Abstract
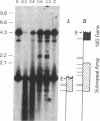
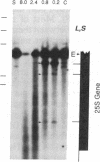
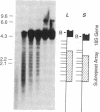
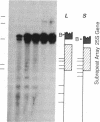
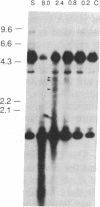
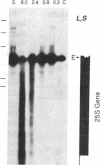
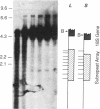
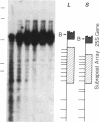
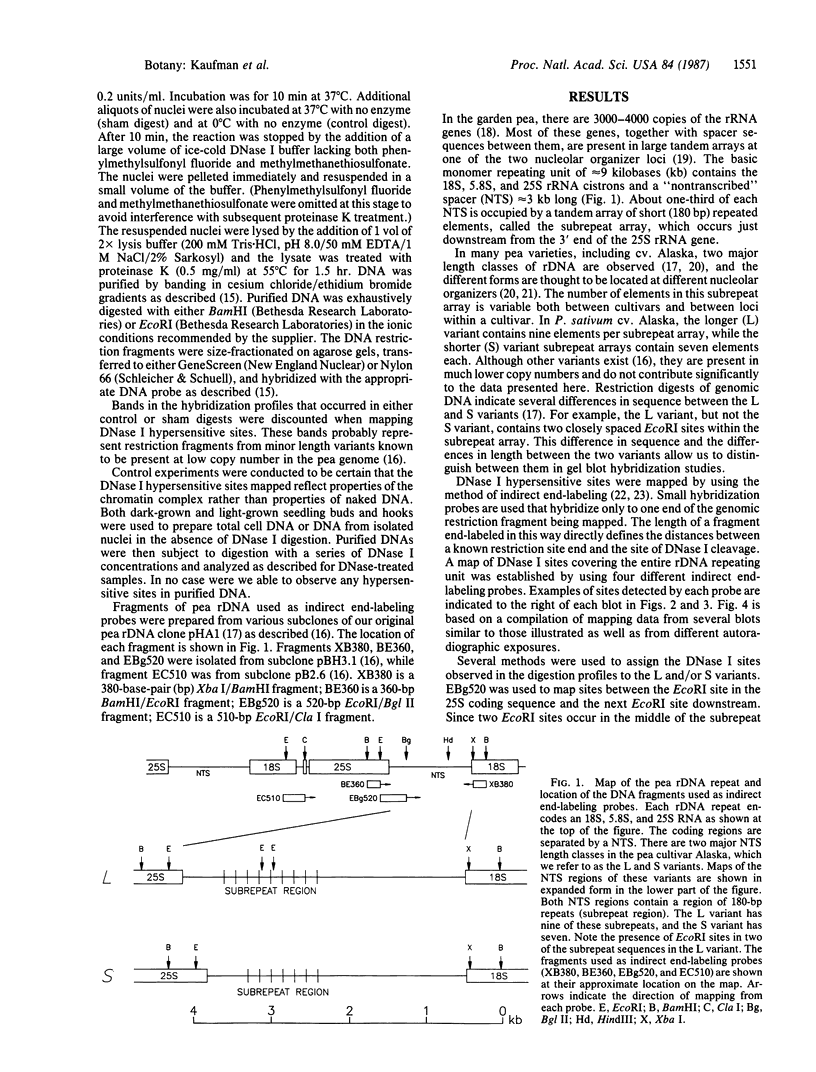
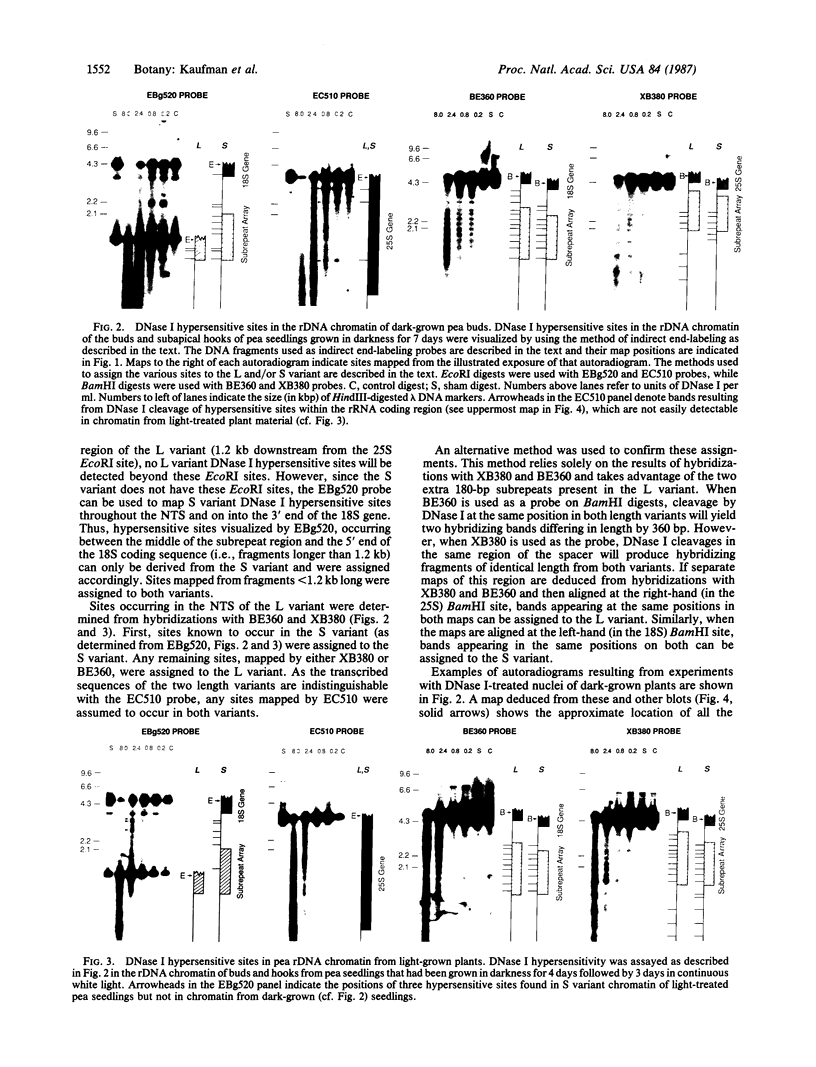
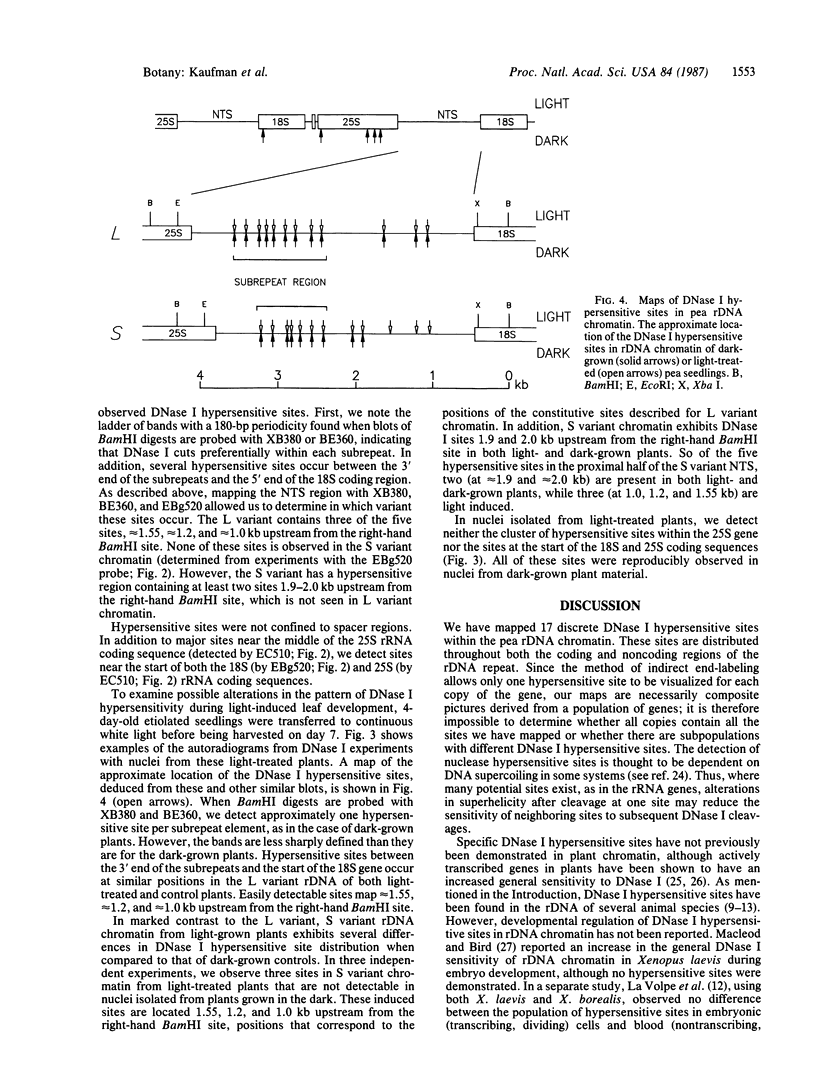
We have examined the rDNA chromatin of Pisum sativum plants grown with or without exposure to light for the presence of DNase I hypersensitive sites and possible developmental changes in their distribution. Isolated nuclei from pea seedlings were incubated with various concentrations of DNase I. To visualize the hypersensitive sites, DNA purified from these nuclei was restricted and analyzed by gel blot hybridization. We find that several sites exist in both the coding and noncoding regions of rDNA repeating units. Several of the sites in the nontranscribed spacer region are present in the light but are absent in the dark. Conversely, the hypersensitive sites within the mature rRNA coding regions are present in the dark but absent in the light. There are two major length variants of the rRNA genes in P. sativum var. Alaska. The sites in the nontranscribed spacer region that appear during the light treatment occur only in the shorter of these two length variants in this cultivar.
Keywords: peas, chromatin, photoregulation, DNA, nucleolar organizer
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonven B., Westergaard O. DNase I hypersensitive regions correlate with a site-specific endogenous nuclease activity on the r-chromatin of Tetrahymena. Nucleic Acids Res. 1982 Dec 11;10(23):7593–7608. doi: 10.1093/nar/10.23.7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchsenius S., Bonven B., Leer J. C., Westergaard O. Nuclease-sensitive regions on the extrachromosomal r-chromatin from Tetrahymena pyriformis. Eur J Biochem. 1981 Jul;117(2):245–250. doi: 10.1111/j.1432-1033.1981.tb06329.x. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Dover G. A. Multiple Pol I initiation sequences in rDNA spacers of Drosophila melanogaster. Nucleic Acids Res. 1982 Nov 11;10(21):7017–7026. doi: 10.1093/nar/10.21.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Gallagher T. F., Ellis R. J. Light-stimulated transcription of genes for two chloroplast polypeptides in isolated pea leaf nuclei. EMBO J. 1982;1(12):1493–1498. doi: 10.1002/j.1460-2075.1982.tb01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle J., Timmis J. N., Sinclair J. The Relationship between Satellite Deoxyribonucleic Acid, Ribosomal Ribonucleic Acid Gene Redundancy, and Genome Size in Plants. Plant Physiol. 1975 Mar;55(3):496–501. doi: 10.1104/pp.55.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman L. S., Briggs W. R., Thompson W. F. Phytochrome control of specific mRNA levels in developing pea buds : the presence of both very low fluence and low fluence responses. Plant Physiol. 1985 Jun;78(2):388–393. doi: 10.1104/pp.78.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Volpe A., Taggart M., McStay B., Bird A. DNaseI-hypersensitive sites at promoter-like sequences in the spacer of Xenopus laevis and Xenopus borealis ribosomal DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5361–5380. doi: 10.1093/nar/11.16.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E. O., Dawid I. B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- Macleod D., Bird A. DNAase I sensitivity and methylation of active versus inactive rRNA genes in xenopus species hybrids. Cell. 1982 May;29(1):211–218. doi: 10.1016/0092-8674(82)90105-2. [DOI] [PubMed] [Google Scholar]
- Moss T., Mitchelson K., de Winter R. The promotion of ribosomal transcription in eukaryotes. Oxf Surv Eukaryot Genes. 1985;2:207–250. [PubMed] [Google Scholar]
- Murtif V. L., Rae P. M. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985 May 10;13(9):3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösinger E., Batschauer A., Schäfer E., Apel K. Phytochrome control of in vitro transcription of specific genes in isolated nuclei from barley (Hordeum vulgare). Eur J Biochem. 1985 Feb 15;147(1):137–142. doi: 10.1111/j.1432-1033.1985.tb08729.x. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Enhancers and ribosomal gene spacers. Cell. 1984 Sep;38(2):349–351. doi: 10.1016/0092-8674(84)90489-6. [DOI] [PubMed] [Google Scholar]
- Silverthorne J., Tobin E. M. Demonstration of transcriptional regulation of specific genes by phytochrome action. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1112–1116. doi: 10.1073/pnas.81.4.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiker S., Murray M. G., Thompson W. F. DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc Natl Acad Sci U S A. 1983 Feb;80(3):815–819. doi: 10.1073/pnas.80.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardy A., Louis C., Han S., Schedl P. Ribosomal RNA genes of Drosophila melanogaster have a novel chromatin structure. J Mol Biol. 1984 May 15;175(2):113–130. doi: 10.1016/0022-2836(84)90470-4. [DOI] [PubMed] [Google Scholar]
- Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985 Oct;42(3):705–711. doi: 10.1016/0092-8674(85)90267-3. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]