Abstract
Spinocerebellar ataxia 3 (SCA3) is a genetic disorder resulting from the expansion of the CAG repeats in the ATXN3 gene. The pathogenesis of SCA3 is based on the toxic function of the mutant ataxin-3 protein, but the exact mechanism of the disease remains elusive. Various types of transgenic mouse models explore different aspects of SCA3 pathogenesis, but a knock-in humanized mouse has not yet been created. The initial aim of this study was to generate an ataxin-3 humanized mouse model using a knock-in strategy. The human cDNA for ataxin-3 containing 69 CAG repeats was cloned from SCA3 patient and introduced into the mouse ataxin-3 locus at exon 2, deleting it along with exon 3 and intron 2. Although the human transgene was inserted correctly, the resulting mice acquired the knock-out properties and did not express ataxin-3 protein in any analyzed tissues, as confirmed by western blot and immunohistochemistry. Analyses of RNA expression revealed that the entire locus consisting of human and mouse exons was expressed and alternatively spliced. We detected mRNA isoforms composed of exon 1 spliced with mouse exon 4 or with human exon 7. After applying 37 PCR cycles, we also detected a very low level of the correct exon 1/exon 2 isoform. Additionally, we confirmed by bioinformatic analysis that the structure and power of the splicing site between mouse intron 1 and human exon 2 (the targeted locus) was not changed compared with the native mouse locus. We hypothesized that these splicing aberrations result from the deletion of further splicing sites and the presence of a strong splicing site in exon 4, which was confirmed by bioinformatic analysis. In summary, we created a functional ataxin-3 knock-out mouse model that is viable and fertile and does not present a reduced life span. Our work provides new insights into the splicing characteristics of the Atxn3 gene and provides useful information for future attempts to create knock-in SCA3 models.
Electronic supplementary material
The online version of this article (doi:10.1007/s12017-010-8137-3) contains supplementary material, which is available to authorized users.
Keywords: Ataxin-3, Mouse model, Knock-in, Knock-out, CAG repeats, Splicing
Background
Spinocerebellar ataxia 3 (SCA3), which is also called Machado–Joseph disease (MJD), is a dominant genetic disorder that results from the expansion of the CAG repeats in the coding sequence of the ATXN3 gene (Coutinho and Andrade 1978; Kawaguchi et al. 1994) (MJD and ATXN3: OMIM 109150 and 607047). The majority of patients suffering from SCA3 carry one allele of ATXN3 with 60–82 CAG repeats and a second allele containing the normal number of repeats, which is usually between 13 and 41 (Kawaguchi et al. 1994; Giunti et al. 1995). Expression of the mutant allele leads to the onset of neurological symptoms, typically in the third or fourth decade of life. SCA3 patients commonly suffer from motor and mental abnormalities, such as gait ataxia, ocular symptoms, and dementia (Coutinho et al. 1982; Ishikawa et al. 2002; Kawai et al. 2004; França et al. 2007, 2008; Riess et al. 2008).
Although the relevant pathogenesis of SCA3 is based on the toxic function of the mutant ataxin-3 protein, the exact mechanism of disease remains elusive (Paulson 2007; Riess et al. 2008). Expanded CAG repeats in the ataxin-3 mRNA result in long polyglutamine (polyQ) tracts within the protein, resulting in protein misfolding. These changes may affect features, such as protein binding, subcellular localization of the protein, and its degradation and elimination from the cell (Chow et al. 2004; Lessing and Bonini 2008). In the misfolded protein, the polyglutamine chains are exposed and can scaffold normal and mutant ataxin-3 as well as important proteins, such as histone acetylases, deacetylases, transcription factors, and the proteasome complex (Evert et al. 2006; Bilen and Bonini 2007; Ferro et al. 2007; Lessing and Bonini 2008; Jia et al. 2008). In turn, the scaffolding and subsequent events can affect gene expression, change the cellular localization of different proteins, and lead to the dysfunction of the protein elimination pathway in cells.
Knock-out, transgenic, and YAC transgenic mouse models have been created to explore various aspects of SCA3 pathogenesis. The transgenic animals in these studies were generated by the random integration of a transgene in the form of an artificial construct that contained either full-length human ataxin-3 cDNA or some fragment of the gene. The transgene was usually driven by a different promoter, such as a Purkinje-specific L7 promoter (Ikeda et al. 1996). Model animals containing the polyQ fragment of ataxin-3 develop the disease phenotype. However, animals with full-length mutant ataxin-3 protein are viable and healthy. To explain this surprising result, the authors hypothesized that the lack of proteolytic cleavage of the mutant transgenic protein is necessary to produce the disease phenotype. Consistent with this hypothesis, transgenic mice expressing higher levels of ataxin-3 with expanded repeats developed a phenotype reminiscent of SCA3 and had a cleavage fragment in brain (Goti et al. 2004). The YAC transgenic animals created by Cemal et al. (2002) expressed human ataxin-3 driven by its native gene promoter. This strategy resulted in a close reproduction of human SCA3 pathology in the mouse model, a result that is probably due to the similar expression pattern of the mutant ataxin-3 protein. This result reveals the importance of non-coding sequences and native protein processing in the onset and progression of SCA3. The impact of the cellular localization of ataxin-3 on the pathology of SCA3 has been investigated using mouse models generated by introducing transgenes coupled to either a nuclear localization signal (NLS) or a nuclear export signal (NES) (Bichelmeier et al. 2007). The resulting animals containing the NLS transgene showed markedly enhanced SCA3 symptoms relative to animals containing the NES transgene. The role of co-chaperones and the ubiquitin ligase C-terminus of the Hsp70-interacting protein in enhancing disease progression and increasing aggregate formation was explored in the SCA3 mouse model (Williams et al. 2009) created by Goti et al. (2004). Recently, Boy et al. (2009) provided evidence that the behavioral SCA3 phenotype could be reversible by creating an inducible model of SCA3 using the Tet-off system. The same group demonstrated the late-onset transgenic model with ataxin-3 driven by huntingtin promoter (Boy et al. 2010). The mouse model where ataxin-3 is driven by the CMV promoter does not show intranuclear inclusions (Silva-Fernandes et al. 2010). In addition, one knock-out model of mouse ataxin-3 has been created. These animals exhibit increased levels of total protein ubiquitination and minor behavioral changes, but they are viable and fertile (Schmitt et al. 2007).
We have created a K300 transgenic mouse model that lacks functional ataxin-3 protein expression confirmed by western blotting and immunohistochemistry. The mouse model was generated as a result of a knock-in strategy wherein human ataxin-3 cDNA was inserted into the mouse ataxin-3 locus. The final mouse model exhibited the presence of several splice forms of ataxin-3 mRNA and the presence of deleted non-functional ataxin-3 proteins that were detected at very low levels only when using a massive excess of protein extract. The correctly spliced mRNA isoform containing mouse exon 1/human exons was detected at very low levels using PCR with increased sensitivity, but the protein of interest was not detected even when using a massive excess of protein extract for immunoblotting. The most prominent mRNA isoforms detected in mut/mut animals were mouse ataxin-3 missing exons 2 and 3 and the mRNA comprising mouse exon 1 spliced with human exon 7. We conclude that the reason for the lack of ataxin-3 protein expression was the occurrence of alternative splicing events at the Atxn3 locus after targeting. In summary, these transgenic mice are functional knock-out of ataxin-3, are viable and fertile, and do not have a reduced life span.
Methods
Animals and Cell Line
The animals were kept under standard conditions with a 12-h light/dark cycle and water and food ad libitum. The animals were marked via a toe-clipping method, and the tissue from this procedure was used for genotyping analyses. The animals were sacrificed by placing them in a 70% CO2 atmosphere. All procedures and animal handling were carried out to minimize animal stress and were approved and monitored by the Local Ethical Commission for Animal Experiments in Poznan. The N171-82Q (Schilling et al. 1999) and B6.129P2-Htttm2Detl/J (Lin et al. 2001) mouse models were obtained from The Jackson Laboratory (Bar Harbor, Maine, USA).
Primary human fibroblasts (cell line number: GM06153) were obtained from the Coriell Institute for Medical Research (Camden, NJ, USA). The cells were propagated in DMEM supplemented with 10% fetal bovine serum (Invitrogen; Carlsbad, USA) at 37°C in a 5% CO2 atmosphere.
Strategy and Generation of the K300 Mouse Model
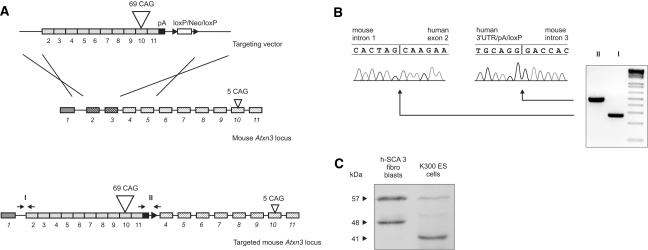
The K300 (ICS internal project number) SCA3 transgenic mouse model was generated at the Institut Clinique de la Souris (ICS) in Strasbourg, France, as part of the “RNA Interference Technology as Human Therapeutic Tool” (RIGHT) project. The human mutant ataxin-3 cDNA sequence containing 69 CAG repeats was cloned from total mRNA isolated from human fibroblasts (GM06153) that were derived from a patient suffering from SCA3. The mouse model was generated using a knock-in strategy that introduced a human cDNA sequence that spanned exons 2 through 11 of the human mutant ataxin-3 mRNA into the mouse ataxin-3 gene. To inactivate the mouse ataxin-3 gene, the strategy was designed to preserve mouse exon 1 and intron 1 in the native gene and introduce human exons 2–11 in place of mouse exons 2 and 3 (Fig. 1). In the final construct used for homologous recombination, an SV40 polyadenylation signal and a loxP-NEOR-loxP cassette were introduced downstream of the human ataxin-3 cDNA. The homologous arms present in the construct were 4.5 and 3.5 kb for the 5′ and 3′ arms, respectively. The construct was subsequently introduced into Sv129/Pas embryonic stem cells, and clones were selected based on neomycin resistance and tested for proper recombination by PCR and sequencing. The clones were then injected into blastocysts of C57BL/6 mice to obtain chimeras, which were then bred to obtain transgenic animals. The loxP-NEOR-loxP cassette was excised by breeding the transgenic animals with animals expressing Cre recombinase.
Fig. 1.
The structure of the mouse ataxin-3 gene and the targeting strategy. The targeting vector containing human exons 2–11 (gray) terminated by the necessary pA signals (black) was inserted into the mouse Atxn3 locus by replacing and deleting mouse exon 2, intron 2, and exon 3 (a). The targeted mouse Atxn3 locus contains mouse exon 1, intron 1, human exons, and mouse exons 4–11. The arrows indicate primers used for the amplification and verification of 5′ and 3′ ends by sequencing. The sequencing analysis of PCR products from the 5′ and 3′ ends revealed correct insertion of the transgene (b). The targeted clones demonstrated the expression of the normal mouse ataxin-3 and human mutant ataxin-3 protein detected by immunoblotting (c). The mouse exons are numbered with italic and human exons with regular font type
Genotyping Strategies
All animals were genotyped using a protocol based on the facts that the mutant allele does not contain an intron between exons 2 and 3 and that the sequences of both exons 2 and 3 are identical between humans and mice. Therefore, primers binding to both human and mouse exons 2 and 3 would generate PCR products that differed only by the length of the intron. The PCR product lengths resulting from this approach would be 376 bp in wild-type (wt/wt) animals, 150 bp in mutant (mut/mut) animals, and both products in wt/mut animals. The sequences of primers for genotyping are as follows: forward exon 2 CAAGGAGAATATTTTAGCCCTGT, reverse exon 3 GCCGCTGTCATCCATATTTCCA.
DNA Isolation and PCR
Total DNA was isolated from mouse toes using an Invisorb tissue mini kit (Invitek; Berlin, Germany) according to the manufacturer’s instructions. DNA concentration was estimated using a NanoDrop (Thermo Scientific; Wilmington, USA) spectrophotometer. PCR was performed with 1 μl of template DNA in a total volume of 10 μl that included 6 pmol of the appropriate primers and 0.25 U of recombinant GoTaq polymerase (Promega; Madison, USA). PCR amplification included 28 cycles of denaturation at 94°C for 30 s, annealing at 58°C for 30 s, and elongation at 72°C for 30 s. Primer sequences and their respective PCR products are listed in Table 1. The reaction products were separated on 1.5% agarose gels in TBE buffer and stained with ethidium bromide.
Table 1.
Primers
| Forward (5′-3′) | Reverse (5′-3′) | Localization of the PCR product* | Product (bp) |
|---|---|---|---|
| CATGGGGCTTACAAAGTTTGTTTTGACTGC | GTGCGATAATCTTCACTAGTAACTCCTCC | 5′ End of the transgene (DNA) | 278 |
| CCCTTCAACTGTATACCGTGTGCTC | CCCCACTCTAAACTGACTGTGGTAG | 3′ End of the transgene (DNA) | 476 |
| CCAGACAAATAAACATGGAG | TGTGGACTATTTTCACACATAC | mE1/mE9 |
823 (wt) 613 (E1/mE4) |
| CCAGACAAATAAACATGGAG | CCCTCTGCAAATCCTCCTCATCT | mE1/hE8,mE8 |
498 (E1/mE4) 234 (E1/hE7) |
| CCAGACAAATAAACATGGAG | CATTCCTGAGCCATCATT | mE1/hE8 |
677 (E1/hE2) 205 (E1/hE7) |
| GCATCGACCAAAACTTAT | TTTCTAAAGACATGGTCACA | CAG repeats mE6,hE6/mE11,hE11 |
674 (human) 473 (mouse) |
The last column shows lengths of the PCR products corresponding to the particular splicing isoform
* m mouse; h human; E# exon number
RNA Isolation and RT–PCR
Total RNA was isolated from whole brain tissue samples using TRI REAGENT (MRC; Cincinnati, USA) according to the manufacturer’s instructions. RNA isolation typically yielded 20 μg of total RNA. The RNA concentration was measured by a NanoDrop spectrophotometer. One microgram of total RNA was digested by DNase I in a volume of 10 μl at room temperature. After 45 min, the digestion reaction was stopped by adding 1 μl of 50 mM EDTA. A total of 10 μl of digested RNA were reverse-transcribed at 42°C for 50 min using 200 U of Superscript II (Invitrogen) and 100 ng of random hexamer primers (Promega) in a final volume of 20 μl. PCR was performed according to the conditions described above. Prior to each primary transcript PCR experiment, the quality of DNase digestion was assessed by PCR using the following templates: 1 μl of digested RNA, 1 μl of undigested RNA, and 100 ng of digested DNA. The quality of the reverse transcription (RT) reaction was assessed by an amplification of the Gapdh gene.
Immunoblotting Analyses
The tissue samples and primary human fibroblasts (GM06153) were lysed in a buffer containing 60 mM TRIS-base, 2% SDS, 10% sucrose, and 2 mM PMSF. Large tissue pieces were chopped and disrupted by sonication. Protein concentration was estimated using a NanoDrop spectrophotometer, and 50 and 200 μg of total protein was diluted in a sample buffer containing 2-mercaptoethanol and boiled for 5 min. The proteins were separated by SDS–polyacrylamide gel electrophoresis (5% stacking/12% resolving gel), transferred to nitrocellulose and stained with Ponceau S solution. The blots were blocked with 5% nonfat milk in TBS/0.05% Tween 20 and subsequently incubated for 24 h at 4°C with anti-ataxin-3 clone 1H9 (1:3,000; wt epitope E214-L233) primary antibody (Millipore; Billerica, USA) and with anti-ubiquitin (1:300) (Dako; Glostrup, Denmark). The blots were probed either with combination of biotinylated antibody and AP-conjugated streptavidin or with the HRP-conjugated secondary antibody (1:3,000) (Jackson Immuno Research, Suffolk, UK). The immunoreaction was detected using the NBT-BCIP substrate (Sigma–Aldrich, St. Louis, USA) or with the ECL substrate (GE Healthcare; Uppsala, Sweden).
Immunohistochemistry
The 10-μm coronal mouse brain sections or sections from testis were cut using a cryostat at -13°C and collected on silanized glass slides. The sections were fixed in cold 4°C acetone for 20 min and stored at −80°C or processed immediately. The sections were permeabilized by incubating in 0.5% Tween 20 in PBS for 10 min in RT and subsequently blocked by incubating in 2% normal goat serum for 30 min in PBS. For immunofluorescent staining of intranuclear inclusions, the K300 and N171/82Q brain sections were incubated overnight at 4°C with primary rabbit anti-ubiquitin (1:300) (Dako). The Cy-3 anti-rabbit fluorescent secondary antibodies (Jackson ImmunoResearch) were incubated at 1:500 for 2 h at RT in PBS-Tween 20. The cryosections from testis were incubated with mouse anti-ataxin-3 antibody (1:500) (Millipore) overnight at 4°C and subsequently detected by 2-h incubation with the appropriate biotinylated secondary Ab and stained using Vectastain Elite ABC (Vector laboratory, Burlingame, CA, USA) and the DAB chromogen. The fluorescent confocal images were acquired using the LSM 510 META system (Zeiss; Poznan, Poland).
Results
Structure of the Targeted Atxn3 Gene and the Expression of Ataxin-3 in Targeted ES Cells
Prior to the electroporation of ES cells, the entire cassette used for homologous recombination was sequenced (ICS; Strasburg). This verified the correct structure and sequence (for a description of the cassette, see the Materials and Methods). Stable ES cell clones were selected, and PCR and sequencing were performed to detect proper transgene integration into the Atxn3 gene locus (ICS; Strasburg). These analyses confirmed that the human cDNA was inserted at mouse exon 2 and that the mouse genomic sequence containing exon 2, intron 2, and exon 3 was deleted. Mouse exon 1 and intron 1 were preserved in the gene structure and were followed by human exons 2–11 and an SV40 polyadenylation signal. The knock-in targeting strategy of the Atxn3 gene is depicted in Fig. 1a. To demonstrate the correct positioning of the transgene in the genome, two distinct pairs of primers were designed to bind mouse intron 1/human exon 2 (5′-flank) and human exon 10/mouse intron 3 (3′-flank). The PCR amplification generated 2 PCR products of 278 bp and 476 bp in length, corresponding to the expected bp count. The PCR products generated from the 5′ and 3′ ends of the construct were sequenced and revealed the correct insertion of the transgene (Fig. 1b). Mouse exons and introns beyond exon 3 were unchanged.
Additionally, to investigate the ataxin-3 protein expression in targeted ES cells, the total protein extract was analyzed by western blotting, and the expression of both the mouse ataxin-3 protein (41 kDa) and human mutant ataxin-3 (57 kDa) was detected (Fig. 1c). For a positive control, the blots of the total protein extracted from GM06153 SCA3 human fibroblasts revealed a mutant ataxin-3 band with 69 CAG repeats that had an estimated molecular weight of 57 kDa.
Animals
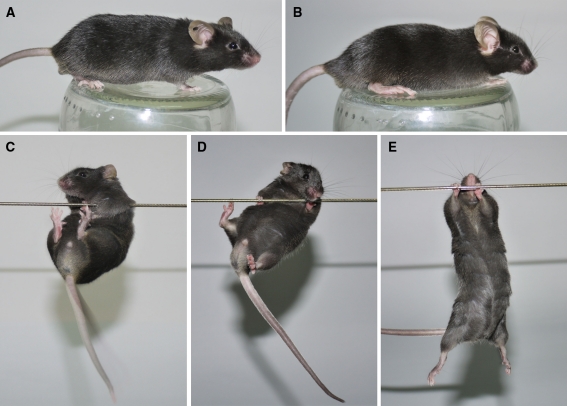
The 6 mut/wt animals were generated at ICS as a result of backcrossing chimeras two times onto the pure C57BL/6J background resulting in mixed background of 75% C57BL/25% Sv129/Pas. These animals were exported to Poznan and used to produce 3 generations of offspring and were withdrawn from breeding. Next, we have generated homozygous mut/mut animals that exhibited normal body posture (Fig. 2a, b) and normal muscle strength assessed by string test (Fig. 2c–e). To assess life span, 5 mut/wt animals initially received from ICS were kept, until they reached 2.5 years of age and then sacrificed.
Fig. 2.
Body posture and string test of K300 mice. The 6-month-old K300 mut/mut exhibited normal body posture with normal pelvis elevation and lack of hunchback (a) like the 6-months-old non-transgenic wt/wt animal (b). The muscle strength was assessed by placing the mouse from above with only front paws on the horizontally stretched string for 2 min. The K300 mut/mut animal (c) and wt/wt animal (d) were able to lift and grab the string with hind limbs. As positive control we used the Huntington’s disease knock-in mouse model (mut/wt; 150CAG) generated by Lin et al. (2001) and supplied by Jackson Laboratory (B6.129P2-Htttm2Detl/J). The HD knock-in mouse at similar age was hardly able to touch the string with hind limbs and often fell down (e)
Analyses of the Ataxin-3 Protein in K300 Mouse Tissues
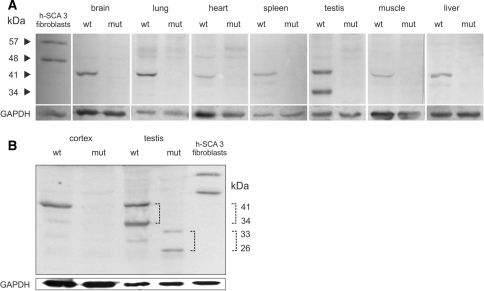
To investigate the expression of the ataxin-3 protein in the transgenic mice, total protein was extracted from the cerebral cortex, skeletal muscles (quadriceps), heart, lungs, liver, spleen, testis, and GM06153 human fibroblasts as a positive control and resolved by 12% SDS–PAGE. In wt/wt animals, the immunoblot analyses revealed a specific band representing mouse ataxin-3 with an estimated molecular weight of 41 kDa that was present at relatively high concentrations in testis, cerebral cortex, lungs, liver, and spleen and was hardly detectable in skeletal muscles and the heart (Fig. 3a). The band detected in wt/wt mice had a lower molecular weight than the positive control band of wt human ataxin-3 (48 kDa). The similar mouse ataxin-3 41-kDa band and the expected human mutant 57-kDa band were not detected in mut/mut animals in any of the tissues analyzed (Fig. 3a). Moreover, none of these immunoreactive bands was observed upon increasing the protein load to 200 μg of mut/mut protein extract from cerebral cortex and testis per lane (Fig. 3b). Additionally, analysis of the ataxin-3 protein in testis from wt/wt animals revealed an additional strongly immunoreactive band of 34 kDa that we describe as ataxin-3 isoform, since it was not present in testis of mut/mut animals (Fig. 3a, b). Upon increasing the total protein extract content to the large amount of 200 μg per lane, we were able to observe two faint immunoreactive bands of 33 and 26 kDa, respectively, that only appeared in mut/mut animals, but exclusively in testis and not in cerebral cortex (Fig. 3b). Note that the apparent 7-kDa molecular weight difference was also present between mouse ataxin-3 (41 kDa) and the 34-kDa ataxin-3 isoform detected in wt/wt testis.
Fig. 3.
Immunoblot analysis of ataxin-3 expression in tissues of K300 mice. Total protein was extracted from brain, lung, heart, spleen, testis, skeletal muscle, and liver samples collected from transgenic (mut/mut) and wild-type (wt/wt) animals as well as from a primary culture of human SCA3 fibroblasts (GM06153), which was used as a positive control. The extracts were resolved by 12% SDS/PAGE, and the blot was stained with a polyclonal antibody for ataxin-3. In wt/wt animals, the analysis revealed a mouse ataxin-3 41-kDa band present at relatively high concentrations in testis, cerebral cortex, lungs, liver, and spleen that was hardly detectable in skeletal muscles and heart. A similar 41-kDa band and the human mutant ataxin-3 57-kDa band were not detected in any tissues from mut/mut animals. Additionally, we detected a 34-kDa band of the ataxin-3 isoform in testis from wt/wt animals. This isoform was not present in the testis of mut/mut animals (a). Upon increasing the total protein load to 200 μg per lane, we were still unable to detect a human mutant ataxin-3 57-kDa band, but we did detect two faint immunoreactive bands of 33 and 26 kDa, only in testis of mut/mut animals. The apparent 7-kDa molecular weight difference between the 33- and 26-kDa bands is the same as that between mouse ataxin-3 (41 kDa) and the 34-kDa ataxin-3 isoform detected in wt/wt testis (b)
The Cellular Phenotype of the K300 Mouse
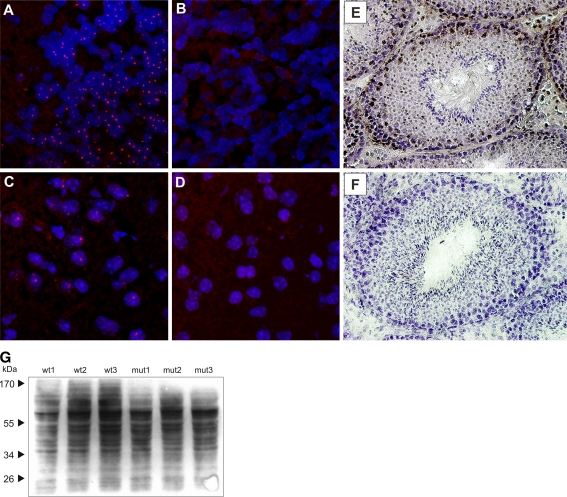
Nuclear inclusions are the hallmark of polyglutamine disorders and are present in both patients and mouse models. Therefore, we examined the brains of K300 animals for the presence of intranuclear inclusions by immunofluorochemistry using anti-ubiquitin antibodies in cerebral cortex and cerebellum of the K300 mut/mut and using N171/82Q animals as positive controls (Huntington’s disease mouse model; Schilling et al. 1999). As expected, the cortex and cerebellum of N171/82Q mice revealed intranuclear inclusions positive for ubiquitin (Fig. 4a, c). Similar structures were not detected in cells from cortex or cerebellum of K300 mut/mut mice stained with anti-ubiquitin antibodies (Fig. 4b, d). To demonstrate the loss of ataxin-3 expression, we cryosectioned the testis from K300 mut/mut animals and wt/wt animals and stained the tissue using anti-ataxin-3 antibodies and the DAB chromogen. The cells in testis of wt/wt animals were positively stained for ataxin-3 (Fig. 4e), and no staining was detected in mut/mut animals (Fig. 4f).
Fig. 4.
The cellular phenotype of the K300 mouse model. The cryosections prepared from cerebellum and cerebral cortex were stained using anti-ubiquitin Ab, and confocal images were acquired. The intranuclear inclusions were not detected in cryosections from K300 mut/mut mouse brain (b, d). The positive control brains from N171/82Q HD animals revealed a large number of intranuclear inclusions positive for ubiquitin in both cerebellum and cerebral cortex (a, c). Additionally, the cryosections from testis of K300 mut/mut and wt/wt animals were stained using anti-ataxin-3 antibodies and the DAB chromogen, and the nuclei were counterstained with hematoxylin dye. Testis from K300 mut/mut animals did not stain positively for ataxin-3, and testis of wt/wt animals exhibited specific, positive brown staining for ataxin-3 in a subset of cells in testis (e, f). The immunoblotting of total proteins with anti-ubiquitin antibodies revealed no difference in the levels of total ubiquitin in mut/mut compared with wt/wt animals (g)
The knock-out model of mouse ataxin-3 previously created by Schmitt et al. (2007) exhibited increased levels of total protein ubiquitination, probably due to the absence of ataxin-3 leading to a partial loss of bulk cellular ubiquitin-specific protease activity. To test whether a similar phenomenon could be observed in K300 animals, we studied the level of ubiquitinated species in brain tissue using western blotting. The immunodetection with anti-ubiquitin antibodies revealed no differences in the total level of ubiquitin in mut/mut compared with wt/wt animals (Fig. 4g).
Expression of Ataxin-3 mRNA in the Brain of K300 Animals
The examination of protein expression and the lack of ataxin-3 staining in tissue provided strong indications that the expression from the targeted Atxn3 allele was defective, leading us to hypothesize that the allele had acquired knock-out properties. To further investigate this issue and the possible reasons for the lack of ataxin-3 protein in mice, we tested Atxn3 mRNA expression. Since the expression of ataxin-3 and the pathology of SCA3 are both located in the brain, we initially examined the expression of ataxin-3 mRNA in brain samples from transgenic animals, including samples of cortex, cerebellum, pons, and basal ganglia. A representative set of RT–PCR results generated using tissue samples from the cerebral cortex was selected for this publication and described below. RT–PCR results from other brain regions yielded similar results (data not shown).
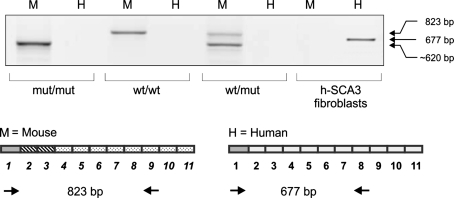
Specific primers were used to selectively detect either human (mut allele) or mouse ataxin-3 mRNA (wt allele) (Fig. 5). A single forward primer was designed for exon 1 to bind mRNA from both wt and mut alleles. The human-specific reverse primer was designed to bind exon 8 and generate a mut-specific PCR product of 677 bp. The mouse-specific reverse primer was designed to bind exon 9 and generate a wt-specific product of 823 bp. Interestingly, the standard 28 PCR cycles did not allow us to detect the mut-specific product (677 bp) using the RT template from mut/mut or wt/mut animals. A detailed description of the detection of the mut-specific product and other splice forms is provided below. Unexpectedly, the wt-specific product was present in mut/mut animals, but was shorter than the wt/wt—specific product (613 bp). Wt-specific RT–PCR of wt/mut animals revealed two products, the 613-bp product described above and the expected 823-bp wt-specific product. As expected, the wt-specific product was present in wt/wt animals (Fig. 5). The presence of the short 613-bp wt product in the mut/mut animals was even more puzzling, as this product appeared to be shortened by the combined length of mouse exons 2 and 3, which had been deleted. Sequencing analysis demonstrated that the sequence of mouse exon 1 was directly linked with mouse exon 4 and subsequent exons (Fig. 6). These results indicate that the aberrant splicing of the mRNA expressed from the targeted allele may be responsible for the lack of ataxin-3 protein expression.
Fig. 5.
Expression of ataxin-3 mRNA in brain samples from transgenic animals. The 677-bp RT–PCR product, which represents mRNA containing mouse exon 1 and human exons of ataxin-3, was initially not detected in the mut/mut and wt/mut animals using the standard 28 PCR cycles (H). The RT–PCR product that was specific for mouse mRNA containing exon 1 was detected in transgenic animals (mut/mut and wt/mut). The product was shorter by the length of exons 2 and 3 (lines) and was 613 bp rather than the predicted 823-bp wt product, suggesting the alternative splicing event. H human ataxin-3 specific PCR; M mouse ataxin-3 specific PCR
Fig. 6.
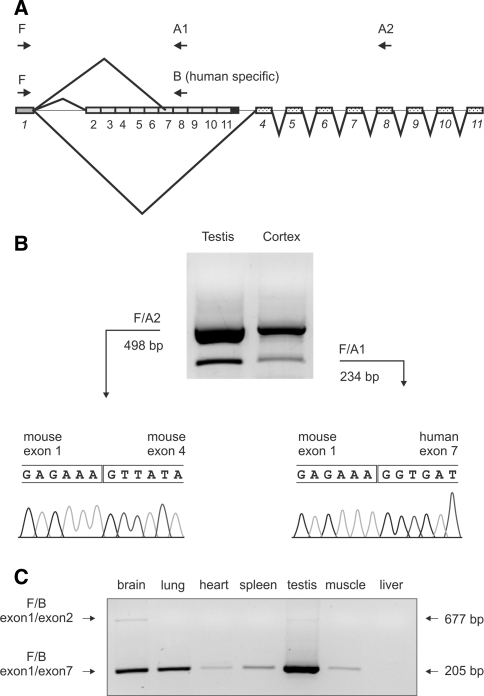
Splicing events in mut-mut K300 animals. The diagram graphically depicts the splicing events, and the primers called F, B, A1, and A2 used to detect the various mRNA isoforms (a). The mouse exon 1/exon 4 and the mouse exon 1/human exon 7 isoforms were detected in cortex, testis, and other tissues (data not shown) and were confirmed by sequencing (b). The exon 1/exon 2 splice variant (677 bp) was detected in the brain and testis at very low levels using primers F/B and using 37 cycles to increase PCR sensitivity. The 205-bp product represents exon 1/human exon 7 isoform (c)
Alternative Splicing Events Present in the K300 Ataxin-3 Knock-Out Mouse
We investigated whether the mouse exon 1/exon 4 splicing form was the only splicing event that hindered the splicing of mouse exon 1 with human exons. We also asked whether a similar splicing isoform was present in the K300 model in tissues other than brain, and for this purpose, we analyzed the total mRNA from skeletal muscles, heart, lungs, liver, spleen, and testis of mut/mut animals. We detected two major splice forms in all analyzed tissues of K300 animals except liver where the exon 1/human exon 7 isoform was not present (Fig. 6). As previously demonstrated for the cerebral cortex, we detected the presence of the mouse exon 1/exon 4 splice variant that was strongly expressed in testis and lungs and was less pronounced in liver, skeletal muscles, heart, and spleen (Fig. 6b; data shown for testis and cortex). Moreover, we detected a PCR product representing a splice variant containing mouse exon 1 and part of human exon 7 (Fig. 6c). This splice variant was spliced at the splicing site that was inside human exon 7, but it was in frame, so that the putative protein would be the exon 2–6 deleted form of ataxin-3. A particularly pronounced expression of the mouse exon 1/human exon 7 isoform was detected in testis, and this isoform was detected in other tissues at lower levels. Because some splicing events and the resulting splice variants may be rare, we decided to increase the sensitivity of our detection by increasing the number of PCR cycles. Indeed, in cortex and testis using 37 PCR cycles, we were able to detect a very faint signal of the 677 bp band representing the mut/mut—specific splice variant containing mouse exon 1 and the human exons. The variety of reverse primers binding to all exons of the ataxin-3 gene (2–11), and the forward primer for mouse exon 1, were used to detect splicing events throughout the primary transcript of the targeted allele. Therefore, Fig. 6 summarizes the experiments detecting splice variants with three primers binding to exon 1, mouse exon 8, and human exon 8 that best reflect the presence of all splice variants in K300 tissues. Based on our experiments, we estimate that the cellular levels of splice variants had the following relationship: the 1/4 splice variant was the most prominent, the level of the 1/7 variant was 10% that of the 1/4 variant, and the 1/2 variant was hardly detectable.
Discussion
The initial goal of this project was to create a mouse model of SCA3 using a knock-in strategy that would introduce human ataxin-3 cDNA into the mouse ataxin-3 gene. In this approach, intron 1, exon 1, the promoter, and other 5′ regulatory sequences were left unmodified, and the human ATXN3 cDNA was inserted into mouse exon 2. This allows the human sequence containing 69 CAG repeats to be expressed from the native mouse ataxin-3 promoter together with other putative natural regulatory sequences, such as CpG methylation islands located in intron 1. After the homologous recombination cassette was checked thoroughly by sequencing, it was electroporated into ES cells. The selected clones were screened for proper homologous cassette recombination, and the clones revealed an exact insertion of human cDNA at exon 2. Moreover, the targeted ES cells revealed the expression of mutant transgene protein, but the mutant protein was expressed at a lower level than the native mutant protein in human fibroblasts and also at a lower level than wt mouse ataxin-3. The mice were generated, and the transgene was validated in animals by PCR amplification and sequencing of 5′ and 3′ insert flanks as well as by PCR amplification of the internal transgress region composed of exons 2–9, confirming correct placement in the genome in vivo. Analyzing brain, lungs, heart, spleen, testis, muscles, and liver from the generated animals, we did not detect the mutant human ataxin-3 protein that was previously detected in targeted ES cells. Therefore, we increased the sensitivity of our western blotting protocol by increasing the protein load up to 200 μg, but we were still unable to detect mutant protein expression. Instead, in testis of mut/mut mice, we detected very low levels of two proteins that were much shorter (33 and 26 kDa) than the wt mouse ataxin-3 protein. The apparent molecular weight difference between the two molecules was 7 kDa, similar to the difference between wt mouse ataxin-3 protein and the second ataxin-3 protein isoform, with the most pronounced expression in testis (41 and 34 kDa, respectively). The two isoforms (33 and 26 kDa) were probably proteins with deleted protein fragments equivalent to the sequences of exons 2 and 3. The presence of splice variant (exon 1/exon 4) equivalent to 33-kDa protein was detected in all tissues. The 33-kDa protein must lack the segment of the catalytic Josephine domain that spans from amino acids Q9–Q78 and includes the C14 residue. The substitution of C14 to alanine (C14A) in wild-type ataxin-3 was previously shown to be sufficient to render ataxin-3 nonfunctional by abolishing ubiquitin-specific protease activity (Burnett et al. 2003).
The presence of the second ataxin-3 protein isoform (wt: 34 kDa; mut: 26 kDa) detected in testis was not confirmed by the detection of an equivalent transcript. Because we screened and designed primers in all exons, the equivalent transcript would have to appear repeatedly as a PCR product resulting from many different reverse primers and the forward primer for exon 1. Therefore, we conclude that this protein is the ataxin-3 isoform (because it is not present in mut/mut mice), that it is a proteolytic fragment, and, being found at relatively high levels, that it is quite stable and accumulates in testis. Pronounced expression of a similar peptide was previously reported in C57BL mouse testis, skeletal muscles, and heart (do Carmo Costa et al. 2004).
Because we did not detect the mutant ataxin-3 protein using western blotting of the mut/mut protein extract, we investigated the expression of this protein using immunochemistry. The intranuclear inclusions often appear in the brains of patients and in mouse models of polyglutamine diseases and are usually strongly ubiquitinated (Cemal et al. 2002; Bichelmeier et al. 2007; Alves et al. 2008). Intranuclear inclusions positive for ubiquitin were not detected in cerebellum or cerebral cortex of K300 animals, indicating the lack of mutant ataxin-3 protein accumulation. Moreover, the direct staining of the testis, the most prominent source of ataxin-3 expression, revealed a complete loss of this protein in mut/mut K300 animals. This strongly indicates that the targeted allele acquired knock-out properties and that the mut/mut K300 mice are knock-out animals. The knock-out of ataxin-3 created by Schmitt et al. (2007) exhibited enhanced total protein ubiquitination, but we did not detect this effect in K300 animals. Since the genome encodes nearly 100 deubiquitinating enzymes, the changes produced by one of them may be compensated for by other DUBs.
To determine the reason for the lack of correct protein expression, we investigated the transcripts of the targeted Atxn3 locus and the level of ataxin-3 mRNA expression. Initially, we examined the brain and were not able to detect a common PCR product representing mRNA that contained mouse exon 1 and the human exons of ataxin-3. Instead, we detected human RNA using PCR for expanded repeats (Suppl. Fig. 1). This indicated that the human transgene was transcribed. Surprisingly, in homozygous transgenic animals, but not in wt animals, we detected a PCR product containing exon 1 that was specific for mouse mRNA. However, this PCR product was shorter than the wt product by the combined length of mouse exons 2 and 3. Sequencing revealed that mouse exon 1 and the remaining mouse exons 4–11 were spliced. The presence of a similar splice variant was recently detected in the blood of healthy human individuals (Bettencourt et al. 2010). Additionally, we characterized the primary and mature transcripts and revealed that the entire locus was expressed, and the cellular levels of both the mouse and the human portions of the transcript in the mut/mut mouse were relatively high. The cellular level of human transcript was lower than the level of mouse transcript, but it was almost twice as high as the level of primary transcript (Suppl. Fig. 2).
The lack of mutant protein expression may result from aberrant mRNA splicing in targeted allele where mouse exons 4–11 are still expressed and induce preferences in splicing that led to the exclusion of the exon 1/human exon 2 isoform. We investigated the splicing aberrations in the targeted allele in more detail and found the presence of another splicing form, the splicing of exon 1 and part of exon 7. Moreover, both exon 1/exon 4 and exon 1/exon 7 splicing forms were also present in other tissues of the K300 model. Additionally, we tried to increase the sensitivity of our PCR detection and were able to detect the expected exon 1/exon 2 splice form at a very low level after 37 PCR cycles. However, we cannot exclude the possibility that other splice variants were produced from this targeted allele. In summary, we detected the presence of three splice variants and observed that the splicing favored distant splicing acceptor sites, such as mouse human exon 7 and exon 4, and virtually eliminated splicing to human exon 2. This situation is surprising since the splicing acceptor site in exon 2 remained identical to the mouse sequence up to the 24th nucleotide and contained only 3 single-nucleotide changes in the first 70 nucleotides of exon 2. We used the SROOGLE webserver, which scores splice sites according to 9 different algorithms (Schwartz et al. 2009). This analysis revealed that combining mouse intron 1 and human exon 2 (like the transgene structure) returned exactly the same splice site characteristics and the same score as the combination of mouse intron 1 and mouse exon 2. Further bioinformatic characterization revealed that the mouse or human intron 1/exon 2 splice site was very weak, with a score of 0.78 (human) and 0.52 (mouse), according to the maximum entropy-based scoring method (Yeo and Burge 2004). Additionally, the algorithms failed to detect the branching site in intron 1. Interestingly, the score for the intron 3/exon 4 splice site was 10.55 (mouse; max. entropy). Therefore, the splicing aberrations present in the targeted allele most likely did not result from changing the properties of the natural intron 1/exon 2 splice site. These aberrations may be attributed to the deletion of splicing sites present in intron 2, exon 3, and intron 3. The lack of these structures and therefore the lack of splicing probably exposed the strong acceptor site in exon 4 to the donor site in exon 1. Moreover, it is possible that the natural low-efficiency splicing between exons 1 and 2 of the ataxin-3 gene may be the limiting step that, at least to some extent, regulates the expression of the protein. These facts provide important lessons for future attempts to create a knock-in mouse model of SCA3. The most important conclusion is to avoid the 5′ region of the Atxn3 gene in future knock-in strategies due to its splicing characteristics, such as week exon 1/exon 2 splicing efficiency.
Conclusions
We have attempted to create a K300 SCA3 mice using the knock-in strategy. The mouse model turned out to be nonfunctional and lacked ataxin-3 protein expression and expressed very low levels of nonfunctional proteins. We have systemically demonstrated the loss of protein expression in all analyzed tissues and concluded that we have produced a knock-out of ataxin-3. We have provided evidence that the reason for the lack of expected protein expression was the occurrence of intensive splicing events present in the targeted locus, since the model (mut/mut) expresses a long primary transcript containing mouse exons and human exons with CAG repeats. The animals do not exhibit changes in fertility or life span relative to their non-transgenic counterparts.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by the EU FP6 project “RNA Interference Technology as Human Therapeutic Tool (RIGHT)”, European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-30-049/09), and Ministry of Science and Higher Education (N N302 299536). We thank Krzysztof Sobczak for useful suggestions and comments on the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alves S, Régulier E, Nascimento-Ferreira I, Hassig R, Dufour N, Koeppen A, Carvalho AL, Simões S, de Lima MCP, Brouillet E, et al. Striatal and nigral pathology in a lentiviral rat model of Machado-Joseph disease. Human Molecular Genetics. 2008;17:2071–2083. doi: 10.1093/hmg/ddn106. [DOI] [PubMed] [Google Scholar]
- Bettencourt C, Santos C, Montiel R, Costa MDC, Cruz-Morales P, Santos LR, Simões N, Kay T, Vasconcelos J, Maciel P, et al. Increased transcript diversity: novel splicing variants of Machado-Joseph disease gene (ATXN3) Neurogenetics. 2010;11:193–202. doi: 10.1007/s10048-009-0216-y. [DOI] [PubMed] [Google Scholar]
- Bichelmeier U, Schmidt T, Hübener J, Boy J, Rüttiger L, Häbig K, Poths S, Bonin M, Knipper M, Schmidt WJ, et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: In vivo evidence. Journal of Neuroscience. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Genome-wide screen for modifiers of ataxin-3 neurodegeneration in Drosophila. PLoS Genetics. 2007;3:1950–1964. doi: 10.1371/journal.pgen.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Schumann U, Grasshoff U, Unser S, Holzmann C, Schmitt I, Karl T, Laccone F, Wolburg H, et al. A transgenic mouse model of spinocerebellar ataxia type 3 resembling late disease onset and gender-specific instability of CAG repeats. Neurobiology of Diseases. 2010;37:284–293. doi: 10.1016/j.nbd.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Boy J, Schmidt T, Wolburg H, Mack A, Nuber S, Böttcher M, Schmitt I, Holzmann C, Zimmermann F, Servadio A, et al. Reversibility of symptoms in a conditional mouse model of spinocerebellar ataxia type 3. Human Molecular Genetics. 2009;18:4282–4295. doi: 10.1093/hmg/ddp381. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Human Molecular Genetics. 2003;12:3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Cemal CK, Carroll CJ, Lawrence L, Lowrie MB, Ruddle P, Al-Mahdawi S, King RHM, Pook MA, Huxley C, Chamberlain S. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Human Molecular Genetics. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- Chow MKM, Ellisdon AM, Cabrita LD, Bottomley SP. Polyglutamine expansion in ataxin-3 does not affect protein stability: implications for misfolding and disease. Journal of Biological Chemistry. 2004;279:47643–47651. doi: 10.1074/jbc.M405799200. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Andrade C. Autosomal dominant system degeneration in Portuguese families of the Azores Islands. A new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology. 1978;28:703–709. doi: 10.1212/wnl.28.7.703. [DOI] [PubMed] [Google Scholar]
- Coutinho P, Guimarães A, Scaravilli F. The pathology of Machado-Joseph disease. Report of a possible homozygous case. Acta Neuropathologica. 1982;58:48–54. doi: 10.1007/BF00692697. [DOI] [PubMed] [Google Scholar]
- do Carmo Costa M, Gomes-da-Silva J, Miranda CJ, Sequeiros J, Santos MM, Maciel P. Genomic structure, promoter activity, and developmental expression of the mouse homologue of the Machado-Joseph disease (MJD) gene. Genomics. 2004;84:361–373. doi: 10.1016/j.ygeno.2004.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evert BO, Araujo J, Vieira-Saecker AM, de Vos RAI, Harendza S, Klockgether T, Wüllner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. Journal of Neuroscience. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro A, Carvalho AL, Teixeira-Castro A, Almeida C, Tomé RJ, Cortes L, Rodrigues A, Logarinho E, Sequeiros J, Macedo-Ribeiro S, et al. NEDD8: a new ataxin-3 interactor. Biochimica et Biophysica Acta. 2007;1773:1619–1627. doi: 10.1016/j.bbamcr.2007.07.012. [DOI] [PubMed] [Google Scholar]
- França MC, D’Abreu A, Friedman JH, Nucci A, Lopes-Cendes I. Chronic pain in Machado-Joseph disease: a frequent and disabling symptom. Archives of Neurology. 2007;64:1767–1770. doi: 10.1001/archneur.64.12.1767. [DOI] [PubMed] [Google Scholar]
- França MC, D’Abreu A, Nucci A, Lopes-Cendes I. Muscle excitability abnormalities in Machado-Joseph disease. Archives of Neurology. 2008;65:525–529. doi: 10.1001/archneur.65.4.525. [DOI] [PubMed] [Google Scholar]
- Giunti P, Sweeney MG, Harding AE. Detection of the Machado-Joseph disease/spinocerebellar ataxia three trinucleotide repeat expansion in families with autosomal dominant motor disorders, including the Drew family of Walworth. Brain. 1995;118(Pt 5):1077–1085. doi: 10.1093/brain/118.5.1077. [DOI] [PubMed] [Google Scholar]
- Goti D, Katzen SM, Mez J, Kurtis N, Kiluk J, Ben-Haiem L, Jenkins NA, Copeland NG, Kakizuka A, Sharp AH, et al. A mutant Ataxin-3 Putative-Cleavage fragment in brains of Machado-Joseph disease patients and transgenic mice is cytotoxic above a critical concentration. Journal of Neuroscience. 2004;24:10266–10279. doi: 10.1523/JNEUROSCI.2734-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Yamaguchi M, Sugai S, Aze Y, Narumiya S, Kakizuka A. Expanded polyglutamine in the Machado-Joseph disease protein induces cell death in vitro and in vivo. Nature Genetics. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- Ishikawa A, Yamada M, Makino K, Aida I, Idezuka J, Ikeuchi T, Soma Y, Takahashi H, Tsuji S. Dementia and delirium in 4 patients with Machado-Joseph disease. Archives of Neurology. 2002;59:1804–1808. doi: 10.1001/archneur.59.11.1804. [DOI] [PubMed] [Google Scholar]
- Jia N, Fei E, Ying Z, Wang H, Wang G. PolyQ-expanded ataxin-3 interacts with full-length ataxin-3 in a polyQ length-dependent manner. Neuroscience Bulletin. 2008;24:201–208. doi: 10.1007/s12264-008-0326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Okamoto T, Taniwaki M, Aizawa M, Inoue M, Katayama S, Kawakami H, Nakamura S, Nishimura M, Akiguchi I. CAG expansions in a novel gene for Machado-Joseph disease at chromosome 14q32.1. Nature Genetics. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- Kawai Y, Takeda A, Abe Y, Washimi Y, Tanaka F, Sobue G. Cognitive impairments in Machado-Joseph disease. Archives of Neurology. 2004;61:1757–1760. doi: 10.1001/archneur.61.11.1757. [DOI] [PubMed] [Google Scholar]
- Lessing D, Bonini NM. Polyglutamine genes interact to modulate the severity and progression of neurodegeneration in Drosophila. PLoS Biology. 2008;6:e29. doi: 10.1371/journal.pbio.0060029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Tallaksen-Greene S, Chien W, Cearley JA, Jackson WS, Crouse AB, Ren S, Li X, Albin RL, Detloff PJ. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Human Molecular Genetics. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Paulson HL. Dominantly inherited ataxias: lessons learned from Machado-Joseph disease/spinocerebellar ataxia type 3. Seminars in Neurology. 2007;27:133–142. doi: 10.1055/s-2007-971172. [DOI] [PubMed] [Google Scholar]
- Riess O, Rüb U, Pastore A, Bauer P, Schöls L. SCA3: Neurological features, pathogenesis and animal models. Cerebellum. 2008;7:125–137. doi: 10.1007/s12311-008-0013-4. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA, Slunt HH, Ratovitski T, Cooper JK, Jenkins NA, et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Human Molecular Genetics. 1999;8:397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Linden M, Khazneh H, Evert BO, Breuer P, Klockgether T, Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochemical and Biophysical Research Communications. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Hall E, Ast G. SROOGLE: Webserver for integrative, user-friendly visualization of splicing signals. Nucleic Acids Research. 2009;37:W189–W192. doi: 10.1093/nar/gkp320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Fernandes A, Costa MDC, Duarte-Silva S, Oliveira P, Botelho CM, Martins L, Mariz JA, Ferreira T, Ribeiro F, Correia-Neves M, et al. Motor uncoordination and neuropathology in a transgenic mouse model of Machado-Joseph disease lacking intranuclear inclusions and ataxin-3 cleavage products. Neurobiology of Diseases. 2010;40:163–176. doi: 10.1016/j.nbd.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Williams AJ, Knutson TM, Colomer Gould VF, Paulson HL. In vivo suppression of polyglutamine neurotoxicity by C-terminus of Hsp70-interacting protein (CHIP) supports an aggregation model of pathogenesis. Neurobiology of Diseases. 2009;33:342–353. doi: 10.1016/j.nbd.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. Journal of Computational Biology. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.