eGFP fluorescence levels were equivalent to that observed in a wild-type host (Fig. 4A-D). However, there was no visible localization or assembly of BclA fusions onto the exosporium in the bxpB mutant (Fig. 4A and B). Examination of free spores (T12) demonstrated the overall lack of BclA incorporation in the bxpB mutant (Fig. 4E-F). Giorno et al. (2009) noted the appearance of BclA solely in the “cap” of developing spores in a bxpB mutant background. In accordance, with prolonged exposure times a small, localized concentration of BclA fusion at one pole of the spores was observed (Fig. 4E-F insets). By comparison, expression of BclA in a wild-type background led to efficient labeling of the entire surface of the released spore (Fig. 4G-H). The overall mean fluorescence of BclA-eGFP fusion-bearing spores in the bxpB mutant background was determined by flow cytometry, and was found to be only 1% of wild-type levels (Fig. 4M). Additionally, localization and incorporation of a BclA NTD 35 amino acid-eGFP fusion was also greatly diminished in a bxpB mutant background (2.1% of wild-type levels). This strongly suggests that BclA assembly and incorporation, through its N-terminal motifs, is dependent upon BxpB for localization and subsequent assembly into the exosporium.
Fig. 4. Micrographs and flow cytometry of BclA and BxpB fusions in mutant backgrounds.
Panels A-D correspond to the T8 time point and panels E-H correspond to free spores (T12). BclA-eGFP immunofluorescence in a bxpB-null mutant background (A and B) and in a wild-type background (C and D). BxpB-mCherry fluorescent images are panels I and K. Brightfield images of sporulating cells (B and D) and free spores (F, H, J, and L). Mean fluorescence of fusion-bearing spores were analyzed by flow cytometry and results for BxpB-mCherry fusions were: BxpB mCherry in ΔSterne, 1.79 (PMF) and BxpB mCherry in the BclA-null mutant, 0.41 (PMF). M) Flow cytometric analysis of ΔSterne (gray area), BclA-eGFP in ΔSterne (green) and BclA NTD-eGFP inΔSterne (black) in comparison to BclA-eGFP (blue) and BclA NTD-eGFP (purple) in the bxpB mutant background. Exposure time for all micrographs A-L – 500 ms, Inset in E – 1800 ms
Because BclA and BxpB assemble into a complex prior to incorporation into the exosporium of B. anthracis, we examined BxpB’s dependency upon BclA for its localization and incorporation. Abundant localization and attachment of BxpB fusions occurred in a wild-type background, enveloping the spores in fluorescence with the exception of one pole (Fig. 4I and J, arrows). Analysis of free spores in a bclA-null mutant background indicated a decrease in overall BxpB fluorescence (Fig. 4K and L). BclA negative spores exhibited only 23.2% of the BxpB-mCherry fluorescence when compared to ΔSterne parent spores, as determined by flow cytometry measurement of mean fluorescence. BxpB can localize and assemble, albeit somewhat less efficiently, in the absence of BclA, but BclA is essential for incorporation of BxpB in spores above a low basal level.
Increased surface exposure was not representative of total protein levels
A previous study suggested increased BxpB protein levels in spores of a bclA null mutant as evidenced by surface antibody labeling of spores during TEM (Steichen et al., 2005). To correlate the increased BxpB antibody labeling in a bclA mutant background with total protein levels in spores, surface exposure of BclA was measured via immunofluorescence microscopy of purified spores with polyclonal rabbit anti-BclA antibodies followed by goat anti-rabbit Alexa Fluor 568 secondary antibodies. BclA antibody reactive levels were visualized throughout the surface of wild-type spores, and were also found on bxpB null spores at decreased levels (23.1% of wild-type levels as assayed by flow cytometry, Fig. 5A-D). The reduced BclA staining on the spore surface was still greater than the relative level of fluorescence observed with the BclA-eGFP fusions (1% of wild-type levels). This finding suggests that the addition of eGFP to the C-terminus of BclA may reduce its competitiveness with wild-type BclA for incorporation onto the spore surface in the absence of BxpB. Since the fusion reporter analysis does not detect this wild-type protein, lower levels of BclA are measured. Alternatively, the BclA antibody reactive signal does not always correlate directly with overall protein levels, only to the number of epitopes exposed to antibody.
Fig. 5. Surface-exposed and total protein levels of BclA and BxpB.
In each fluorescent micrograph, the normalized mean fluorescence (fluorescence minus secondary control fluorescence) of each sample obtained via flow cytometric analysis is indicated in the bottom right hand corner. The surface-exposure levels of BclA assayed via immunolabeling with polyclonal anti-BclA antibody was performed in ΔSterne (A and B), the bxpB-null mutant (C and D), and the bclA-null mutant (E and F). Surface exposure levels of BxpB were determined by immunolabeling with polyclonal anti-BxpB antibodies in ΔSterne (G and H), bxpB mutant (I and J), and bclA mutant (K and L). The restoration of BxpB levels in a bclA mutant background can be observed by a comparison of bclA mutant spores (K and L), bclA mutant spores expressing the NTD of BclA-eGFP (P and Q) and bclA mutant spores with full-length BclA-eGFP (R and S) fusions. M, N, and O) Western blot analysis of extracts from 5×107 spores with anti-BclA (M), anti-BxpB (N), or anti-GFP antibodies (O). The filter was sequentially probed with one of the antisera and then stripped for re-probing with the next antiserum. Lane 1) ΔSterne, lane 2) bclA null mutant, 3) bclA null mutant with BclA-eGFP fusion, 4) bclA null mutant with BclA NTD-eGFP fusion, 5) bxpB null mutant 6) bxpB null mutant with BxpB-mCherry fusion, 7) ΔSterne with BclA NTD-eGFP fusion.
We then examined levels of surface exposed BxpB by immunofluorescence microscopy using rabbit anti-BxpB polyclonal antibodies. BxpB was readily detected on wild-type ΔSterne spores, but antibody reactive levels decreased in a bclA mutant background (20.7% of wild-type levels, Fig. 5G-H and K-L). This decreased labeling of BxpB in the bclA mutant background differs from the TEM results reported by Steichen et al. (2005) in which BxpB was more readily detected by immunogold labeling with the BclA-negative spores. Differences in antigen accessibility with sectioned spores (by Steichen et al.) versus intact spores (immunofluorescence microscopy) and loss of antigenicity that would accompany sample preparation for electron microscopy likely account for the different results obtained. The heterogenic surface antibody labeling of free spores seen with both the anti-BclA and anti-BxpB antibodies is likely due to differences in nap density along the spore surface resulting in different accessibility of antibodies to their epitopes, as we described previously (Thompson et al. 2007).
We next examined total protein levels to determine overall concentrations of extractable BclA in the various spore types. As expected, the bxpB null spores exhibited decreased overall BclA levels and no BxpB reactivity when compared to wild-type spores (Fig. 5M and N, lanes 1 and 5; Steichen et al., 2005). The presence of the BxpB-mCherry fusion in the bxpB null mutant was found to increase BclA levels in the spores, but did not fully complement the mutant phenotype as shown by modest levels of BclA protein extracted from spores in the presence of the spore-associated BxpB-mCherry (Fig. 5M and N, lane 6). BclA incorporation into these spores was approximately 3-fold higher than that observed with bxpB-null spores as observed by western blot, immunofluorescence, and with BclA-eGFP fusions (Supplemental Fig. 1).
We observed a pronounced decline in BxpB-extractable material in the bclA-null mutant background when compared to wild-type levels, again suggesting the co-dependency of BclA and BxpB incorporation (Fig. 5M and N, lane 2). Less extractable BxpB was obtained than would be expected based on the immunofluorescence signal (Fig. 5K, 5N lane 2). The loss of the BclA-nap layer in these spores may have provided better access of the anti-BxpB antibodies to the BxpB epitopes giving a higher signal than would be expected based upon the actual overall protein levels.
Interactions of BxpB and the N-terminus of BclA
The introduction of the BclA NTD 35 amino acid fusion in the bclA-null mutant background greatly enhanced the surface exposed levels of BxpB (2.9-fold higher than wild-type, 14.2-fold higher than bclA-null spores by immunofluorescence labeling followed by flow cytometry, Fig. 5P-Q). This indicated the N-terminal domain of BclA alone is sufficient for fulfilling the BclA requirement to permit BxpB incorporation into the high molecular weight complexes and ultimately into the exosporium. Introduction of the full length BclA fusion in the bclA null mutant background also increased the BxpB surface exposed levels, but at a diminished rate compared to the BclA N-terminal complementation, likely due to blocking of BxpB epitopes by the restored BclA naplayer (83.0% of wild-type by flow cytometric analysis, Fig. 5R-S). This demonstrates that the BclA-eGFP and BclA NTD-eGFP fusions are functional and can complement the bclA defect in this assay.
The presence of just the first 35 amino acids of BclA, which contain the localization and attachment domains, was also sufficient to restore BxpB protein levels in bclA null spores as evident from the western blot analysis (Fig. 5N lanes 2 and 4), although the levels were lower than that observed with full length BclA-eGFP (Fig. 5N, lane 3). The N-terminal 35 amino acid domain of BclA is cleaved to 16 residues when incorporated into the exosporium and this short sequence reacts poorly with polyclonal BclA antibodies. This explains the lack of appreciable signal in the anti-BclA probed sample (Fig. 5M, lanes 4 and 7) and the presence of the 30 kDa BclA NTD-eGFP species identified with anti-GFP antibodies (Fig. 5O, lane 7). Addition of the full-length BclA-eGFP fusion into a bclA mutant background also restored BxpB reactivity, found in several protein complex species (Fig. 5N, lane 3).
Extracts from wild-type spores containing the BclA NTD 35 amino acid-eGFP fusion contained complexes between BxpB and the N-terminal domain of BclA (220, 130 kDa, 85 kDa, and 55 kDa, Fig. 5N and O, lane 7, each position denoted by an asterisk). These complexes contain BxpB molecules in association with the BclA NTD-eGFP fusion molecules as evidenced by reactivity to both anti-BxpB and anti-eGFP sera.
Lysed protein samples of sporulating ΔSterne cells at T6 containing the 35 N-terminal BclA residues fused to eGFP were taken and subjected to immunoprecipitation using anti-GFP polyclonal antibodies. T6 corresponds to the time point where BclA and BxpB are initiating their co-localization to the exosporium layer in the mother cell. The immunoprecipitated proteins were resolved by SDS-PAGE and subjected to immunoblotting with anti-BxpB, anti-BclA, and anti-GFP antibodies (Fig. 6A-C.) A 55 kDa band consistent with the expected size of a complex of the BclA NTD-eGFP fusion and BxpB was present in each of the three blots. Because BxpB does not interact with eGFP (data not shown), the complex must form between BxpB and the N-terminus of BclA. In addition, there were larger molecular weight species containing BxpB (Figure 6A) suggesting that BxpB either self-associates or binds to other exosporium proteins when in complex with the BclA N-terminal domain. No BclA full length or larger species were identified with the anti-BclA serum, suggesting that the BclA-NTD fusion protein does not associate with the wild-type BclA protein. The likeliest explanation for this observation is that the BclA NTD species can interact with BxpB via the NTD, but because the NTD protein cannot trimerize (owing to a lack of the collagen-like repeat domain), it does not capture wild-type BclA in the complexes immunoprecipitated with the anti-GFP serum.
Fig. 6. Western blots from pull down assays on mid-stage sporulating extracts.
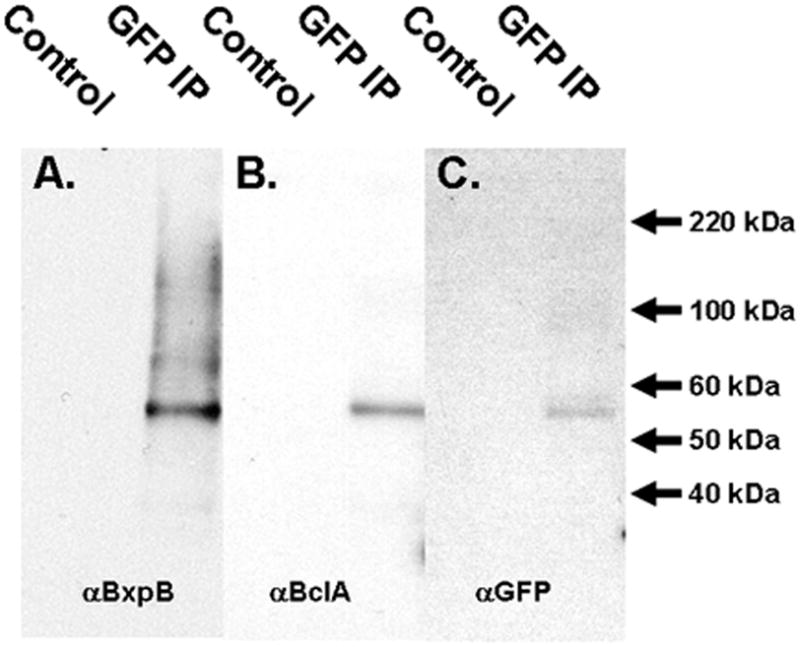
A) Mid-stage (T6)sporulating extracts of ΔSterne cells expressing the BclA NTD-eGFP fusion were immunoprecipitated with anti-GFP antibodies (GFP IP) or with control pre-immune serum (Control). Immunoprecipitated proteins were subjected to SDS-PAGE and immobilized on filters. The filters were probed for the presence of BxpB (panel A), BclA (B), and GFP (C). The exposure time for panel B was extended to compensate for the relatively weak reactivity by the anti-BclA antiserum with the BclA NTD.
Role of BclA residues 17–35 in the assembly of BclA and BxpB
Because BxpB associates with the localization and attachment domain on the N-terminus of BclA in the BclA NTD-eGFP fusion protein, we systematically replaced each residue of BclA amino acids 17–35 to alanine (and alanines to serines) and then examined the impact of each residue change in the BclA-BxpB interaction. Fluorescence of spores expressing the fusion constructs was quantified via flow cytometry. An example of a representative experiment demonstrating the quantification of the native BclA fusion (control) and the P35A and L30A mutation fusions is shown in Fig. 7A. Quantification of fusion spores was done in triplicate with spores from three independent clones of each fusion type.
Fig. 7. The role of individual residues in the N-terminus of BclA in localization to the spore, assembly, and binding to BxpB.
A) Flow cytometry results from spores expressing representative BclA NTD fusions to DsRed. Gray area denotes control spores, black line denotes control unaltered NTD, blue line denotes results with the P35A mutant, and green line denotes results with the L30A mutant. B) Quantitative results of mean fluorescence of three independent clones of each mutation in triplicate via flow cytometry. Residues mutated are listed on the X-axis. C) Quantitative results of mean fluorescence of various mutations in a bxpB mutant background and bclA mutant background. D) Model of the N-terminal 35 amino acids of BclA with the attachment and localization motifs identified in Thompson and Stewart (2008). The arrow denotes the cleavage site in BclA after the 19th residue. The critical residues for BclA localization and attachment to the exosporium are denoted in purple. E) Micrographs of late stage sporulating cells and free spores of select mutations. The top two rows are fluorescent images and the bottom row is consists of bright field images. (* p-value <0.5, *** p-value <0.001; students paired t test).
The most significant losses in fluorescence on spores resulted from the S19A, D22A, L25A, G27A, P28A, T29A, L30A, and P31A substitutions (Fig. 7B). These residues were crucial for the proper incorporation of BclA onto the spore surface. Critical residue mutations were also analyzed in a bclA-null mutant background, to eliminate any competition between the N-terminal fusions and wild-type BclA molecules. Although the wild-type NTD control fusion in a bclA null mutant background only exhibited ~50% of the fluorescence found in wild-type spores, fusions expressing the alanine mutants in this bclA null mutant background were not as severely affected as those expressed in wild-type spores, likely due to the loss of competition from the full length wild-type BclA (Fig. 7C). However, the overall patterns of relative efficiency of BclA NTD-fusion incorporation into the spore were the same in both the bclA-positive and bclA-null host backgrounds.
BclA incorporation into the exosporium occurs in two distinct steps, the first being positioning of BclA at the spore surface site of the developing exosporium and the second step is the stable incorporation of BclA which is associated with a cleavage event removing the N-terminal 19 amino acids. We previously demonstrated two distinct motifs in the N-terminal 35 amino acids of BclA (Thompson and Stewart, 2008). There is an 11 amino acid region (residues 25–35) that positions BclA to the developing exosporium preceded by a 5–7 amino acid sequence that is critical for stable attachment of BclA into the exosporium (Fig. 7D). To determine if alanine-replacement fusions exhibited the same defects (positioning around the developing spore and stable incorporation), we imaged late stage sporulating (T8) cells of the severely affected fusion types and resultant free spores. The S19A and D22A mutations, affecting residues in the attachment domain, permitted the proper positioning of BclA fusions at the exosporium layer (Fig. 7E). However, the resultant spores were not highly fluorescent, indicative of poor attachment of the fusion protein (Fig. 7E). Mutations of residues L25A and G27A, within the localization domain, resulted in reduced localization of the fusions to the exosporium as well as poor attachment of the BclA fusions (Fig. 7E). Mutations in the latter part of the 35 amino acid N-terminus of BclA, such as P28A and P31A, resulted in no observable localization of the BclA fusions to the exosporium (fluorescence remains dispersed in the cytoplasm and the developing spore remains dark) and very little attachment to the released spores as a consequence of the poor positioning. This mutation analysis supported the identity of the two distinct motifs, one for the positioning of BclA at the exosporium (residues 25–35) and one for the proper attachment of BclA (upstream of the positioning domain and including the proteolytic cleavage site between residues 19 and 20). Both of these domains are crucial for proper BclA assembly into the exosporium.
Expression of the mutated BclA NTD-fusion constructs in a bxpB null mutant background resulted in limited detectable fluorescence on the spores. The wild-type and P35A results are presented in Fig. 7C.
The role of ExsFB in the incorporation of BclA
ExsFB shares substantial sequence similarity with ExsFA/BxpB and likely plays a role in BclA incorporation (Sylvestre et al., 2005). Therefore we constructed an ExsFB-mCherry fusion expressed under the native exsFB promoter elements and monitored it in sporulating cultures of wild-type, bxpB-null, and bclA-null strains of B. anthracis. During late stage sporulation (T7), ExsFB fusions concentrate at the mother cell-central pole of the spore (Fig. 8A-C, arrowheads). This likely represents the presence of ExsFB in the cap region of the exosporium (Steichen et al., 2007). In the bclA mutant, the ExsFB localization to the spore is aberrant with an increased polar presence (Fig. 8C). When released, wild-type spores expressing the ExsFB-mCherry fusion were imaged and only a weak incorporation of the fusion was observed (Fig. 8D and G). ExsFB-mCherry incorporation in free spores was polar with the majority of the fluorescence at the cap. In contrast to wild-type spores, spores of the bxpB null mutant (Fig. 8E and H) and the bclA null mutant (Fig. 8F and I) displayed a dramatic increase in the amount of ExsFB fusion incorporated, especially in the bxpB mutant background. When this difference in ExsFB-mCherry incorporation was quantified by flow cytometry a 3.4-fold increase was found in a bclA mutant background and a 9.4-fold increase in a bxpB mutant background. The overall polar distribution of ExsFB remained in these mutant backgrounds. To further examine the kinetics of ExsFB expression, cells containing both the ExsFB-mCherry fusion and a full-length BclA-eGFP fusion were allowed to sporulate in synchronous culture. Examination of cultures at T7 indicated that both ExsFB and BclA were incorporated into the exosporium. However, in contrast with BxpB, the ExsFB fusions did not completely co-localize with BclA fusions (Fig. 8J-M). The majority of ExsFB appeared to localize to the two poles of the spore, with a primary concentration at the cap (arrowheads).
Fig. 8. A role for ExsFB in the assembly of BclA.
A-C) Micrographs of T7 cultures of wild-type ΔSterne (A), the bxpB-null mutant (B), and the bclA-null mutant (C). Arrowheads denote cap localization pattern at the mother cell central pole of the spore. D-I) Free spores of ΔSterne (D and G), the bclA mutant (E and H), and the bxpB mutant (F and I) expressing the ExsFB-mCherry fusions. A-F and J-L are fluorescence images and G-I and M are bright field images. Arrows denote the concentration of ExsFB fusions at the mother cell-central pole of the spore in panels D-F. J-M) Micrographs of the ExsFB-mCherry fusion and the BclA-eGFP fusion co-expressed in T7 sporulating cells. J) ExsFB-mCherry, K) BclA-eGFP, L) merged image of J and K, M) bright field image. Arrowheads denote polar concentration of ExsFB at the mother cell central pole. N) Western blot analysis of extracts from ExsFB-mCherry fusion containing spores from a wild-type (lane 1), bxpB mutant (lane 2), and bclA mutant (lane 3) host strains. O) Expression of BxpB over time in synchronous sporulating cultures via western blot. P) Identification of ExsFB-mCherry with anti-GFP antibodies in the same samples as panel O.
The presence of ExsFB-mCherry in extracts from wild-type spores, bclA mutant spores, and bxpB mutant spores containing the ExsFB-mCherry fusion were determined by western blot analysis using anti-mCherry antibodies. Wild-type spores contained too little extractable ExsFB-fusion protein to be detected (Fig. 8N, lane 1). However, loss of BxpB or BclA resulted in a significant increase in the amount of ExsFB-mCherry fusion extracted from the spores (Fig. 8N, lanes 2–3). This suggested the absence of BxpB-BclA complexes might facilitate an increased spore incorporation of ExsFB. To further examine this possibility, synchronous cultures of sporulating cells expressing the ExsFB-mCherry fusion were subjected to western blot analysis for a comparison of the timing of ExsFB expression relative to that of BclA and BxpB. BxpB initially appeared by western blot at T6, increasing in amount through T8 (Fig. 8O). ExsFB appeared very weakly at T6, increased in amount from T7-T8 (Fig. 8P). This indicated near identical timing of expression between ExsFB, BxpB, and BclA.
The ExsFB fusion results differ from that reported by Giorno et al. (2009) in that we observed a solid ring of fluorescence as opposed to the ovoid pattern described in their report. We suggest that the ExsFB is located within the exosporium. Differences between the two studies may be due to the higher fluorescence of our mCherry reporter compared to GFP. Both studies describe similar expression patterns for ExsFB and BxpB.
Discussion
Possible interactions between BxpB and BclA have been suggested in previous studies (Sylvestre et al., 2005; Steichen et al., 2005). We have demonstrated these two proteins were co-expressed, localized to the developing exosporium together, and were concurrently assembled. The BclA NTD and BxpB were found to be in close proximity to each other. At T8, two hours after initial BclA/BxpB expression and a time point which corresponds to the almost complete envelopment of the spore with BxpB and BclA complexes, there is an increase in the size of the complexes which corresponded with the appearance of glycosylation. This time point also corresponds to the timing of processing and release of the N-terminal 19 amino acids of BclA, resulting in stable incorporation of this protein in the exosporium (Thompson and Stewart, 2008). Taken together, this suggests that BclA and BxpB are co-expressed, immediately complex, and localize to the mother cell-central pole of the spore. Later in sporulation, as the complexes finish their polymerization around the developing exosporium surface, BclA is glycosylated and the N-terminal 19 amino acid peptide of BclA is cleaved and released. Thus the interactions between BclA and BxpB appear to be direct and the two proteins are co-dependent for their proper assembly onto the spore.
Because the BclA NTD is sufficient to restore BxpB incorporation into the spore, the interaction between BclA and BxpB does not require BclA trimerization or glycosylation. Mutations in the bclA NTD coding sequence that affect either the positioning of BclA around the developing exosporium or stable incorporation onto the spore surface also prevent BxpB incorporation into the spore. These observations suggest a model for BclA and BxpB assembly. BclA and BxpB complex in the mother cell cytoplasm and are positioned around the spore using the BclA NTD signals. BxpB, presumably in conjunction with other exosporium proteins, provides the proper scaffold for subsequent stable incorporation of BclA, a process involving the NTD proteolytic cleavage event. Absence of BclA would limit BxpB spore incorporation because of the lack of the localization signal. Loss of BxpB would have a somewhat lesser effect on BclA incorporation into the spore by preventing proper positioning of the BclA complexes. This would be analogous to the effect seen with BclA NTD positioning domain mutants, where the BclA protein does not correctly assemble around the spore, but some residual incorporation is observed (this study and Thompson and Stewart, 2008).
Interestingly, in sporulating cells bearing both full length BclA and the N-terminal BclA domain fusions, BxpB binds to both, but the native BclA and NTD fusion of BclA are not found in the same extractable protein complexes. This is likely due to the inability of the BclA NTD-fusion to trimerize and thus capture wild-type BclA in its complexes. Mutant spores lacking BxpB are profoundly deficient in BclA incorporation. Conversely, BclA-lacking mutant spores are less profoundly deficient in BxpB incorporation. Because BxpB interacts with the BclA NTD sequences, it is possible that other proteins may, at least partially, compensate for a lack of BclA. Other collagen-like proteins have been identified bearing the NTD localization sequences (Thompson and Stewart, 2008). BclB or another similar collagen-like protein with similar N-terminal motifs may be able to substitute for BclA and permit BxpB’s incorporation into the spore.
BxpB was found to be associated in complex with BclA NTD-eGFP by immunoprecipitation. Larger complexes of BxpB likely contain increased numbers of BxpB molecules and/or other exosporium-associated proteins. We did not, however, find larger SDS-resistant complexes with the BclA NTD-eGFP molecule. This finding suggests either this fusion cannot form the larger complexes seen with native BclA, or that this BclA NTD-eGFP and BxpB complex is more easily extracted from larger complexes with our SDS+8M urea extraction process.
To better define which specific residues within the BclA N-terminal domain are required for BclA incorporation into the exosporium and BxpB binding, we created a series of single alanine replacement mutations along the BclA NTD coding sequence. This study confirmed the presence of the positioning and attachment domains we first described (Thompson and Stewart, 2008), but suggested some overlap in function at residues 25–27. The critical residues found in this study agree in part with those found using bclA deletions complementing a bclA mutant background from Tan et al. (2010). However, we found the aspartate residue at position 22 to be critical for BclA incorporation and which was not identified as such in the deletion analysis. The use of alanine replacements, rather than a deletion approach, more likely permits maintenance of overall protein conformations and thus simplifies interpretation of the results. A deletion analysis approach is also complicated because the deletion positions downstream residues in the place of the deleted residues and the specific amino acids so repositioned can impact the overall results.
Mutations at BclA NTD residues found to be imperative for incorporation in a wild-type spore are able to partially attach in bclA mutant spores. This decrease in the defect severity likely resulted from a lack of competition from wild-type BclA in this background. In the bclA mutant background, differences in the composition of the complexes formed in the absence of wild-type BclA illustrate the importance of conducting the localization domain studies in the presence of native BclA molecules, and not solely in a bclA-null mutant host strains.
The ExsFB protein has been described as playing a minor role in the assembly of BclA (Sylvestre et al., 2005). In an exsFB-null mutant, there was no difference found in overall BclA levels by western blot analysis but a slight reduction in the hair-like nap filaments. In the dual bxpB and exsFB mutant background, no BclA was found on free spores (Sylvestre et al., 2005). This suggests that ExsFB may be able to compensate and partner with BclA to permit its incorporation into the spore in the absence of BxpB. We found that in the absence of BxpB or BclA, an increased amount of ExsFB was incorporated into the mature spore. When the BclA and BxpB complex cannot form, ExsFB can likely compensate for this loss of BxpB by complexing with BclA and leading to the incorporation of both proteins into the exosporium. However, the localization pattern of ExsFB on a developing spore is dramatically different from BxpB and BclA in wild-type spores. ExsFB is incorporated into the spore in an asymmetrically bipolar pattern whereas BxpB is incorporated in uniform pattern covering 75% of the spore. Although highly similar in sequence, differences do exist between BxpB and ExsFB which likely affect their interactions with the exosporium assembly apparatus. The polar localization of ExsFB may indicate its presence in the “cap” of the spores (Steichen et al., 2007). The presence of ExsFB in the “cap” and the assembly of BclA solely onto the cap in a bxpB mutant may suggest that ExsFB can form complexes with BclA to deliver BclA to the spore pole and thus form the cap structure (Giorno et al., 2009). In wild-type spores, we find ExsFB in the cap and a diminished presence of BxpB at one pole. This may indicate a switch from ExsFB-BclA to BxpB-BclA complexes as the exosporium develops beyond the “cap” structure and envelops the rest of the spore.
Although BxpB permits the localization of BclA to the developing spore, the identity of protein(s) which act to affect the cleavage of the BclA N-terminal 19 amino acids remain unknown. Moreover, the additional proteins within high MW BclA/BxpB/ExsFB-containing complexes and their role in the assembly of the exosporium have yet to be determined.
Experimental Procedures
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Supplemental Table 1. Descriptions on the creation of the allele replacement mutant strains can be found in Giorno et al. 2007 (bxpB-null) and Thompson et al. 2008(bclA-null). E. coli was grown at 37°C with shaking (225 rpm) in Luria Burtani broth. B. anthracis was grown at 37°C with shaking (225 rpm) in Tryptic Soy Broth (Difco). Sporulation was induced by growth on nutrient agar plates at 30°C with appropriate antibiotic selection. Sporulation was essentially complete (>95%) after 72 hr. The degree of sporulation was assessed by phase contrast microscopy. Spores were harvested from 7-day old nutrient agar plates to ensure complete sporulation, washed 3 times in PBS, and stored at room temperature. When required, media were supplemented with 100 μg/ml ampicillin, 5 μg/ml erythromycin, 10 μg/ml chloramphenicol, or 100 μg/ml spectinomycin.
Tiger broth is a modified version of ModG medium (Bergman et al., 2006) that permits better synchronous growth and sporulation in liquid culture. Tiger broth matches the recipe for ModG but with the addition of a phosphate buffering capacity (1.44 g/L of Na2HPO4 and 0.48 g/L of KH2PO4, pH 7.3). Overnight cultures were grown in 5 ml of BHI broth with the addition of appropriate antibiotics. 1 ml of the overnight culture was used to inoculate 50 ml of pre-warmed Tiger broth cultures to achieve a starting OD600 of less than 0.1. Cultures were measured spectophometrically at A600 until the plot of the growth kinetics deviated from linearity (defined as T0). This more traditional Bacillus subtilis-based T0 corresponds to a 2 hour difference from our previously defined T0 (Thompson and Stewart, 2008). The average time from T0 to free spores is 10 hours under these conditions.
Creation of fusion constructs
Fusion constructs were created by PCR amplification of the intact ORF and the native promoter elements followed by splicing by overlapping extension to attach the ORF in-frame to the fluorescent reporter ORF (Ho et al., 1989; Horton et al., 1989). Correct fusions contained intact promoter regions including σK elements, as well as native RBS and start codons. Fusion constructs were then subcloned into the shuttle plasmid pMK4 (Sullivan et al., 1984), plasmid propagated in a dam-minus E. coli host, followed by electroporation into the various strains of Bacillus anthracis. Correct transformants were screened by antibiotic selection, followed by DNA sequencing of plasmids isolated from vegetative cells. Additionally, when two fusions constructs were utilized, the second fusion (always BclA or BclA NTD eGFP) was cloned into pHPS. pHPS is a derivative of pHP13 with the chloramphenicol cassette replaced by a spectinomycin cassette. pHPS and pMK4 are compatible in their coexistence in Bacillus spp.
Spore analysis by flow cytometry and immunofluorescence microscopy
1×107 purified spores were resuspended in 500 μl of 4% paraformaldehyde in PBS and incubated for 2 hours at room temperature. The spores were washed 4X with PBS and resuspended in StartingBlock (Pierce) and incubated with mixing at room temperature for 45 minutes. The spores were pelleted and resuspended in StartingBlock. Rabbit polyclonal antiserum against rBclA (1:250 dilution) or against rBxpB (1:250 dilution) was added and incubated with mixing at room temperature for 45 minutes. The spores were then washed 3X in StartingBlock PBS and then incubated with mixing with goat anti-rabbit Alexa Fluor 568 (Invitrogen) and incubated for 45 minutes at room temperature. The spores were washed 3X with StartingBlock, followed by 2X with PBS and then processed on a FACScan flow cytometer (Beckton Dickinson Biosciences) or a MoFlo XDP Flow Cytometer (Beckton Dickinson Biosciences). Data were analyzed using Cell Quest analysis software or Summit 5.2 (Beckton Dickinson Biosciences).
Micrographic images
Samples from sporulating cells collected from timed Tiger broth cultures were harvested by centrifugation, fixed in 4% paraformaldehyde for 15 minutes, and then washed 3X with PBS. Samples from each time point were pelleted and resuspended in 10 μl of PBS containing 0.4% DABCO (Diazabicyclooctane, Acros Organics) anti-fade reagent. All images were obtained using a Nikon E600 epi-fluorscence microscope using a 60× objective.
Western Blots
Protein samples for western blots were collected in two ways. For synchronous Tiger broth protein samples, 1 ml of sporulating culture was harvested by centrifugation and resuspended with 100 μl of 8M urea-containing SDS sample buffer (Thompson et al. 2007) and immediately boiled. For blots containing spore extracts obtained from purified spores, 107 spores were spun down and resuspended in 100 μl urea-containing SDS sample buffer and boiled for 5 minutes. All samples were microfuged (13,000 × g for 1 minute) and 50 μl of extract was loaded into each well. Samples were electrophoresed in 4–20% Tris-glycine Fast gels (BioRad). Blots were reacted with primary antibodies as follows: polyclonal anti-rBclA (1:20,000), polyclonal anti-rBxpB (1:100,000), polyclonal anti-GFP (1:1,000) (Molecular Probes), polyclonal anti-DsRed (1:10,000) (Clontech, for mCherry). Blots were exposed using a Fujifilm LAS-3000 Intelligent Dark Box imaging machine.
Glycoprotein Stain
Samples for the glycoprotein stain were processed identically to those used in the western blot section above. Glycoproteins were identified using the Krypton Glycoprotein Staining Kit (Thermo Scientific).
Immunoprecipitation assays
To obtain protein samples for immunoprecipitation, BclA-eGFP fusion-expressing sporulating cultures in Tiger broth at T5 were harvested by centrifugation, and the cells lysed by bead-beating using 0.1 mm glass beads (Biospec Products). After the lysate was centrifuged to remove residual intact cells and cell debris, the remaining lysate was subjected to pulldown isolation using anti-eGFP polyclonal antibody (Molecular Probes) as per the Crosslink Immunoprecipitation Kit (Thermo Scientific). Reactions included a no eGFP antibody control and also a sample without lysate control. All samples were pre-cleared using control beads as per the manufacturer’s suggestion. All assays were performed in triplicate.
Alanine Replacement mutations of the N-terminal domain of BclA
A derivative of the shuttle vector pMK4 (pSP4039) was created that contained the BclA promoter, ribosome binding site, and first 13 bclA codons followed by an EcoRV recognition sequence (digestion with EcoRV creates a blunt end after the 13th codon) and an in-frame DsRed coding sequence. Insert DNA consisting of double stranded annealed oligonucleotides (Supplemental Table 2) were ligated into EcoRV-digested vector. Insertion of the DNA fragment resulted in expression of the 35 amino acid BclA NTD bearing a single amino acid substitution. A series of sequential alanine replacements for amino acids 17–35 of the BclA NTD were created. Correct clones were verified by DNA sequence analysis and electroporated into B. anthracis. Transformants were cultured and allowed to sporulate on N agar plates at 30°C for 7 days. At select time points, samples were taken and imaged to determine the extent of BclA localization and attachment to the free spores. A control was constructed with the wild-type sequence of 35 BclA codons in-frame with the DsRed reporter. At least three independent clones were tested for each construct, and each construct was introduced into B. anthracis at least in duplicate.
Supplementary Material
Acknowledgments
We thank Adam Driks (Loyola University, Chicago, IL) for the bxpB mutant strain and Sean Peters for technical support. B. Thompson was supported in part by a Veterinary Postdoctoral Fellowship through the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Disease Research.
References
- Albertazzi L, Arosio D, Marchetti L, Ricci F, Beltram F. Quantitative FRET analysis with the EGFP-mCherry Fluorescent Protein Pair. Photochem Photobiol. 2009;85:287–297. doi: 10.1111/j.1751-1097.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- Banning C, Votteler J, Hoffman D, Koppensteiner H, Warmer M, Reimer R, Kirchhoff F, Schubert U, Hauber J, Schindler M. A flow cytometry-based FRET Assay to identify and analyse protein-protein interactions in living cells. PLoS ONE. 2010;5:e9344. doi: 10.1371/journal.pone.0009344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boydston JA, Yue L, Kearney JF, Turnbough CL., Jr The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J Bacteriol. 2006;188:7440–7448. doi: 10.1128/JB.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozue J, Moody KL, Cote CK, Stiles BG, Friedlander AM, Welkos SL, Hale ML. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect Immun. 2007;75:4498–4505. doi: 10.1128/IAI.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt TN, Janes BK, Stibitz ES, Darnell SC, Sanz P, Rasmussen SB, O’Brien AD. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect Immun. 2007;75:5233–5239. doi: 10.1128/IAI.00660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton S, Moir AJG, Ballie L, Moir A. Characterization of the exosporium of Bacillus cereus. J Appl Micro. 1999;87:241–245. doi: 10.1046/j.1365-2672.1999.00878.x. [DOI] [PubMed] [Google Scholar]
- Daubenspeck JM, Zeng H, Chen P, Dong S, Steichen CT, Krishna NR, Pritchard DG, Turnbough CL., Jr Novel oligosaccharide side chains of the collagen-like region of BclA, the major glycoprotein of the Bacillus anthracis exosporium. J Biol Chem. 2004;279:30945–30953. doi: 10.1074/jbc.M401613200. [DOI] [PubMed] [Google Scholar]
- Driks A. Maximum shields: the assembly and function of the bacterial spore coat. Trends Microbiol. 2002;10:251–254. doi: 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- Giorno R, Bozue J, Cote C, Wenzel T, Moody KS, Mallozzi M, Ryan M, Wang R, Zielke R, Maddock JR, Friedlander A, Welkos S, Driks A. Morphogenesis of the Bacillus anthracis spore. J Bacteriol. 2007;189:691–705. doi: 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorno R, Mallozzi M, Bozue J, Moody K, Slack A, Qui D, Wang R, Friedlander A, Welkos S, Driks A. Localization and assembly of proteins comprising the outer structures of the Bacillus anthracis spore. Microbiology. 2009;155:1133–1145. doi: 10.1099/mic.0.023333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachisuka Y, Kojima K, Sato T. Fine filaments on the outside of the exosporium of Bacillus anthracis spores. J Bacteriol. 1966;91:2382–2384. doi: 10.1128/jb.91.6.2382-2384.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haima P, Bron S, Venema G. The effect of restriction on shotgun cloning and plasmid stability in Bacillus subtilis Marburg. Mol Gen Genet. 1987;209:335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr Structure, assembly, and function of the spore surface layers. Ann Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Hilbert DW, Piggot PJ. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol Mol Biol Rev. 2004;68:234–262. doi: 10.1128/MMBR.68.2.234-262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Johnson MJ, Todd SJ, Bell DA, Shepherd AM, Sylvestre P, Moir A. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J Bacteriol. 2006;188:7905–7913. doi: 10.1128/JB.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MJ, Roth IL. Ultrastructural differences in the exosporium of the Sterne and Vollum strains of Bacillus anthracis. Can J Microbiol. 1968;14:1297–1299. doi: 10.1139/m68-217. [DOI] [PubMed] [Google Scholar]
- Lai E, Phadke ND, Kachman MT, Giorno R, Vazquez S, Vazquez JA, Maddock JR, Driks A. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol. 2003;185:1443–1454. doi: 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohye DF, Murrell WG. Exosporium and coat formation in Bacillus cereus. J Bacteriol. 1973;115:1179–1190. doi: 10.1128/jb.115.3.1179-1190.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva CR, Swiecki MK, Griguer CE, Lisanby MW, Bullard DC, Turnbough CL, Jr, Kearney JF. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc Natl Acad Sci USA. 2008;105:1261–1266. doi: 10.1073/pnas.0709321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond C, Ballie LWJ, Hibbs S, Moir AJG, Moir A. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology. 2004;150:355–363. doi: 10.1099/mic.0.26681-0. [DOI] [PubMed] [Google Scholar]
- Severson KM, Mallozzi M, Bozue J, Welkos SL, Cote CK, Knight KL, Driks A. Roles of the Bacillus anthracis spore protein ExsK in exosporium maturation and germination. J Bacteriol. 2009;191:7587–7596. doi: 10.1128/JB.01110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Steichen C, Chen P, Kearney JF, Turnbough CL., Jr Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol. 2003;185:1903–1910. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen CT, Kearney JF, Turnbough CL., Jr Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol. 2005;187:5868–5876. doi: 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steichen C, Kearney JF, Turnbough CL., Jr Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol Micro. 2007;64:359–367. doi: 10.1111/j.1365-2958.2007.05658.x. [DOI] [PubMed] [Google Scholar]
- Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- Swiecki MK, Lisanby MW, Shu F, Turnbough CL, Jr, Kearney JF. Monoclonal antibodies for Bacillus anthracis spore detection and functional analyses of spore germination and outgrowth. J Immunol. 2006;176:6076–6084. doi: 10.4049/jimmunol.176.10.6076. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. A collagen-like surface glycoprotein is a structural component of the Bacillus anthracis exosporium. Mol Microbiol. 2002;45:169–178. doi: 10.1046/j.1365-2958.2000.03000.x. [DOI] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. Polymorphism in the collagen-like region of the Bacillus anthracis BclA protein leads to variation in exosporium filament length. J Bacteriol. 2003;185:1555–1563. doi: 10.1128/JB.185.5.1555-1563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre P, Couture-Tosi E, Mock M. Contribution of ExsFA and ExsFB proteins to the localization of BclA on the spore surface and to the stability of the Bacillus anthracis exosporium. J Bacteriol. 2005;187:5122–5128. doi: 10.1128/JB.187.15.5122-5128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Turnbough CL., Jr Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis. J Bacteriol. 2010;192:1259–1268. doi: 10.1128/JB.01003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd SJ, Moir AJG, Johnson MJ, Moir A. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J Bacteriol. 2003;185:3373–3378. doi: 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BM, Waller LN, Fox KF, Fox A, Stewart GC. The BclB glycoprotein of Bacillus anthracis is involved in exosporium integrity. J Bacteriol. 2007;189:6704–6713. doi: 10.1128/JB.00762-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BM, Stewart GC. Targeting of the BclA and BclB proteins to the Bacillus anthracis spore surface. Mol Microbiol. 2008;70:421–434. doi: 10.1111/j.1365-2958.2008.06420.x. [DOI] [PubMed] [Google Scholar]
- Waller LN, Stump MJ, Fox KF, Harley WM, Fox A, Stewart GC, Shahgholi M. Identification of a second collagen-like glycoprotein produced by Bacillus anthracis and demonstration of associated spore-specific sugars. J Bacteriol. 2005;187:4592–4597. doi: 10.1128/JB.187.13.4592-4597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.