Abstract
Background
Clostridium acetobutylicum, a gram-positive and spore-forming anaerobe, is a major strain for the fermentative production of acetone, butanol and ethanol. But a previously isolated hyper-butanol producing strain C. acetobutylicum EA 2018 does not produce spores and has greater capability of solvent production, especially for butanol, than the type strain C. acetobutylicum ATCC 824.
Results
Complete genome of C. acetobutylicum EA 2018 was sequenced using Roche 454 pyrosequencing. Genomic comparison with ATCC 824 identified many variations which may contribute to the hyper-butanol producing characteristics in the EA 2018 strain, including a total of 46 deletion sites and 26 insertion sites. In addition, transcriptomic profiling of gene expression in EA 2018 relative to that of ATCC824 revealed expression-level changes of several key genes related to solvent formation. For example, spo0A and adhEII have higher expression level, and most of the acid formation related genes have lower expression level in EA 2018. Interestingly, the results also showed that the variation in CEA_G2622 (CAC2613 in ATCC 824), a putative transcriptional regulator involved in xylose utilization, might accelerate utilization of substrate xylose.
Conclusions
Comparative analysis of C. acetobutylicum hyper-butanol producing strain EA 2018 and type strain ATCC 824 at both genomic and transcriptomic levels, for the first time, provides molecular-level understanding of non-sporulation, higher solvent production and enhanced xylose utilization in the mutant EA 2018. The information could be valuable for further genetic modification of C. acetobutylicum for more effective butanol production.
Background
High oil prices, growing concerns over national security and climate change are driving investment and innovation in the renewable alternative fuels [1,2]. Among various potentially alternatives, butanol has been proposed as an excellent substitute or supplement for gasoline, and has been demonstrated to work in some vehicles designed for use with gasoline without any engine modification [1]. In addition to manufacture from petroleum through chemical refinery process, industry production of butanol is typically through a so-called ABE fermentation process employing gram-positive, spore forming and anaerobic organism Clostridium acetobutylicum [2]. C. acetobutylicum is capable of producing a mixture of acetone (A), butanol (B) and ethanol (E) from a variety of carbohydrate substrates such as starch [3]. According to an estimate in 2008, butanol accounted for a 7-8.4 billon US dollar market worldwide and has a projected market expansion of 3% per year in the near future [4].
Significant efforts have been spent on physiological and genetic characterization of solvent-producing C. acetobutylicum in the past decades [5-8], and tools for genetic manipulation of C. acetobutylicum were also developed [9-11]. In 2001, the whole genome of well studied C. acetobutylicum ATCC 824 was sequenced, revealing a 3.94 Mb chromosome which encodes 3740 open reading frames (ORF), and a 192 Kb megaplasmid which encodes 178 ORFs [12]. Afterwards, a series of studies employing global approaches have been performed [13-16], and the genome-scale metabolic model of C. acetobutylicum was also constructed [17-19]. These efforts have improved the understanding of regulatory and metabolic networks of this industry significant species.
However, most of the C. acetobutylicum strains are not optimized systems for butanol production because their spore-forming life cycle decreases the efficiency of industrial fermentation, and the ABE fermentation process also creates a number of by-products, such as H2, acetic, lactic and propionic acids, acetone, isopropanol and ethanol [20]. As a result, the butanol yield is difficult to control and a significant amount of energy is wasted in these by-products. Moreover, it also increases the cost of downstream butanol purification. To address these issues, various modification approaches, such as mutagenesis by chemical or radiation agents, and genetic engineering, have been performed to improve the butanol production [10,21]. Our laboratory has previously obtained a high butanol producing strain, C. acetobutylicum EA 2018, through butanol resistance screening of N-methyl-N-nitro-N-nitrosoguanidine (NTG) treated Clostridium strain isolated from soil [22]. Preliminary results in a 100-ton continuous fermenter showed that butanol ratio and starch conversion rates of EA 2018 strain were 10% and 5% higher than those reported in recent literature [23]. To explore the genetic difference between EA 2018 and ATCC 824, in this study, the C. acetobutylicum EA 2018 genome was sequenced using Roche 454 pyrosequencing together with traditional Sanger sequencing. In addition, comparative genomic and transcriptomic analyses of EA 2018 and ATCC 824 were also performed. The study, for the first time, provides a molecular-level understanding of higher solvent production, enhanced xylose utilization and non-sporulation in the mutant EA 2018. The information could be valuable for further genetic modification of C. acetobutylicum for more effective butanol production.
Results and Discussion
Characterization of isolate EA 2018
The original solvent producing strain was isolated by our laboratory previously [22]. After several rounds of mutagenesis using NTG (N-methyl-N'-nitro-N-nitrosoguanidine), we obtained a hyper butanol-producing strain designated as EA 2018. This strain was later identified as Clostridium acetobutylicum by the China Center for Type Culture Collection (CCTCC) and was kept in CCTCC under the preservation No. CCTCC M_94061. In this work, the 16S rDNA of C. acetobutylicum EA 2018 was cloned and sequenced. The 1399 bp 16S rDNA sequence of C. acetobutylicum EA 2018 was 100% identical to that of the type strain C. acetobutylicum ATCC 824 (Accession number NC_003030 for ATCC 824 genome sequence) [12]. Furthermore, the sol operon involved in butanol production was also cloned from C. acetobutylicum EA 2018 and sequenced, the comparative analysis showed that the sol operon of C. acetobutylicum EA 2018 was also 100% identical to that of C. acetobutylicum ATCC 824 (Accession number NC_001988 for ATCC 824 mega-plasmid sequence) [24]. The analysis demonstrated that EA 2018 and ATCC 824 belong to the same species.
Fermentation experiments were performed to compare the solvent production patterns of C. acetobutylicum EA 2018 and ATCC 824. EA 2018 exhibited higher solvent formation capacity than ATCC 824 strain in either 6% (w/v) glucose or xylose media (Figure 1A, B). After 48 h fermentation, 8.3 g/L glucose remained in the EA 2018 culture, while 16.5 g/L glucose still remained in the ATCC 824 culture after 72 h fermentation (Figure 1A). After 96 h fermentation, 23.6 g/L xylose was present in the EA 2018 culture, while 35.7 g/L xylose was still remained in the ATCC 824 culture (Figure 1B). In most C. acetobutylicum strains, solvent formation is always coupled with initiation of sporulation [10]. However, after fermentation of 72 h, there was no spore found in EA 2018 cultures, while significant spores were found in ATCC 824. With its higher solvent production and non-spore forming characteristics, EA 2018 strain could be an excellent strain for industrial application.
Figure 1.
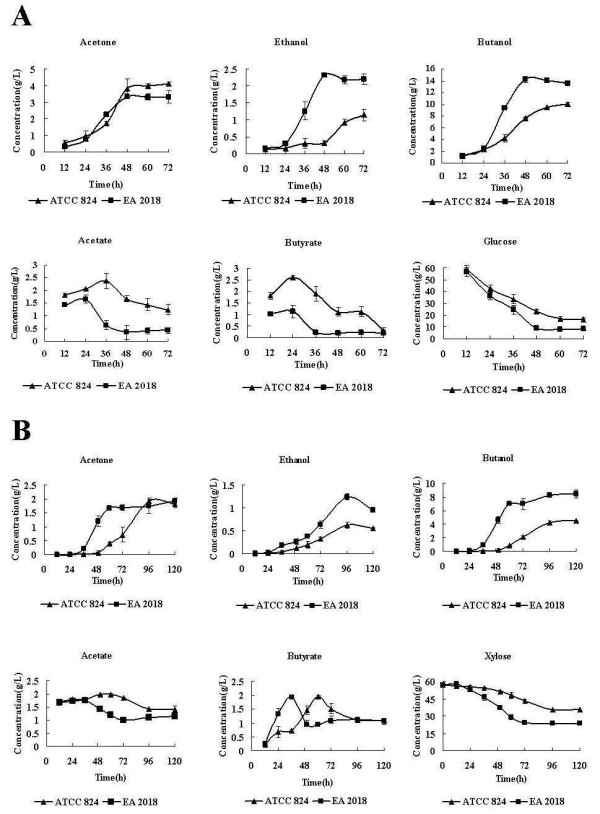
Solvent and acid production, sugar utilization of C. acetobutylicum EA 2018 versus C. acetobutylicum ATCC 824 in P2 medium contained with 6% glucose (A) and 6% xylose (B).
Overview of C. acetobutylicum EA 2018 genome
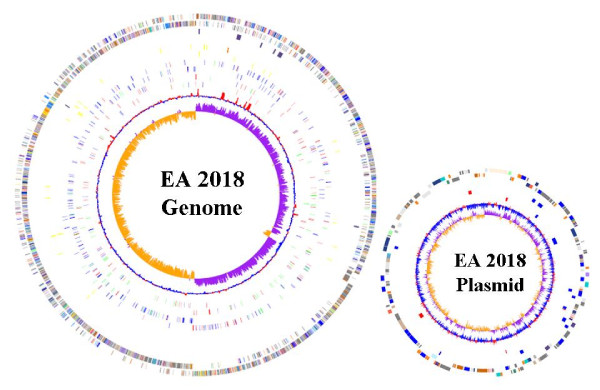
For better understanding of the genetic basis of improved butanol producing characteristics in EA 2018, the whole genome of EA 2018 was sequenced. The genome has a circular chromosome consisting of 3,940,230 bp with an average G+C content of 30.93% and a circular megaplasmid of 191,996 bp with an average G+C content of 30.91%. The genome finishing procedures were listed in Additional file 1, 2, 3. A total of 3923 protein coding sequence (CDS) including 3,746 in chromosome and 176 in megaplasmid were indentified in the EA 2018 genome, representing 86.8% of the genome and 83.8% of the megaplasmid, respectively. The functional classification of all EA 2018 genes was listed in Table 1. There are 11 copies of rDNA operons and a total of 75 tRNA genes scattered over the EA 2018 genome. Genomic comparison with type strain ATCC 824 revealed the highly conserved gene content and gene order between these two strains. The base numbering start point of EA 2018 were chosen as the same site in ATCC 824 (Figure 2).
Table 1.
Function Classification of EA 2018 genes
| Function | Numbers in plasmid | Numbers in genome | Function | Numbers in plasmid | Numbers in genome |
|---|---|---|---|---|---|
| Energy production and conversion | 12 | 121 | Cell envelope biogenesis, outer membrane | 10 | 182 |
| Cell division and chromosome partitioning | 3 | 38 | Cell motility and secretion | 1 | 92 |
| Amino acid transport and metabolism | 1 | 212 | Posttranslational modification, protein turnover, chaperones | 2 | 78 |
| Nucleotide transport and metabolism | 1 | 73 | Inorganic ion transport and metabolism | 6 | 93 |
| Carbohydrate transport and metabolism | 22 | 221 | Secondary metabolites biosynthesis, transport and catabolism | 3 | 27 |
| Coenzyme metabolism | 1 | 110 | General function prediction only | 16 | 321 |
| Lipid metabolism | 6 | 76 | Function unknown | 5 | 258 |
| translation,ribosomal structure and biogenesis | 0 | 159 | Signal transduction mechanisms | 4 | 126 |
| transcription | 22 | 243 | Intracellular trafficking and secretion | 0 | 14 |
| DNA replication, recombination and repair | 5 | 137 | Defense mechanisms | 5 | 106 |
| Not in this system | 55 | 1059 |
Figure 2.
Atlas of the chromosome and the megaplasmid of C. acetobutylicum EA 2018 and its comparison with C. acetobutylicum ATCC 824. Moving inside, each concentric circle represents genomic data for C. acetobutylicum EA 2018 and its comparison with C. acetobutylicum ATCC 824. For chromosome atlas, the outer circle illustrates predicted coding sequences on the plus and minus strands, respectively, colored by functional categories according to COG classification. The 2nd circle represents EA 2018 variation genes compared with ATCC 824. The 3rd circle displays IS elements in EA 2018. The 4th circle shows rDNA genes in EA 2018, distinguished by plus strand (gold) and minus strand (red). The 5th circle shows tRNA genes in EA 2018, distinguished by plus strand (pink) and minus strand (blue). The 6th circle represent GC content, red for GC content above average and blue for GC content below average. The 7th circle (innermost) represents GC skew (G-C)/(G+C) calculated using a 100 kb window. For megaplasmid atlas, the outer circle illustrates predicted coding sequences on the plus and minus strands, respectively, colored by functional categories according to COG classification. 2nd circle represents EA 2018 variation genes compared with ATCC 824. 3th circle represent GC content, red for GC content above average and blue for GC content below average. The 4th circle (innermost) represents GC skew (G-C)/(G+C) calculated using a 100 kb window.
Comparative genomic analysis of EA 2018 and ATCC 824
The size of the EA 2018 chromosome is 650 bp smaller than that of ATCC 824, and the size of the EA 2018 megaplasmid is 4 bp smaller than that of ATCC 824. Compared with ATCC 824, a total of 46 deletion sites and 26 insertion sites were found across the EA 2018 genome, including 1 deletion site in the megaplasmid (Additional file 4). Among them, 55 sites are single nucleotide indel (i.e. insertion or deletion), and only 7 indel sites are larger than 100 bp. The largest insertion is 1768 bp in 1276337-1278091 which is located within a hypothetical protein gene (CEA_G1125, corresponding to CAC1113 in ATCC 824), while the largest deletion is 1812 bp in 1112030-1112031 which is located within a predicted member protein gene (CEA_G0978, corresponding to CAC0966 in ATCC 824). Apart from those indels, 440 single nucleotide variations (SNVs) were identified between the two chromosomes, and 11 SNVs were identified between the megaplasmid of EA 2018 and ATCC 824 (Additional file 5). The 523 variations (including 72 indels and 451 SNVs) affected a total of 229 protein encoding genes (including 10 genes in megaplasmid) and 38 predicted promoters (including 1 in megaplasmid) in the EA 2018 genome. Excluding the synonymous substitution, a total of 189 proteins had amino acids changes, while 47 of those proteins were with similar amino acid variations (Additional file 6). The 38 genes with their putative promoters affected were listed in Additional file 7. In Table 2 and 3, variations within putative promoters and genes related to solvent formation, xylose utilization, and spore formation (i.e. three key aspects related to enhanced butanol production in EA 2018) were listed.
Table 2.
EA 2018 gene variations associated with key phenotypes
| Gene locus in EA 2018 | Gene locus in ATCC 824 | Gene variation sites in EA 2018 | Gene variation sites in ATCC 824 | Protein variation sites in EA 2018 | Protein variation sites in ATCC 824 | Product name |
|---|---|---|---|---|---|---|
| Solvent formation related genes | ||||||
| CEA_G1048 | CAC1036 | 899(T) | 899(C) | 300(V) | 300(A) | Pyruvate kinase |
| CEA_G1755 | CAC1742 | 391(A) | 391(G) | 131(N) | 131(D) | phosphotransacetylase |
| CEA_G2463 | CAC2449 | 256(A) | 256(G) | 86(S) | 86(G) | Predicted flavoprotein |
| CEA_G2485 | CAC2471 | 439(A) | 438-439(-) | 147-150(R,L,P,I) | 147-150 (G,C,L,Stop codon) | Transcriptional regulator, TetR/AcrR family |
| CEA_G2556 | CAC2543 | 241-249 (G,T,A,G, A,T,C,A,T) | 240-241(---) | 81-83(V,D,H) | 80-81(---) | Electron-transferring flavoprotein large subunit |
| CEA_G2806 | CAC2798 | 286(A) | 286(G) | 96(M) | 96(V) | NADH:flavin oxidoreductase |
| CEA_P0058 | CA_P0059 | 754(A) | 754(C) | 252(N) | 252(H) | Alcohol dehydrogenase |
| CEA_P0077 | CA_P0078 | 91(A) | 91(G) | 31(T) | 31(A) | acetyl-CoA acetyltransferase |
| CEA_P0140 | CA_P0141 | 566(T) | 566(C) | 189(I) | 189(T) | Periplasmic hydrogenase small subunit, dehydrogenase |
| Substrate utilization related genes | ||||||
| CEA_G0239 | CAC0234 | 1072(T) | 1072(C) | 358(Stop codon) | 358(Q) | PTS system, fructoso-specific IIBC component |
| CEA_G1333 | CAC1319 | 5(T) | 5(C) | 2(I) | 2(T) | Glycerol uptake facilitator protein, GLPF |
| CEA_G1472 | CAC1456 | 974(T) | 974(A) | 325(M) | 325(K) | Sugar-binding periplasmic protein |
| CEA_G2622 | CAC2613 | 270(T) | 270(G) | 90(C) | 90(W) | Transcriptional regulators of NagC/XylR family |
| CEA_G2919 | CAC2912 | 97(A) | 97(C) | 33(T) | 33(P) | Sugar-binding periplasmic protein |
| CEA_P0052 | CA_P0053 | 317(C) | 317(T) | 106(P) | 106(L) | Xylanase, glycosyl hydrolase family 10 |
| Sporulation related genes | ||||||
| CEA_G0080 | CAC0080 | 1160(A) | 1159-1160(-) | 387-392(N,I,Q,D,L,Stop codon) | 387-391(I,Y,K,I,Y,K) | Histidine kinase-like ATPase |
| CEA_G0656 | CAC0644 | 1226(T) | 1226(G) | 409(V) | 409(G) | Spore germination protein gerKA |
| CEA_G0710 | CAC0699 | 416(C) | 416(T) | 139(T) | 139(I) | Spore photoproduct lyase, splB |
| CEA_G2066 | CAC2052 | 688(A) | 687-688(-) | 230-248 (18 amino acid) | 230-238 (7 amino acid and a Stop Codon) | DNA-dependent RNA polymerase sigma subunit |
| CEA_G3736 | CAC3729 | 374(T) | 374(C) | 125(L) | 125(P) | Stage 0 sporulation J, ParB family of DNA-binding proteins |
| CEA_P0016 | CA_P0017 | 11,245(G,A) | 11,245(T,C) | 4,82(E,G) | 4,82(A,V) | Spore germination protein, GRKB |
| CEA_P0019 | CA_P0020 | 1120(C) | 1120(T) | 407(A) | 407(V) | Spore germination protein, GRKA |
| CEA_P0021 | CA_P0022 | -151(T) | 104(C) | 1(---) | 1-85(85 amino acids insertion) | Spore germination protein, GRKB |
Numbers in gene or protein variation sites lines indicated the variation sites in genes; the letters in bracket means the corresponding variation bases or amino acid; the symbol "-" means deletion in genes; the symbol "---" means consecutive deletion in genes; "No" means no amino acid variation; "Stop codon" means this site is mutated to stop codon.
Table 3.
EA 2018 putative promoter variations related to key phenotypes
| Gene locus in EA 2018 | Gene locus in ATCC 824 | Gene variation sites in EA 2018 | Gene variation sites in ATCC 824 | Product Name |
|---|---|---|---|---|
| Solvent formation related genes | ||||
| CEA_G0028 | CAC0028 | -12(T) | -12(C) | Hydrogen dehydrogenase |
| CEA_P0034 | CA_P0035 | -84(T) | -84(C) | Aldehyde-alcohol dehydrogenase, ADHEII |
| Substrate utilization related genes | ||||
| CEA_G3043 | CAC3037 | -80(T) | -80(C) | Catabolite control protein, LacI family transcriptional regulator |
| CEA_G3455 | CAC3451 | -84(T) | -84(G) | Sugar/Na+(H+) simporter |
| CEA_G1086 | CAC1075 | -56(T) | -56(G) | Beta-glucosidase family protein |
| CEA_G1365 | CAC1351 | -97(T) | -97(C) | Periplasmic sugar-binding protein |
| Sporulation related genes | ||||
| CEA_G1634 | CAC1620 | -136(T) | -136(G) | Small acid-soluble spore protein |
| CEA_G3742 | CAC3735 | -(8-7)(-) | -7(C) | Predicted RNA-binding protein Jag, SpoIIIJ-associated |
Numbers in gene or protein variation sites lines indicated the variation sites in genes; the letters in bracket means the corresponding variation bases or amino acid; the symbol "-" means deletion in genes.
Comparative transcriptomic analysis of EA 2018 and ATCC 824
To further explore the molecular mechanism of enhanced butanol production in the EA 2018 strain, DNA microarray of C. acetobutylicum were manufactured and used for a comparative analysis between EA 2018 and ATCC 824. The complete set of DNA array data was available in Additional file 8. Microarray analysis showed that a total of 2215 genes were differentially regulated at transcriptional level in at least 1 cultivation time point. Among them, differentially regulated genes related to some important metabolic pathways were listed in Table 4. And some putative promoter variation genes such as adhEII were found with differential expression level in the EA 2018 strain (Table 5). The detailed comparative analysis of transcriptomic data, along with genomic data (i.e. variation gene sequence) and biochemical phenotypes will be provided below from three key aspects related to enhanced butanol production (i.e. solvent formation, xylose utilization, and spore formation).
Table 4.
List of differentially regulated genes in key functional groups.
| Gene locus in EA 2018 | Gene locus in ATCC 824 | 9 h (Log2Ratio 2018/824) | 13 h (Log2Ratio 2018/824) | 17 h (Log2Ratio 2018/824) | 21 h (Log2Ratio 2018/824) | 24 h (Log2Ratio 2018/824) | 30 h (Log2Ratio 2018/824) | Product Name |
|---|---|---|---|---|---|---|---|---|
| Carbohydrate transportant and metabolism | ||||||||
| CEA_G0343 | CA_C0332 | -2.97436 | -3.22501 | -2.37758 | -2.08064 | -2.57556 | -1.49158 | Beta-mannanase |
| CEA_G0501 | CA_C0490 | 2.331505 | 2.410311 | 2.244172 | 2.361906 | 2.703307 | 2.474026 | sugar kinase, N-terminal region - uncharacterized protein |
| CEA_G0552 | CA_C0539 | -3.20909 | -3.00324 | -2.93013 | -2.29917 | -3.34939 | -3.01922 | ChW repeat-containing mannanase ManB |
| CEA_G0553 | CA_C0540 | -3.097 | -3.03398 | -2.7227 | -2.28306 | -3.3665 | -2.72934 | ChW repeat-containing mannanase ManB |
| CEA_G1677 | CA_C1664 | 1.744408 | 2.013214 | 2.53088 | 1.134795 | 1.816259 | 2.062687 | glycogen phosphorylase |
| CEA_G2012 | CA_C1997 | 3.052977 | 3.503142 | 3.004392 | 2.8712 | 2.075025 | 1.372718 | glycosyltransferase |
| CEA_G2022 | CA_C2007 | 3.179932 | 3.308867 | 2.812136 | 2.509044 | 1.656072 | 1.958591 | glycosyltransferase |
| CEA_G2528 | CA_C2514 | 2.588809 | 2.748177 | 3.625871 | 4.329215 | 2.708288 | 1.042021 | Beta galactosidase |
| CEA_G2815 | CA_C2807 | 2.40568 | 2.124155 | 2.476058 | 2.653992 | 2.808752 | 1.737289 | endo-1,3(4)-beta-glucanase family protein 16 |
| CEA_G2818 | CA_C2810 | 1.835243 | 2.287891 | 2.800041 | 3.929157 | 3.739976 | 4.256289 | glucoamylase family protein |
| CEA_G3051 | CA_C3045 | -2.41601 | -2.05151 | -2.19775 | -2.37262 | -2.3321 | -2.16444 | PHP family hydrolase |
| CEA_G3060 | CA_C3054 | -3.63818 | -4.38617 | -4.64833 | -5.02922 | -5.12371 | -3.50751 | phosphoheptose isomerase |
| CEA_G3426 | CA_C3422 | -1.07396 | -2.79221 | -3.22016 | -2.15086 | -2.69786 | -1.37931 | sugar:proton symporter (xylulose) |
| CEA_P0052 | CA_P0053 | 2.477697 | 2.868013 | 3.649329 | 3.695172 | 3.721454 | 5.987476 | xylanase |
| CEA_P0053 | CA_P0054 | 4.25084 | 2.735168 | 2.447001 | 3.424303 | 3.598697 | 5.185082 | xylanase/chitin deacetylase family protein |
| CEA_P0065 | CA_P0066 | 3.062167 | 1.145145 | 2.159875 | 4.902259 | 3.81378 | 2.953822 | mannose-specific phosphotransferase system component IIAB |
| CEA_P0066 | CA_P0067 | 2.925021 | 1.579742 | 2.174524 | 4.883877 | 3.683632 | 2.826095 | mannose/fructose-specific phosphotransferase system component IIC |
| CEA_P0067 | CA_P0068 | 3.006142 | 1.74298 | 2.611356 | 4.717371 | 3.502957 | 2.456002 | mannose-specific phosphotransferase system component IID |
| CEA_P0115 | CA_P0116 | 1.777221 | 2.342324 | 2.902238 | 3.126694 | 3.321994 | 5.378643 | xylanase |
| Amino acid transport and metabolism | ||||||||
| CEA_G0180 | CA_C0176 | 2.56498 | 2.063235 | 2.36635 | 2.08086 | 3.251555 | 3.705152 | oligopeptide-binding protein, periplasmic component |
| CEA_G0327 | CA_C0316 | -4.09229 | 6.429377 | 7.957548 | 6.881528 | 4.185199 | 2.037144 | ornithine carbomoyltransferase |
| CEA_G0390 | CA_C0380 | -4.23921 | 5.06975 | 5.365591 | 5.142555 | 3.394778 | 2.099342 | periplasmic amino acid-binding protein |
| CEA_G0984 | CA_C0973 | -4.67305 | 6.390879 | 8.78089 | 7.141168 | 3.889621 | 2.522783 | argininosuccinate synthase |
| CEA_G0985 | CA_C0974 | -4.03279 | 6.033303 | 8.738086 | 7.66813 | 4.662246 | 3.462605 | argininosuccinate lyase |
| CEA_G2392 | CA_C2377 | -4.81354 | -4.50847 | -4.89131 | -5.25303 | -5.6598 | -6.94396 | oligopeptide ABC-type transporter, periplasmic binding component (frameshift) |
| CEA_G2403 | CA_C2388 | -3.653 | 6.58659 | 9.490741 | 7.214547 | 3.839977 | 1.277684 | acetylornithine aminotransferase |
| CEA_G2405 | CA_C2390 | -4.029 | 7.044851 | 9.332151 | 8.593887 | 5.225523 | 1.729156 | N-acetyl-gamma-glutamyl-phosphate reductase |
| CEA_G2406 | CA_C2391 | -3.63924 | 6.659273 | 9.430206 | 8.041522 | 4.889782 | 1.658329 | bifunctional ornithine acetyltransferase/N-acetylglutamate synthase protein |
| CEA_G2531 | CA_C2517 | 2.601974 | 2.875402 | 3.244092 | 4.199853 | 5.407919 | 5.659197 | extracellular neutral metalloprotease, NPRE |
| CEA_G3059 | CA_C3053 | -3.80847 | -4.46351 | -4.85998 | -5.20027 | -5.64176 | -4.16398 | histidinol phosphatase related enzyme |
| CEA_G3625 | CA_C3618 | -2.33371 | 4.975742 | 4.579403 | 4.356899 | 3.79613 | 1.400104 | ABC-type polar amino acid transport system, ATPase component |
| CEA_G3626 | CA_C3619 | -2.49499 | 4.94909 | 4.797836 | 4.831871 | 4.227636 | 1.349231 | amino acid ABC transporter permease |
| CEA_G3627 | CA_C3620 | -2.79225 | 4.869307 | 2.77014 | 4.039679 | 3.923914 | 1.658129 | amino acid ABC transporter periplasmic-binding protein |
| CEA_G3629 | CA_C3622 | 3.744723 | 1.616914 | -1.60643 | -2.30924 | -3.70552 | -4.20925 | benzoyl-CoA reductase/2-hydroxyglutaryl-CoA dehydratase |
| CEA_G3648 | CA_C3641 | 3.877563 | 5.273677 | 6.532841 | 7.05381 | 6.150331 | 2.645978 | oligopeptide ABC transporter, ATPase component |
| CEA_G3649 | CA_C3642 | 3.862767 | 5.064748 | 6.213521 | 7.100308 | 6.123778 | 2.328286 | oligopeptide ABC transporter, ATPase component |
| CEA_G3651 | CA_C3644 | 3.540737 | 4.731217 | 4.796332 | 6.751584 | 6.151055 | 3.301733 | oligopeptide ABC transporter, permease component |
| Lipid transport and metabolism | ||||||||
| CEA_P0077 | CA_P0078 | -1.03663 | -2.39938 | -2.90422 | -5.61457 | -6.37659 | -6.669 | acetyl-CoA acetyltransferase |
| CEA_G0500 | CA_C0489 | 2.843362 | 2.727643 | 3.104514 | 2.258672 | 2.789674 | 2.994569 | 4'-phosphopantetheinyl transferase |
| CEA_G2024 | CA_C2009 | 3.02793 | 3.412819 | 2.753976 | 2.880761 | 2.465438 | 1.317947 | 3-hydroxyacyl-CoA dehydrogenase |
| CEA_G2027 | CA_C2012 | 3.139119 | 3.519089 | 2.760385 | 2.466674 | 1.747722 | 1.295802 | enoyl-CoA hydratase |
| CEA_G2023 | CA_C2008 | 3.21547 | 3.393795 | 2.65468 | 2.624962 | 1.849978 | 1.696553 | 3-oxoacyl-(acyl-carrier-protein) synthase |
| CEA_G0825 | CA_C0814 | 3.447389 | 2.690101 | 3.341654 | 3.492821 | 2.738719 | 2.617536 | 3-oxoacyl- |
| CEA_G3630 | CA_C3623 | 3.78405 | 1.698615 | -1.62094 | -2.13197 | -3.66189 | -4.47732 | 2-hydroxyglutaryl-CoA dehydratase activator |
| Coenzyme transport and metabolism | ||||||||
| CEA_G2539 | CA_C2526 | -5.80423 | -4.29934 | -2.18936 | -3.20732 | -2.04208 | -2.09088 | 6-pyruvoyl-tetrahydropterin synthase related protein |
| CEA_G0110 | CA_C0109 | -1.93293 | -1.10289 | -4.51818 | -4.22606 | -3.40598 | -5.95312 | sulfate adenylyltransferase subunit 2 |
| CEA_G2240 | CA_C2226 | 1.175604 | 1.630568 | 1.933437 | 3.44313 | 4.132037 | 3.011011 | branched-chain amino acid aminotransferase |
| CEA_G2817 | CA_C2809 | 1.909506 | 2.362089 | 1.59545 | 3.297164 | 2.183049 | 1.029092 | HD superfamily hydrolase |
| CEA_G2037 | CA_C2022 | 2.45013 | 1.945668 | 2.521958 | 2.427533 | 2.187431 | 3.238565 | molybdopterin biosynthesis protein MoaB |
| CEA_G3633 | CA_C3626 | 2.627562 | 1.339719 | -1.38437 | -2.32524 | -3.57634 | -5.06491 | GTP cyclohydrolase I |
| CEA_G2036 | CA_C2021 | 2.849463 | 2.144804 | 2.666979 | 4.297463 | 3.319713 | 3.863173 | molybdopterin biosynthesis protein MoeA |
| CEA_G2009 | CA_C1994 | 3.136184 | 3.75827 | 3.408469 | 2.84809 | 1.881193 | 1.268359 | molybdopterin biosynthesis protein MoaB |
| CEA_G2035 | CA_C2020 | 3.146626 | 3.515592 | 5.182665 | 4.677354 | 4.692789 | 4.012931 | molybdopterin biosynthesis protein MoeA |
| CEA_G3631 | CA_C3624 | 3.416604 | 1.490697 | -1.38827 | -2.23319 | -3.83641 | -6.15768 | 6-pyruvoyl-tetrahydropterin synthase |
| Signal transduction | ||||||||
| CEA_G0078 | CA_C0078 | -4.99228 | -8.71284 | -9.43594 | -9.21854 | -9.14927 | -7.97564 | putative accessory gene regulator protein |
| CEA_G3328 | CA_C3325 | -1.00804 | -2.85267 | -4.3502 | -5.19446 | -4.39209 | -5.05262 | periplasmic amino acid binding protein |
| CEA_G0921 | CA_C0909 | -2.84748 | -1.88833 | -1.93256 | -1.50557 | -2.64087 | -2.89144 | methyl-accepting chemotaxis protein |
| CEA_G2085 | CA_C2071 | 2.455498 | 2.263411 | 2.413649 | 1.803983 | 2.20351 | 2.550339 | Spo0A protein |
| CEA_G2422 | CA_C2407 | 3.784844 | 1.475039 | 1.927948 | 1.823945 | 2.526403 | 2.502499 | CheY-like domain-containing protein |
| CEA_G0448 | CA_C0437 | 2.192653 | 1.378372 | 2.665966 | 1.827148 | 2.252286 | 2.342264 | sensory transduction histidine kinase |
| CEA_G3025 | CA_C3019 | -2.65448 | 3.491201 | 3.449102 | 2.735229 | 1.336433 | 1.266833 | sensory transduction protein |
| CEA_G0296 | CA_C0289 | 1.792323 | 1.978035 | 2.438903 | 2.751395 | 3.435751 | 3.052853 | response regulator |
| CEA_G2782 | CA_C2774 | 1.625932 | 1.895382 | 3.060796 | 2.846554 | 2.092881 | 1.726376 | methyl-accepting chemotaxis protein |
| CEA_G0334 | CA_C0323 | 3.362504 | 2.662176 | 2.652177 | 3.222052 | 4.952742 | 2.037345 | sensory transduction histidine kinase |
| CEA_G3627 | CA_C3620 | -2.79225 | 4.869307 | 2.77014 | 4.039679 | 3.923914 | 1.658129 | amino acid ABC transporter periplasmic-binding protein |
| CEA_G0390 | CA_C0380 | -4.23921 | 5.06975 | 5.365591 | 5.142555 | 3.394778 | 2.099342 | periplasmic amino acid-binding protein |
| Energy production and convertion | ||||||||
| CEA_G1083 | CA_C1072 | 3.271648 | 3.230747 | 4.254617 | 2.59071 | 2.723209 | 3.766376 | Fe-S oxidoreductase |
| CEA_G2012 | CA_C1997 | 3.052977 | 3.503142 | 3.004392 | 2.8712 | 2.075025 | 1.372718 | glycosyltransferase |
| CEA_G2015 | CA_C2000 | 2.963078 | 3.638083 | 2.745804 | 2.983214 | 2.587284 | 2.204476 | indolepyruvate oxidoreductase subunit beta |
| CEA_G2016 | CA_C2001 | 2.832561 | 3.688549 | 3.007107 | 3.355002 | 2.718563 | 2.206461 | indolepyruvate ferredoxin oxidoreductase, subunit |
| CEA_G2022 | CA_C2007 | 3.179932 | 3.308867 | 2.812136 | 2.509044 | 1.656072 | 1.958591 | glycosyltransferase |
| CEA_G2025 | CA_C2010 | 3.282342 | 3.533526 | 2.84142 | 3.003615 | 2.50221 | 2.005231 | Fe-S oxidoreductase |
| CEA_G2555 | CA_C2542 | 8.22595 | 5.876659 | 5.423124 | 5.135223 | 4.224852 | 4.280766 | FAD/FMN-containing dehydrogenase |
| CEA_G2556 | CA_C2543 | 8.451735 | 6.100494 | 5.401602 | 5.24343 | 4.426238 | 3.987954 | electron-transferring flavoprotein large subunit |
| CEA_G2557 | CA_C2544 | 7.904188 | 6.040402 | 4.848558 | 4.578106 | 4.214698 | 3.969443 | electron-transferring flavoprotein small subunit |
| CEA_G3411 | CA_C3408 | -2.71993 | -3.64684 | -4.53359 | -4.7967 | -4.89809 | -4.45863 | NADH oxidase |
| Cell mobility | ||||||||
| CEA_G0921 | CA_C0909 | -2.84748 | -1.88833 | -1.93256 | -1.50557 | -2.64087 | -2.89144 | methyl-accepting chemotaxis protein |
| CEA_G3572 | CA_C3565 | 1.783589 | 1.966743 | 1.626123 | 1.303128 | 1.522492 | 1.56009 | cell adhesion domain-containing protein |
| CEA_G3091 | CA_C3085 | 1.669031 | 1.879947 | 1.641572 | 3.329325 | 2.811493 | 2.558158 | TPR repeat-containing cell adhesion protein |
| CEA_G3092 | CA_C3086 | 1.689695 | 1.7646 | 1.823344 | 3.167841 | 2.629106 | 2.423856 | cell adhesion domain-containing protein |
| CEA_G2782 | CA_C2774 | 1.625932 | 1.895382 | 3.060796 | 2.846554 | 2.092881 | 1.726376 | methyl-accepting chemotaxis protein |
| CEA_P0159 | CA_P0160 | 2.878477 | 3.739495 | 3.515178 | 3.50976 | 3.19802 | 3.547315 | cell-adhesion domain-containing protein |
Table 5.
DNA and transcriptional variations of putative promoter variations between EA 2018 and ATCC 824
| Gene locus in EA 2018 | Gene locus in ATCC 824 | Gene varition site in 2018 | Gene varition site in 824 | 9 h (Log2Ratio 2018/824) | 13 h (Log2Ratio 2018/824) | 17 h (Log2Ratio 2018/824) | 21 h (Log2Ratio 2018/824) | 24 h (Log2Ratio 2018/824) | 30 h (Log2Ratio 2018/824) | Product Name |
|---|---|---|---|---|---|---|---|---|---|---|
| CEA_G0334 | CA_C0323 | -96(A) | -(97-96)(-) | 3.362504 | 2.662176 | 2.652177 | 3.222052 | 4.952742 | 2.037345 | Sensory transduction histidine kinase |
| CEA_G0390 | CA_C0380 | -135,-131(A,A) | -135,-131(T,T) | -4.23921 | 5.06975 | 5.365591 | 5.142555 | 3.394778 | 2.099342 | Periplasmic amino acid-binding protein |
| CEA_G2504 | CA_C2490 | -(13-12),-53(-,T) | -12,-(54-53)(C,-) | -2.39555 | -3.35613 | -3.80832 | -2.2032 | -2.7562 | -1.10185 | Xre family DNA-binding domain and TPR repeats containing protein |
| CEA_G3701 | CA_C3694 | -43(T) | -43(C) | 4.045439 | 3.430925 | 3.859781 | 3.69948 | 4.131629 | 3.75554 | TPR-repeat-containing protein |
| CEA_P0034 | CA_P0035 | -84(T) | -84(C) | 3.331902 | 5.740285 | 6.007483 | 8.056546 | 6.576379 | 7.664691 | Aldehyde-alcohol dehydrogenase, AdhEII |
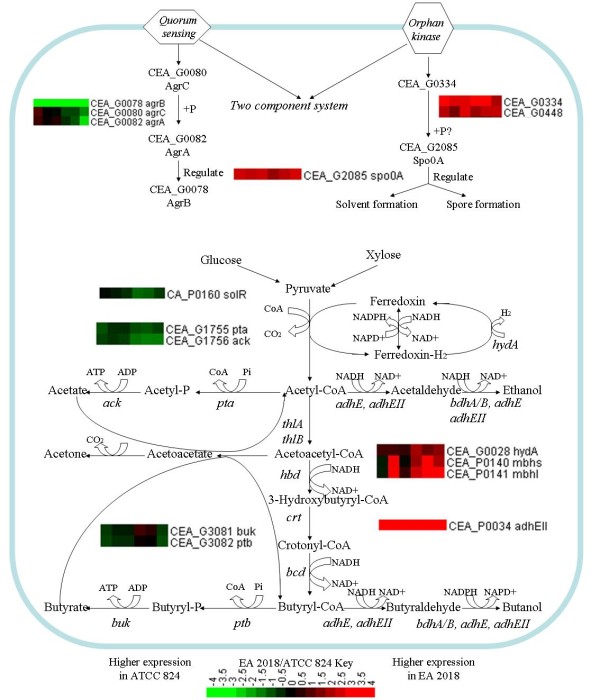
Changes in expression of adhEII, spo0A and hydrogenase gene may contribute to enhanced solvent formation in EA 2018
Comparative genomic analysis identified a set of solvent-relevant genes with variations within their coding sequences (Table 2). And most of which were SNV variations, such as genes encoding phosphotransacetylase and acetyl-CoA acetyltransferase. In addition, comparative genomics analysis also identified some variations up-stream of solvent-relevant genes (Table 3), which could potentially affect expression level of these genes. For example, a SNV site was found 84 bases upstream of the start codon of the AdhEII encoded gene CEA_P0034 (CA_P0035 in ATCC 824), under the s1 transcription start point [25]. Consistent with the variations at genomic level, we also found that transcription level of solvents formation genes, such as adhEII were highly expressed in EA 2018 relative to ATCC 824(Table 5 and Figure 3).
Figure 3.
Expression profiles of variation genes involved in sporulation related two-component system and solvent formation pathway of C. acetobutylicum EA 2018 versus ATCC 824.
Spo0A is a central regulator of sporulation and solvent formation in C. acetobutylicum. Previous studies showed that C. acetobutylicum spo0A inactivation mutant stops producing spores and solvent, while over-expression of spo0A gene can enhance solvent production since 0A binding boxes have been identified in the promoter regions of solvent formation genes [26]. Therefore, it is speculative that the higher expression of spo0A could be one of the reasons for the higher butanol formation in EA 2018. Comparative transcriptomic analysis confirmed our speculation that higher transcriptional level of spo0A was found in EA 2018 (Table 4 and Figure 3). There were three ways for Spo0A to be phosphorylated (i.e. by a sensory kinase; through a novel phosphorylation system; by butyryl-P or acetyl-P) [27]. Using trans-membrane domain along with transcriptional analysis, four orphan kinases (CAC0437, CAC0323, CAC0903, and CAC2730) were identified as plausible kinases that might phosphorylate Spo0A in B. subtilis [27]. In our study, we also found that the transcriptional level of an orphan kinase CEA_G0344 (corresponding to CAC0323 in ATCC 824) was higher in EA 2018 (Figure 3), and the result is consistent well with the spo0A gene expression data.
During solvent fermentation process in C. acetobutylicum, a considerable amount of NADH was consumed by hydrogenase via reduced Fd (FeH2) to form hydrogen [28]. Previous reports showed that butanol production by C. acetobutylicum can be elevated by inhibiting hydrogen formation through adding viologen dyes or increasing hydrogen partial pressure [28], and knockdown of hupCBA cluster which encoded hydrogen uptake genes in C. saccharoperbutylacetonicum strain N1-4 decreased butanol formation (to 75.6% compared to the control strain) successfully [29]. Biochemical analysis showed that hydrogen formation in EA 2018 was nearly 29% lower than in ATCC 824 (Table 6). Interestingly, comparative genomic analysis also revealed SNVs in NiFe-hydrogenase coded gene CEA_P0140 (CA_P0141 in ATCC 824) and in the promoter of Fe-only hydrogenase coded gene CEA_G0028 (CAC 0028 in ATCC 824) (Table 2, 3). The variation site of CEA_G 0028 was located on the 12th base upstream of the start codon of hydA and altered the ribosome binding site (RBS) of this important gene (GGGAGG in ATCC 824 versus AGGAGG in EA 2018). In addition, the higher expression level of hydrogen uptake genes mbhs and mbhl were also revealed in EA 2018 (Figure 3). The result showed that hydrogen uptake could be an important factor for butanol formation, and increased expression level of hydrogen uptake gene mbhs and mbhl was closely correlated to the lower hydrogen formation in EA 2018, which can eventually help balance the NAD(P)H needed for higher production of butanol.
Table 6.
Hydrogen production of C. acetobutylicum EA 2018 and C. acetobutylicum ATCC 824 in 6% glucose contained P2 medium
| Fermentation Time (h) |
Glucose consumed (mM) |
Hydrogen production (mM) |
Hydrogen formation ratio (mM/100 mM glucose) |
|
|---|---|---|---|---|
| EA 2018 | 72 | 260.6 ± 3.7 | 244.1 ± 1.3 | 93.9 ± 1.6 |
| ATCC 824 | 72 | 239.4 ± 5.0 | 315.4 ± 0.5 | 131.7 ± 3.1 |
Using non-replicating plasmid pO1X, putative solvent formation repressor solR gene was inactivated in ATTC 824, and its fermentation experiment revealed that more solvent were produced in the solR inactivation mutant [5]. Although there are different speculations on the function of SolR [25], it has been confirmed that low expression of solR will enhance solvent formation [26]. Transcriptomic analysis revealed a lower expression level of solR, especially in the solventogenic phase in EA 2018, which might be related to hyper-butanol formation (Figure 3).
It has been suggested that the onset of solvent production is closely related to the accumulation of acid end products [30], and the addition of acetate and butyrate might result in a rapid induction of solventogenesis [31]. For example, it was reported that the concentration of undissociated butyric acid might play an important role in the induction of solventogenesis [32]. Transcriptomic analysis showed that expression of ack, pta, buk and ptb were all lower in EA 2018 than in ATCC 824 (Figure 3), consistent with the biochemical analysis (Figure 1). In addition, the results also suggested that the transition to the solventogenesis took place at a lower acetate and butyrate acid concentration in EA 2018 compared to ATCC 824.
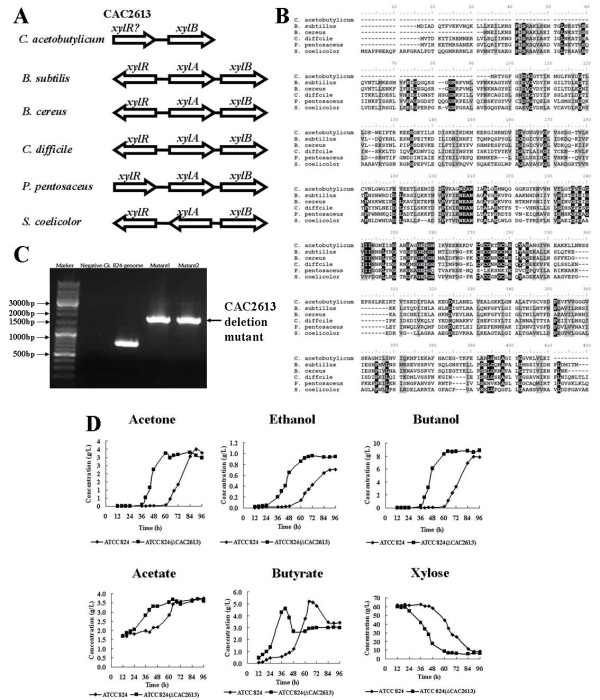
Analysis of substrate utilization genes and inactivation of CAC2613 revealed genetic bases of better xylose utilization in EA 2018
Solvent production from agriculturally based lignocellulosic substrates (i.e. cellulose or hemicellulose) was studied previously and results showed that a large part of the lignocellulosic substrates were hydrolyzed into glucose and xylose [33]. Therefore, utilization of these substrates, especially xylose, can be important in determining the efficiency of solvent production. Comparative genomic analysis identified several mutations in the putative promoters and within the coding region of genes which might be involved in substrates utilization (Table 2, 3). Among them, three out of seven mutated genes encode sugar-binding periplasmic proteins. One interesting gene was CEA_G2622 (CAC2613 in ATCC 824), which encodes a transcriptional regulator of NagC/XylR family and the sequence variation could cause a putative W90C substitution. The gene is located on the upstream of xylB (xylulose kinase) gene (Figure 4A). Since most of the known xylR genes in other AT-rich gram-positive species such as B. subtillus and C. difficile were located upstream of xyl operon [34,35], we speculated that this gene (CEA_G2622) may function similarly as xylR in EA 2018 (Figure 4B). Transcriptomic analysis showed that even in the glucose-based medium, the expression level of xylB was higher in EA 2018 (Figure 3). However, evidence is still needed to confirm the direct regulatory function of CEA_G2622 on xyl operon. To do so, we disrupted CAC2613 gene in C. acetobutylicum ATCC 824 (corresponding to CEA_G2622 in EA 2018) using Targetron system (Figure 4C). Batch fermentation showed that xylose utilization in CAC2613 disrupted mutant was faster than ATCC 824 (constant pH 5.0). In addition, the time of butanol formation and acids reassimilation in the mutant were 24 h earlier than ATCC 824 strain, although the final concentration of end products and xylose were nearly the same (Figure 4D). The similarities, in terms of the time of butanol formation and acid reassimilation, between EA 2018 and the CAC2613 disrupted ATCC 824 derived mutant, suggested that better xylose utilization in EA 2018 could be related to the mutation in CEA_G2622 (CAC2613 in ATCC 824).
Figure 4.
Inactivation of putative xylR CAC2613 in ATCC 824 leaded to faster xylose utilization in C. acetobutylicum. A: location of CAC2613 in C. acetobutylicum and xylR in different organisms; B: Alignment of CAC2613 and xylR in different organisms; C: CAC2613 inactivation in ATCC 824 using Targetron system; D: Accurate pH-controlled fermentation profile of mutant strain versus wild type in 6% xylose contained P2 medium.
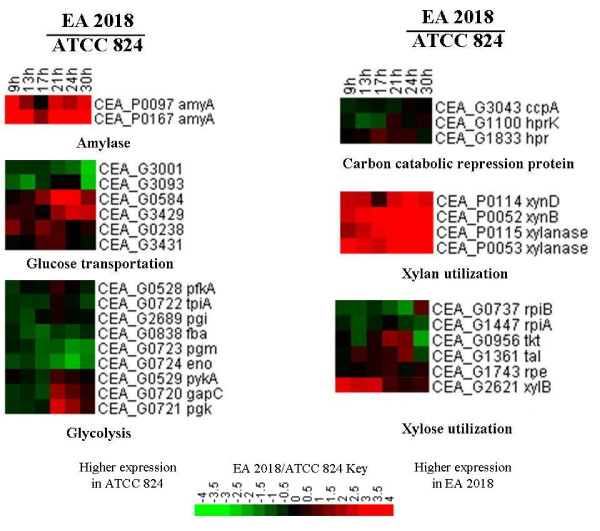
Previous study showed that ATCC 824 harbors extracellular and cell bound xylanase activities when grown under xylose or glucose-based media, and most of the putative xylanase encoded genes were located on the megaplasmid [36]. Two endoxylanase genes, thermostable xylanase 10A gene (CA_P0053 in ATCC 824 and CEA_P0052 in EA 2018) and xylanase 10B gene (CA_P0116 in ATCC 824 and CEA_P0115 in EA 2018) located on mega-plasmid were identified in EA 2018 [37]. In addition, transcriptomic analysis showed higher expression level of those two genes in EA 2018 (Figure 5). Xylan is the major component of hemicelluloses. The higher expression level of xylanase in EA 2018 could make it suitable for hemicellulosic fermentation, and could offer potential economic benefits in the future [1].
Figure 5.
Expression profiles of substrate utilization related genes. Gene functions were shown below the expression profile. Red and green indicated higher or lower expression, respectively.
Among all putative promoter variations, there were 4 sites which may affect substrate utilization in EA 2018 (Table 3). Among them, CEA_G3043 (CAC3037 in ATCC 824) gene that encodes a catabolite control protein (CcpA) has a variation site located on 80 bp upstream of the start codon. CcpA play an important role in catabolite repression and inactivation of this gene will release catabolic repression in many gram-positive organisms [8,38,39]. However, no significant regulation was observed for the expression level of CcpA gene in EA 2018, and the potential effect of the variation in ccpA promoter region still needs further investigation.
agrC and sigma factor variations may involve in spore formation in EA 2018
Comparative genomic analysis identified several genes which may be accounted for difference in terms of spore formation (Table 2). Among these muted genes, CEA_G2066 (CAC2052 in ATCC 824) encodes a putative sigma factor. It has been known that the transcription of this gene was closely related to spore-formation in ATCC 824 [40]. CEA_G2066 has a single nucleotide (A) insertion site in 687-688th bases, which altered the C-terminal protein sequence. Transcriptomic analysis showed that the transcription level of CEA_G2066 and other 4 putative sporulation related sigma factors [40] was lower in EA 2018 at 21 h, 24 h and 30 h (Figure 6), which might contribute to non-sporulation property in EA 2018.
Figure 6.
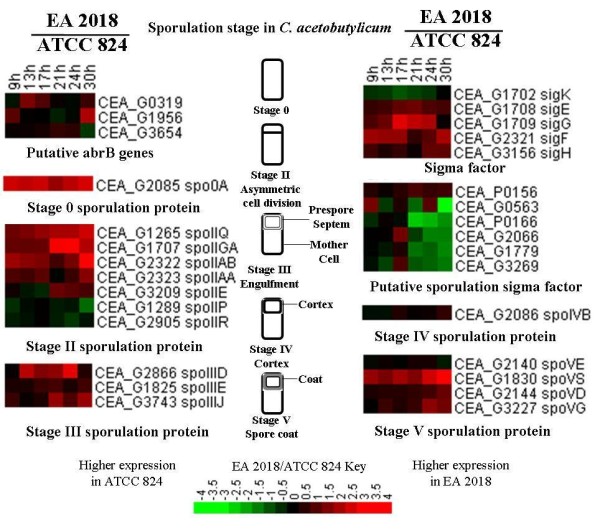
Expression profiles of sporulation related genes. Gene functions were shown below the expression profile. Red and green indicated higher or lower expression, respectively.
Quarum sensing is related to some important characteristics of bacteria, such as sporulation, virulence, and biofilm formation [41]. In C. perfringens, the virulence gene was regulated by agr system, and agrBD knockout mutant did not express theta-toxin gene, and transcription of the alpha- and kappa-toxin genes was also significantly decreased in the mutant strain [42]. A two-component system gene, CEA_G0080 (CAC0080 in ATCC 824), encoding a histidine kinase-like ATPase (AgrC), has a single nucleotide (A) insertion site in 1159-1160th bases when compared with agrC gene in ATCC 824, which may truncate the protein encoding sequence. In addition, the expression level of agrB (CEA_G0078) was constantly lower in EA 2018 (Figure 3). Early studies suggested that deletion of any agr system genes could result in no spore formation in ATCC 824 [43]. Therefore, mutation in agrC gene and lower expression of agrB gene in EA 2018 might be responsible for to the non-sporulation property in EA 2018.
Conclusion
A hyper-butanol, non spore-forming C. acetobutylicum EA 2018 strain we isolated previously can produce 10% more butanol than the type strain C. acetobutylicum ATCC 824 [23]. To seek molecular basis of these characteristics in EA 2018 strain, we completed the genome sequencing of this strain using 454 GS FLX pyrosequceing and performed a detailed genomic comparison with a C. acetobutylicum type strain ATCC 824. Although EA 2018 was found more than 99.8% identical to ATCC 824, 72 indels (i.e. insertions and deletions) and 451 SNVs were identified, some of which may be related to the enhanced butanol production in EA 2018. In addition, we performed a comparative transcriptomic analysis of C. acetobutylicum EA 2018 and ATCC 824 using oligonucleotide microarrays. The results showed that increased expression of several key genes related to solvent formation, and decreased expression of the acid formation related genes may be related to the enhanced butanol production in EA 2018. Furthermore, the results also showed that the variation in CEA_G2622 (CAC2613 in ATCC 824), a putative transcriptional regulator involved in xylose utilization, may be able to accelerate utilization of substarte xylose. The comparisons of hyper-butanol EA 2018 and type strain ATCC 824 at both genomic and transcriptomic levels not only improved our understanding of the hyper butanol-producing, xylose utilization as well as non-spore formation properties in EA 2018 strain, but also provided some useful clues for the future genetic modification of C. acetobutylicum to produce solvents, especially butanol more effectively.
Methods
Bacteria strain and genome sequencing
C. acetobutylicum EA 2018 (CCTCC M 94061) used for this study [22] is deposited in China Center for Type Culture Collection (http://www.cctcc.org/). C. acetobutylicum ATCC 824 was the wild type strain we purchased from American Type Culture Collection (ATCC) [23]. The C. acetobutylicum EA 2018 was grown anaerobically (Thermo Electron Crop., San Jose, USA). Colonies picked from Clostridia growth medium (CGM) plate were inoculated into 5 ml liquid CGM and cultured at 37°C overnight [44], and then the cells were transferred into 100 ml CGM and incubated at 37°C for 16-20 h until they reach late-exponential phase. Cells collected were used for chromosomal DNA isolation as described previously [45]. Roche 454 GS FLX pyrosequencing was used to sequence the DNA. A total of 60.3 Megabases was generated, with an average read length of 200 bp. The GS FLX reads were assembled into a total of 198 contigs using a GS de novo assembler, among them, 157 contigs are larger than 500 bp. The gaps were closed by PCR procedure using the ATCC 824 genome sequence as reference. The large PCR products were sequenced via primer walking. The whole sequence was assembled by using the software phredPhrap (http://www.phrap.org) and was visualized by Consed [46]. The low-quality sequences were verified by PCR resequencing using ABI 3730 (Applied Biosystem Inc.). The sequence accuracy of the final genome was 99.9919%. All the variation between EA 2018 and ATCC 824 were verified by PCR resequencing using ABI 3730.
Genome annotation and bioinformatic analysis
CDSs were identified by combining the results of ZCURVE 1.0 [47] and Glimmer 3.2 (http://www.cbcb.umd.edu/software/glimmer). Transfer RNA genes were predicted by tRNAscan-SE [48]. Functional annotation of CDSs was performed through comparison with NCBI non-redundant protein database using BLASTP, followed by manual curation. Comparative genomic analysis was performed by using the Artemis Comparison Tool (ACT; http://www.sanger.ac.uk/resources/software/act/). The atlas of the genome is drawn by using GenomeViz1.1 [49].
Nucleotide sequence accession number
The annotated genome sequence has been deposited into GenBank under accession no. CP002118 (Chromosome) and no. CP002119 (Plasmid).
Oligonucleotide microarray experiments
Cells for RNA isolation were grown on P2 medium and collected at 9 h, 13 h, 17 h, 21 h, 24 h and 30 h by centrifugation at 4°C and 4500 × g for 10 min. Total RNA was extracted and purified by using Trizol (Invitrogen, Carlsbad, CA, USA) and RNeasy cleanup kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's protocol. The total RNA yield was quantified by spectrophotometric analysis (NanoDrop Technology, Cambridge, UK) and the quality was verified by gel electrophoresis. Agilent oligonucleotide microarrays technology was used for monochromic analysis, in which probes (size: 60 bp; three replicates for each ORF) from the two groups were labeled by incorporation of cyanine 3 (Cy3) (Agilent Technologies, Palo Alto, CA, USA). The experiment procedures and data normalization were performed using the methods described previously [50]. Average linkage hierarchical clustering was performed using Cluster 3.0, and gene clusters were visualized in Treeview [51].
Oligonucleotide microarray accession number
The Oligonucleotide microarray data has been deposited into GEO under accession no. GSE23071.
Gene disruption
Gene disruption in C. acetobutylicum ATCC 824 was performed as described previously [11]. The disruption procedures were shown in Additional file 9. The selected site for CAC2613 disruption was 532/533a, where the group II intron will insert into gene CAC2613 coding region between amino acid 532 and 533 sites in the antisense direction. The primers (CAC2613-532-533-IBS, CAC2613-532-533-EBS1d and CAC2613-532-533-EBS2), for retargeting the RNA portion of the intron for C. acetobutylicum CAC2613 gene disruption, are listed in Additional file 10. The 350 bp targetron fragment was obtained by PCR based on the plasmid pACD4K-C and protocol provided by the TargeTron™ Gene Knockout System Kit (Sigma-Aldrich, St Louis, MO, USA). The 350 bp PCR fragment was digested with XhoI and BsrGI, and then inserted into pSY6 [11] digested with the same restriction enzymes, to generate the plasmid pSY6-2613. The plasmid pSY6-2613 was methylated in E. coli ER2275 (pANS1) first [52], and then electroporated into C. acetobutylicum ATCC 824. Cells were plated on CGM agar containing 50 μg/mL erythromycin and incubated at 37°C for about 2-3 days. The positive transformants containing the inserted intron were identified by colony PCR, using primers CAC2613-ID-fw and CAC2613-ID-rev (Additional file 10).
Fermentation conditions
Solvent production and sugar utilization of C. acetobutylicum ATCC 824 and C. acetobutylicum EA 2018 were determined when the cultures were grown on P2 medium [53]. 6% Glucose or xylose contained P2 solution I (840 ml) and KH2PO4 (0.5 g/L), K2HPO4 (0.5 g/L), CH3COONH4 (2.2 g/L) contained P2 solution II (100 ml) were boiled for 20 min, then cooled by flushing O2-free N2 gas, and autoclaved at 121°C for 15 min separately. After autoclaving, 100 ml P2 solution II, 10 ml filter-sterilized P2 medium stock solution III (MgSO4·7H2O, 20 g/L; MnSO4·H2O, 1 g/L; FeSO4·7H2O, 1 g/L; NaCl, 1 g/L) and 1 ml solution IV (Para-amino-benzoicacid, 0.1 g/L; thiamin, 0.1 g/L; biotin, 0.001 g/L) were added into P2 solution I. The stock solutions were filter sterilized through a 0.2 μm pore-size filter. An inoculum of 5% from a CGM grown culture was typically used. Batch fermentation was carried out in 250 ml sealed bottle with 100 ml medium. 1 ml samples were taken every 12 h and analyzed for solvent and sugar. Accurate pH-controlled fermentations were carried out in BioFlo 110 bioreactors with 1.5 L working volume (New Brunswick Scientific, Edison, NJ). The pH control was achieved by using 9% (w/v) aqueous ammonia. Anaerobic conditions of fermentors were maintained through aeration of filtered nitrogen.
Analytical methods
The surface morphology of C. acetobutylicum EA 2018 was studied using a JSM-6360 scanning electron microscope (JEOL Co. Ltd. Japan). Spore formation analysis was performed by growing the cells in P2 medium. After 48 h fermentation, cells were collected by centrifuging at 10,000 × g for 5 min, and stained with crystal violet for imaging analysis using a U-CTR30-2 microscope (Olympus Optical Co. Ltd. Japan). Butanol, acetone, ethanol, acetic acid and butyric acid were determined using a gas chromatograph (7890A, Agilent, Wilmington, DE, USA) equipped with a capillary column (Alltech EC™-WAX) and a flame ionization detector. The analysis was carried out under the following conditions: oven temperature, programmed from 80 to 140°C at a rate of 25°C/min; injector temperature, 200°C; detector temperature, 200°C; nitrogen (carrier gas) flow rate, 13 ml/min; hydrogen flow rate, 20 ml/min; air flow rate, 140 ml/min. Total solvent was defined as the sum of ABE. Isobutylalcohol and isobutyric acid were used as the internal standards for ABE and acid determination, respectively. Glucose and xylose were determined using a HPLC system (Model 1200, Agilent) equipped with a sugar-pak I column (Waters) and a refractive index detector. The analysis was carried out with water as mobile phase at a rate of 0.6 ml/min, and the column temperature was set up at 70°C. The composition of the gas produced (mainly H2 and CO2) was measured using the method described previously [54].
Abbreviations
ABE: Acetone-Butanol-Ethanol; ATCC: American Type Culture Collection; CCTCC: China Center for Type Culture Collection; ORF: Open Reading Frames; NTG: N-methyl-N-nitro-N-nitrosoguanidine; SNVs: Single Nucleotide Variations; RBS: Ribosome Binding Site; CcpA: Catabolite Control Protein A; CGM: Clostridia Growth Medium.
Authors' contributions
SYH, YLY, GPZ, SY, WHJ conceived of the study. SYH performed the genome sequencing, microarray samples preparation and gene disruption. SYH and YG performed the strain fermentation and analysis. JBZ performed the spore formation analysis. SYH, HJZ and SYW performed the genome annotation and analysis work. SYH and WWZ drafted the manuscript. All authors contributed to and approved the final manuscript.
Supplementary Material
Genome finishing procedures of EA 2018. The file describes the genome finishing process including gap closure, weak regions and homopolymer sites verification.
Weak region verification primers. The file lists the primers used in genome weak region verification procedure.
Variation places between EA 2018 and ATCC 824 verification primers. This file lists the verification primers for all of the indels sites and SNVs sites found between EA 2018 and ATCC 824.
Deletion and insertion sites. The file lists a total of 46 deletion sites and 26 insertion sites between EA 2018 and ATCC 824.
SNV sites. The file lists 440 single nucleotide variations between EA 2018 and ATCC 824.
Protein variations. The file shows the protein sequence variations affected by deletion, insertion or SNVs between EA 2018 and ATCC 824.
Predicted promoter variations. The file shows the predicted promoter variations between EA 2018 and ATCC 824.
DNA array data. The file lists the complete set of DNA array data of EA 2018 versus ATCC 824.
The scheme of targeted gene disruption using group II intron. The file displays the gene disruption procedure using group II intron.
Targeted gene disruption and verification primers. The file lists the primers for retargeting the RNA portion of the intron for C. acetobutylicum CAC2613 gene disruption and for identifying the insertion mutant..
Contributor Information
Shiyuan Hu, Email: husy@sibs.ac.cn.
Huajun Zheng, Email: zhenghj@chgc.sh.cn.
Yang Gu, Email: ygu02@sibs.ac.cn.
Jingbo Zhao, Email: jbzhao@sibs.ac.cn.
Weiwen Zhang, Email: Weiwen.Zhang@asu.edu.
Yunliu Yang, Email: ylyang@sibs.ac.cn.
Shengyue Wang, Email: wangsy@chgc.sh.cn.
Guoping Zhao, Email: gpzhao@sibs.ac.cn.
Sheng Yang, Email: syang@sibs.ac.cn.
Weihong Jiang, Email: whjiang@sibs.ac.cn.
Acknowledgements
This work was supported by National Basic Research Program of China (2007CB707803, 2011CBA00806), Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-G-1, KSCX2-EW-J-12), National Natural Science Foundation of China (31070075) and Key Program of Science and Technology Commission of Shanghai Municipality (08DZ1207100).
We thank Prof. Peter Duerre for providing the manipulation methods on Clostridium acetobutylicum. We thank Jun Chen, Huiqi He, Lijun Shao, Cong Ren, Yongqiang Zhu, Qiuping Hu, Yiwei Yang and Qing Tang for their kindly help on this project.
During the review process we realized there is a concern from one of the reviewers regarding the origin of EA 2018 used in this research due to its high genome sequence similarity with ATCC 824. Although all the historical records we know about indicated that this is an independently isolated strain [23], at this moment we would like to keep the discussion regarding its origin ongoing until further evidences emerge.
References
- Durre P. Biobutanol: an attractive biofuel. J Biotechnol. 2007;2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- Durre P. New insights and novel developments in clostridial acetone/butanol/isopropanol fermentation. Appl Microbiol Biotechnol. 1998;49:639–648. doi: 10.1007/s002530051226. [DOI] [Google Scholar]
- Demain AL, Newcomb M, Wu JHD. Cellulase, Clostridia, and Ethanol. Microbiol Mol Biol Rev. 2005;69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Fermentative butanol production by Clostridia. Biotechnol Bioeng. 2008;101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- Nair RV, Green EM, Watson DE, Bennett GN, Papoutsakis ET. Regulation of the sol locus genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 by a putative transcriptional repressor. J Bacteriol. 1999;181:319–330. doi: 10.1128/jb.181.1.319-330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorwa MF, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of the putative hydrogenase gene of Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:2668–2675. doi: 10.1128/jb.178.9.2668-2675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton ZL, Bennet GN, Rudolph FB. Cloning, sequencing, and expression of clustered genes encoding beta-hydroxybutyryl-coenzyme A (CoA) dehydrogenase, crotonase, and butyryl-CoA dehydrogenase from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1996;178:3015–3024. doi: 10.1128/jb.178.11.3015-3024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueck CJ, Kraus A, Schmiedel D, Hillen W. Cloning, expression and functional analyses of the catabolite control protein CcpA from Bacillus megaterium. Mol Microbiol. 1995;16:855–864. doi: 10.1111/j.1365-2958.1995.tb02313.x. [DOI] [PubMed] [Google Scholar]
- Sillers R, Al-Hinai MA, Papoutsakis ET. Aldehyde-alcohol dehydrogenase and/or thiolase overexpression coupled with CoA transferase downregulation lead to higher alcohol titers and selectivity in Clostridium acetobutylicum fermentations. Biotechnol Bioeng. 2009;102:38–49. doi: 10.1002/bit.22058. [DOI] [PubMed] [Google Scholar]
- Papoutsakis ET. Engineering solventogenic clostridia. Curr Opin Biotechnol. 2008;19:420–429. doi: 10.1016/j.copbio.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Shao L, Hu S, Yang Y, Gu Y, Chen J, Yang Y, Jiang W, Yang S. Targeted gene disruption by use of a group II intron (targetron) vector in Clostridium acetobutylicum. Cell Res. 2007;17:963–965. doi: 10.1038/cr.2007.91. [DOI] [PubMed] [Google Scholar]
- Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao SM, Luo YAM, Zhang TR, Li JS, Bao GAH, Zhu Y, Chen ZG, Zhang YP, Li Y, Ma YH. Proteome Reference Map and Comparative Proteomic Analysis between a Wild Type Clostridium acetobutylicum DSM 1731 and its Mutant with Enhanced Butanol Tolerance and Butanol Yield. J Proteome Res. 2010;9:3046–3061. doi: 10.1021/pr9012078. [DOI] [PubMed] [Google Scholar]
- Shi Z, Blaschek HP. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 and the hyper-butanol-producing mutant BA101 during the shift from acidogenesis to solventogenesis. Appl Environ Microbiol. 2008;74:7709–7714. doi: 10.1128/AEM.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes CJ, Senger RS, Spath WS, Borden JR, Sillers R, Papoutsakis ET. A general framework for designing and validating oligomer-based DNA microarrays and its a application to Clostridium acetobutylicum. Appl Environ Microbiol. 2007;73:4631–4638. doi: 10.1128/AEM.00144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L, Bennett GN. Proteome analysis and comparison of Clostridium acetobutylicum ATCC 824 and Spo0A strain variants. J Ind Microbiol Biotechnol. 2006;33:298–308. doi: 10.1007/s10295-005-0050-7. [DOI] [PubMed] [Google Scholar]
- Lee J, Yun H, Feist AM, Palsson BO, Lee SY. Genome-scale reconstruction and in silico analysis of the Clostridium acetobutylicum ATCC 824 metabolic network. Appl Microbiol Biotechnol. 2008;80:849–862. doi: 10.1007/s00253-008-1654-4. [DOI] [PubMed] [Google Scholar]
- Ryan S, Senger ETP. Genome-Scale Model for Clostridium acetobutylicum. Part 2: Development of Specific Proton Flux States and Numerically-Determined Sub-Systems. Biotechnol Bioeng. 2008;101:1053–1071. doi: 10.1002/bit.22009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S, Senger ETP. Genome-scale model for Clostridium acetobutylicum. Part 1: Metabolic network resolution and analysis. Biotechnol Bioeng. 2008;101:1036–1052. doi: 10.1002/bit.22010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durre P, Fischer RJ, Kuhn A, Lorenz K, Schreiber W, Sturzenhofecker B, Ullmann S, Winzer K, Sauer U. Solventogenic enzymes of Clostridium acetobutylicum: catalytic properties, genetic organization, and transcriptional regulation. FEMS Microbiol Rev. 1995;17:251–262. doi: 10.1016/0168-6445(95)00006-X. [DOI] [PubMed] [Google Scholar]
- Annous BA, Blaschek HP. Isolation and characterization of Clostridium acetobutylicum mutants with enhanced amylolytic activity. Appl Environ Microbiol. 1991;57:2544–2548. doi: 10.1128/aem.57.9.2544-2548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen J, Yang Y, Jiao R. Breeding high-ration butanol strains of Clostridium acetobutylicum and application to industrial production. Industrial Microbiology (in Chinese) 1996;26:1–6. [Google Scholar]
- Chiao JS, Sun ZH. History of the Acetone-Butanol-Ethanol Fermentation Industry in China: Development of Continuous Production Technology. J Mol Microbiol Biotechnol. 2007;13:12–14. doi: 10.1159/000103592. [DOI] [PubMed] [Google Scholar]
- Fischer RJ, Helms J, Durre P. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol. 1993;175:6959–6969. doi: 10.1128/jb.175.21.6959-6969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Meynial-Salles I, Girbal L, Yang XH, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris LM, Welker NE, Papoutsakis ET. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes CJ, Alsaker KV, Papoutsakis ET. A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol. 2005;3:969–978. doi: 10.1038/nrmicro1288. [DOI] [PubMed] [Google Scholar]
- Rao G, Mutharasan R. Altered Electron Flow in Continuous Cultures of Clostridium acetobutylicum Induced by Viologen Dyes. Appl Environ Microbiol. 1987;53:1232–1235. doi: 10.1128/aem.53.6.1232-1235.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Si, Kosaka T, Hirakawa H, Matsuura K, Yoshino S, Furukawa K. Metabolic engineering for solvent productivity by downregulation of the hydrogenase gene cluster hupCBA in Clostridium saccharoperbutylacetonicum strain N1-4. Appl Microbiol Biotechnol. 2008;78:483–493. doi: 10.1007/s00253-007-1323-z. [DOI] [PubMed] [Google Scholar]
- Haggstrom L. Acetone-butanol fermentation and its variants. Biotechnol Adv. 1985;3:13–28. doi: 10.1016/0734-9750(85)90003-5. [DOI] [PubMed] [Google Scholar]
- Grupe H, Gottschalk G. Physiological Events in Clostridium acetobutylicum during the Shift from Acidogenesis to Solventogenesis in Continuous Culture and Presentation of a Model for Shift Induction. Appl Environ Microbiol. 1992;58:3896–3902. doi: 10.1128/aem.58.12.3896-3902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monot F, Engasser JM, Petitdemange H. Influence of pH and undissociated butyric acid on the production of acetone and butanol in batch cultures of Clostridium acetobutylicum. Appl Microbiol Biotechnol. 1984;19:422–426. doi: 10.1007/BF00454381. [DOI] [Google Scholar]
- Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008;10:295–304. doi: 10.1016/j.ymben.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Kreuzer P, Gartner D, Allmansberger R, Hillen W. Identification and Sequence-Analysis of the Bacillus-Subtilis W23 Xylr Gene and Xyl Operator. J Bacteriol. 1989;171:3840–3845. doi: 10.1128/jb.171.7.3840-3845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA, Gelfand MS. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol Lett. 2001;205:305–314. doi: 10.1111/j.1574-6968.2001.tb10965.x. [DOI] [PubMed] [Google Scholar]
- Lee SF, Forsberg CW, Gibbins LN. Xylanolytic Activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:1068–1076. doi: 10.1128/aem.50.4.1068-1076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MK, Rudolph FB, Bennett GN. Characterization of thermostable Xyn10A enzyme from mesophilic Clostridium acetobutylicum ATCC 824. J Ind Microbiol Biotechnol. 2005;32:12–18. doi: 10.1007/s10295-004-0192-z. [DOI] [PubMed] [Google Scholar]
- Kraus A, Hillen W. Analysis of CcpA mutations defective in carbon catabolite repression in Bacillus megaterium. FEMS Microbiol Lett. 1997;153:221–226. doi: 10.1111/j.1574-6968.1997.tb10485.x. [DOI] [PubMed] [Google Scholar]
- Chauvaux S. CcpA and HPr(ser-P): mediators of catabolite repression in Bacillus subtilis. Res Microbiol. 1996;147:518–522. doi: 10.1016/0923-2508(96)84006-X. [DOI] [PubMed] [Google Scholar]
- Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 2008;9:R114. doi: 10.1186/gb-2008-9-7-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni NT, Li MY, Wang JF, Wang BH. Inhibitors and Antagonists of Bacterial Quorum Sensing. Med Res Rev. 2009;29:65–124. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Yuan Y, Hassan S, Wang R, Wang Y, Shimizu T. Virulence Gene Regulation by the agr System in Clostridium perfringens. J Bacteriol. 2009;191:3919–3927. doi: 10.1128/JB.01455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer K. BBSRC China partnering award workshop on improving biobutanol production by solventogenic clostridia. Shanghai China; 2009. Quorum sensing in solventogenic clostridia. [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Thiolase from Clostridium acetobutylicum ATCC 824 and Its Role in the Synthesis of Acids and Solvents. Appl Environ Microbiol. 1988;54:2717–2722. doi: 10.1128/aem.54.11.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Garcia MJ, Lathigra R, Allen B, Moreno C, Vanembden JDA, Young D. Alterations in the Superoxide-Dismutase Gene of an Isoniazid-Resistant Strain of Mycobacterium tuberculosis. Infect Immun. 1992;60:2160–2165. doi: 10.1128/iai.60.6.2160-2165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Guo FB, Zhang CT. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics. 2006;7:9. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Hain T, Chakraborty T. GenomeViz: visualizing microbial genomes. BMC Bioinformatics. 2004;5:198. doi: 10.1186/1471-2105-5-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SY, Shen XY, Wu CY, Pan F, Shen YY, Sheng HH, Chen XM, Gao HJ. Analysis of whole genomic expression profiles of Helicobacter pylori related chronic atrophic gastritis with IL-1B-31CC/-511TT genotypes. Digest Dis. 2009;10:99–106. doi: 10.1111/j.1751-2980.2009.00367.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein LD, Papoutsakis ET. In vivo methylation in Escherichia coli by the Bacillus subtilis phage Φ3T I methyltransferase to protect plasmids from restriction upon transformation of Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1993;59:1077–1081. doi: 10.1128/aem.59.4.1077-1081.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annous BA, Blaschek HP. Regulation and localization of amylolytic enzymes in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1990;56:2559–2561. doi: 10.1128/aem.56.8.2559-2561.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao YZ, He YL, Wu YQ, Liu FH, Li XF, Zong WM, Zhou ZH. Characteristics of a new photosynthetic bacterial strain for hydrogen production and its application in wastewater treatment. Int J Hydrogen Energy. 2008;33:963–973. doi: 10.1016/j.ijhydene.2007.11.021. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genome finishing procedures of EA 2018. The file describes the genome finishing process including gap closure, weak regions and homopolymer sites verification.
Weak region verification primers. The file lists the primers used in genome weak region verification procedure.
Variation places between EA 2018 and ATCC 824 verification primers. This file lists the verification primers for all of the indels sites and SNVs sites found between EA 2018 and ATCC 824.
Deletion and insertion sites. The file lists a total of 46 deletion sites and 26 insertion sites between EA 2018 and ATCC 824.
SNV sites. The file lists 440 single nucleotide variations between EA 2018 and ATCC 824.
Protein variations. The file shows the protein sequence variations affected by deletion, insertion or SNVs between EA 2018 and ATCC 824.
Predicted promoter variations. The file shows the predicted promoter variations between EA 2018 and ATCC 824.
DNA array data. The file lists the complete set of DNA array data of EA 2018 versus ATCC 824.
The scheme of targeted gene disruption using group II intron. The file displays the gene disruption procedure using group II intron.
Targeted gene disruption and verification primers. The file lists the primers for retargeting the RNA portion of the intron for C. acetobutylicum CAC2613 gene disruption and for identifying the insertion mutant..