Abstract
The complete genomic sequence of an isolate (PRI-509) of the C strain of Potato virus Y (PVYC), which was originally isolated from potato in 1938, was elucidated. The genomic RNA of PRI-509 consists of 9699 nucleotides, with the capacity to encode a polyprotein of 3061 amino acids with a molecular mass of 337 kDa.
This is the first full-length sequence of a PVY C isolate from potato that belongs to the C1 phylogenetic subgroup, which was previously thought to exclusively contain non-potato isolates.
Electronic supplementary material
The online version of this article (doi:10.1007/s00705-010-0853-3) contains supplementary material, which is available to authorized users.
Potato virus Y (PVY), the type member of the genus Potyvirus, was described for the first time in the early 1930s as the causal agent of a serious disease in potato [26]. PVY is one of the most common viruses of potato (Solanum tuberosum L.) and causes significant economic losses worldwide, especially in the production of seed potatoes.
The species Potato virus Y includes several different strains. Initially, PVYO, PVYC and PVYN [8] were distinguished based on their biological properties (symptomatology on hosts and on test plants and resistance-breaking capability). In the Netherlands, PVYO and PVYC were predominant until the 1950s [2]. In 1957, PVYN was reported for the Netherlands by De Bokx [9]. More recently, new PVY strains have been found worldwide that appear to be genetic recombinants bearing genetic material of the PVYO and PVYN strains. Examples of such strains are PVYNTN and PVYN-Wi [12, 16, 24, 25].
PVYO and PVYC can be distinguished in potato cultivars bearing the resistance genes Ny tbr and Nc [7, 25]. PVYC isolates are defined by the typical hypersensitive response they induce in potato cultivars carrying the Nc gene. PVYC causes systemic mosaic or stipple streak symptoms in potato [8] and is generally not considered a serious problem, since many potato cultivars appear to display a hypersensitive response (field resistance) to this strain [6]. A survey in France in 1985 showed an infection rate of less than 5% with PVYC, while for PVYO and PVYN, infection levels of 80 and 12%, respectively, were reported [13]. In the Netherlands, field surveys conducted from 1994 to 2007 showed similar results [28].
PVY also causes diseases and production loss in pepper (Capsicum annuum L.) [22], tomato (Solanum lycopersicum L.) and tobacco crops [25]. For some isolates from pepper, a high level of sequence identity to PVYC from potato was found [11, 15, 22]. Based on biological data, RFLP restrictotypes and coat protein sequence data, these non-potato isolates were proposed to form a genetic cluster separate from the potato isolates [22].
The plant virus collection at Plant Research International (PRI) was established in the early 1950s and includes several historical isolates assigned to different PVY strains. The PVYC isolate described in this study (PRI-509) was isolated from the Dutch potato variety ‘Zeeuwse Blauwe’ in the Netherlands in 1938 [10, 23] and was maintained only in potato. It causes a very distinctive pattern of systemic necrotic lines and lesions, known as stipple streak, along the veins of certain potato varieties carrying the Nc resistance gene, such as ‘Duke of York’ (‘Eersteling’). This host response distinguishes PVYC isolates from PVYN and PVYO isolates. Similar to PVYO, it causes a distinct systemic mosaic on Nicotiana tabacum L. and mosaic and necrosis on Physalis floridana L. [8].
Virus indexing of seed potatoes is done mainly by DAS-ELISA, employing a broad-spectrum antiserum that does not distinguish between PVY strains. Many RT-PCR tests have been described for PVY detection, including tests for detection of recombinant strains [16, 21, 24], but none of them can distinguish PVYC isolates easily.
Schubert et al. [24] determined the entire sequence of the French PVYC isolate Adgen from potato. Moury [19] showed that the biological characteristics of this Adgen isolate are typical of a PVYC isolate, but its serological characteristics remain to be determined. In their study, Schubert et al. [24] compared full-length sequences of 37 PVY strains and variants, including the Adgen isolate and three related non-potato PVYC isolates. These PVY isolates, nnp [11], SON41 and LYE84.2 [18], were isolated from other solanaceous hosts (C. annuum, Solanum nigrum L. and S. lycopersicum, respectively), but based on sequence identity, they were found to be closely related to the Adgen isolate from potato, with the nnp isolate shown to be a PVYO/PVYC/PVYN recombinant [24].
In addition, coat protein (CP) sequences of many PVY isolates, including eleven isolates assigned to strain group C, are currently available. Unfortunately, four of these eleven PVYC CP sequences are partial (EF192312-192314 and DQ000990). Based on alignments of CP-cistron and full-length sequences, the C isolates group separately from the PVYN and PVYO strains. In addition, the C isolates are subdivided into two different groups (C1 and C2) based on CP sequence alignments [4, 19, 22].
Throughout its history, PVYC isolate PRI-509, previously described as isolate ‘Zeeuwse Blauwe’ [23], was maintained in the plant virus collection at PRI (formerly Research Insitute for Plant Protection [IPO-DLO]) by replanting infected potato tubers yearly in an insect-proof glasshouse (18°C/16°C, day length 16 h). Aphid transmissibility of the isolate was confirmed by transmission to healthy potato plants every five years using Myzus persicae. Infection of the tubers was regularly confirmed by DAS-ELISA using a broad-spectrum antiserum to PVY (Prime Diagnostics, Wageningen, the Netherlands) and TAS-ELISA with a PVYo/c monoclonal antibody (Neogen Europe Ltd, Glasgow, Scotland).
To confirm the hypersensitive response, a biological property characteristic of PVYC isolates, PRI-509 was mechanically inoculated on the potato cultivars ‘Eersteling’ and ‘Désirée’, using potato as a source plant. In ‘Eersteling’, PRI-509 evoked the hypersensitive ‘stipple streak’ response with no systemic infection, which is typical for the interaction with the Nc gene, while in ‘Désirée’, inoculation resulted in green rings on the inoculated leaves and a systemic infection, as expected. Another PVYC isolate, PRI-503 (‘Gelderse Rode’), included earlier as a standard PVYC isolate by Kerlan et al. [14], showed comparable but milder symptoms.
Total RNA for reverse transcriptase PCR (RT-PCR) was isolated from leaves of infected potato plants using an RNeasy Plant Mini Kit (QIAGEN) according to the manufacturer’s instructions. RT-PCR amplicons covering the complete RNA genome were produced using primer pairs Y3end (2)/Y5-6300, Y3-7560/Y5-3000, and 5end (2)/Y3-4270 [24] in the Access RT-PCR System (Promega). Amplicons were purified (QIAquick PCR Purification System, QIAGEN) and sequenced directly with the PCR primers and additional PVY sequence-specific primers using an Applied Biosystems 3100 Genetic Analyser, with a DYEnamic ET Terminator Cycle Sequencing Kit (Amersham). Additionally, PVYC-specific primers were used for RT-PCR to create smaller specific PCR products for further sequencing. The 5’ terminus of the PVYC sequence was determined using a 5’ RACE kit (Roche) according to the manufacturer’s instructions. The obtained amplicon was purified and sequenced directly. Nucleotide and amino acid sequence data were analyzed and assembled using the DNASTAR package (Lasergene).
Sequence comparisons with other viruses were performed with programs from the PHYLIP package. Multiple alignments and phylogenies were performed with the CLUSTAL_X program after bootstrapping with 1000 replicates. Neighbour- joining consensus phylogenies were viewed using the NJplot program [27] and printed by using TreeView [20].
The complete RNA genome sequence of the PRI-509 isolate was determined. It contains 9699 nucleotides (nt), excluding the poly(A) tail with an open reading frame (ORF) of 9183 nt encoding a predicted polyprotein of 3061 amino acids (aa) with a molecular mass of 337 kDa. The first in-frame AUG is found at nt positions 185-187. The ORF has an UGA stop codon at nt positions 9368-9370. All typical PVY consensus amino acid sequences are present, including an EVHHQ/A NIb/CP cleavage site, the aphid-transmission-related DAG triplet in the N-terminus of the CP region (aa 6-8 of the CP) [1] and the aphid-transmission-related KITC and PTK motifs in the helper component protein (HC-Pro) region (aa 49-52 and aa 307-309 of the HC-Pro) [3].
The G1-2A6-7 motif (GGAAAAAAA; nt 2914-2922) is found in the P3 cistron. This motif indicates the beginning of the “Pretty Interesting Potyviridae ORF” (pipo), which is translated in the +2 reading-frame [5]. The ORF terminates with UAA (nt 3145-3147 and encodes a predicted protein of 77 aa.
Nucleotide sequence and derived amino acid sequence comparisons to the Adgen PVYC isolate and the three closely related non-potato isolates (nnp, SON41 and LYE84.2) show high levels of overall identity (92-95% at the nt level and 95-97% at the aa level). Based on CP sequence identity, PRI-509 is most closely related to the non-potato isolates LYE84.2 and SON41 (97 and 98% identity at the nt level and 96 and 97% identity at the aa level). This relationship was also reflected in the levels of both nt and aa sequence identities in other regions of the genome. An analysis using the different programs included in the RDP software package (version 3) [17] revealed no recombination sites within the PRI-509 full-length sequence.
Blanco-Urgoiti et al. [4] identified two different ‘genetic strains’ of PVYC (PVYC1 and PVYC2), which were separated on the basis of genetic distances, host range, reactions to the monoclonal antibody 10E3 and the coat protein processing site. Several authors have extended this distinction to include the ability to infect pepper (C1) or potato (C2) [4, 19, 22]. Remarkably, the PVYC isolate PRI-509, which had already been isolated in 1938 from, and has been maintained ever since in, potato, showed the highest levels of sequence identity in the CP region with the non-potato isolates (Figure 1). Phylogenetic analysis using either full-length sequences or those from the HC-Pro region consistently placed PRI-509 in the non-potato group C1 (trees not shown).
Fig. 1.
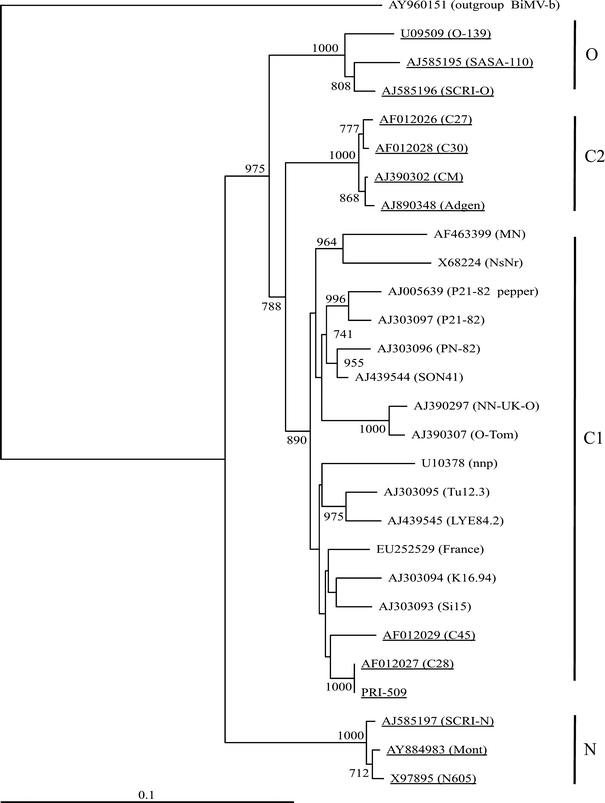
Phylogenetic analysis of all PVYC isolates and three PVYN and three PVYO isolates based on the nucleotide sequence alignment of the CP cistron (nt 8567-9367 PRI-509). Only bootstrap values greater than 70% are shown. The bar represents a p-distance of 0.1. Strain groups are marked at the right side of the figure: groups PVYN and PVYO are indicated as N and O, respectively. The two different phylogenetic PVYC subgroups are referred to as C1 and C2, respectively. The CP sequence of bidens mosaic virus isolate b (BiMV-b) was used as an outgroup. Sequence identifiers are accession numbers, with isolate names given in brackets and potato isolates underlined
In the phylogenetic analysis of CP sequences, two isolates within the subgroup C1 appear closely related to PRI-509. These two isolates (C28 and C45), sharing 100 and 97.6% sequence identity, respectively, with PRI-509, were designated as non-potato isolates by Blanco-Urgoiti et al. [4]. This prompted us to look into the origin of these isolates. They were obtained from the plant virus collection of G. Adam (at that time at DSMZ Plant Virus Division, Braunschweig, Germany) in which these isolates (and many others) were included as part of a PVY Diagnostics Ring Test performed in 1991 in the framework of COST-88 in Braunschweig. Our institute (formerly IPO-DLO) participated in this ring test. Table 2 in this ring test report, which is added to this paper as an Electronic Supplementary Table, lists all PVY isolates used in the ring test including their original host plant. Nearly all 48 PVY isolates originated from potato, while only three came from pepper (no. 4, 5 and 6) and only one each from tomato (no. 52) and tobacco (no. 51). The ring test isolate numbers 28 and 45 refer to PVYC potato isolates PRI-505 (‘Lichte Rode Star’) from the Netherlands and the PVY-cIR2 from Northern Ireland (‘Red Pentland’), respectively. Erroneously, they were later on indicated as the non-potato PVYC isolates C28 and C45 [4].
The isolates PRI-509 (‘Zeeuwse Blauwe’) and PRI-503 (‘Gelderse Rode’) were also included in the COST-88 ring test and designated as isolates 26 and 30, respectively. The serological data of the ring test confirm the PVYC status of these two isolates. (Final report about the PVY-Ringtest 1991 in the Framework of COST-88, unpublished results, available upon request).
This is the first report of a historical PVYC isolate that phylogenetically groups with isolates of the C1 subgroup of PVY, which, up to now, was erroneously thought to exclusively contain non-potato isolates.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table 2 of the “Final report about the PVY-Ringtest 1991 in the Framework of COST-88”. List of PVY-isolates used as antigens during the ring test. (TIFF 28963 kb)
Acknowledgements
This research was financially supported by the Dutch Ministry of Agriculture, Nature and Food Quality, the Dutch General Inspection Service (NAK) and the Product Board for Agriculture (PA). We thank Prof. Dr. G. Adam (University of Hamburg) for the opportunity to add Table 2 of the “Final report about the PVY-Ringtest 1991 in the Framework of COST-88” as an Electronic Supplementary Table to this paper.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
The nucleotide sequence reported in this article is available under GenBank accession number: EU563512.
References
- 1.Atreya CD, Raccah B, Pirone TP (1990) A point mutation in the coat protein abolishes aphid transmissibility of a potyvirus. Virology 178: 161-165 [DOI] [PubMed]
- 2.Beemster ABR, Rozendaal A (1972 ) Potato viruses: properties and symptoms. Viruses of potato and seed-potato production. Ed JA de Bokx Wageningen, the Netherlands. Pudoc: 115-143
- 3.Blanc S, Ammar ED, Garcia-Lampasona S, Dolja VV, Llave C, Baker J, Pirone TP (1998) Mutations in the potyvirus helper component protein: Effects on interactions with virions and aphid stylets. J Gen Virol 79: 3119-3122 [DOI] [PubMed]
- 4.Blanco-Urgoiti B, Sánchez F, Pérez De San Román C, Dopazo J, Ponz F (1998) Potato virus Y group C isolates are a homogeneous pathotype but two different genetic strains. J Gen Virol 79: 2037-2042 [DOI] [PubMed]
- 5.Chung BYW, Miller WA, Atkins JF, Firth AE (2008) An overlapping essential gene in the Potyviridae. Proc Natl Acad Sci USA 105: 5897-5902 [DOI] [PMC free article] [PubMed]
- 6.Cockerham G (1943) The reactions of potato varieties to viruses X, A, B and C. Ann Appl Biol 30: 338-344
- 7.Cockerham G (1970) Genetical studies on resistance to potato viruses X and Y. Heredity 25: 309-348
- 8.De Bokx J, Huttinga H (1981) Potato Virus Y. CMI/AAB Description of plant viruses Nr 242, Kew, Surrey
- 9.De Bokx JA (1961) Hostplants of the potato virus YN (tobacco veinal necrosis virus). T Pl-ziekten 67: 273-277
- 10.De Bokx JA, Kratchanova B, Maat DZ (1975) Some properties of a deviating strain of potato virus Y. Potato Res 18: 38-51
- 11.Fanigliulo A, Comes S, Pacella R, Harrach B, Martin DP, Crescenzi A (2005) Characterisation of Potato virus Y nnp strain inducing veinal necrosis in pepper: A naturally occurring recombinant strain of PVY. Arch Virol 150: 709-720 [DOI] [PubMed]
- 12.Glais L, Tribodet M, Kerlan C (2002) Genomic variability in Potato potyvirus Y (PVY): Evidence that PVYNW and PVYNTN variants are single to multiple recombinants between PVYO and PVYN isolates. Arch Virol 147: 363-378 [DOI] [PubMed]
- 13.Kerlan C, Robert Y, Perennec P, Guillery E (1987) Mise au point sur l'incidence du virus Yoo et Méthodes de lutte mises en oeuvre en France pour la production de semences de pommes de terre. Potato Res 30: 651-667
- 14.Kerlan C, Tribodet M, Glais L, Guillet M (1999) Variability of potato virus Y in potato crops in France. J Phytopathol 147: 643-651
- 15.Kerlan C, Moury, B (2008) Potato Virus Y. Encyclopedia of Virology Third edition
- 16.Lorenzen JH, Piche LM, Gudmestad NC, Meacham T, Shiel P (2006) A multiplex PCR assay to characterize Potato virus Y isolates and identify strain mixtures. Plant Dis 90: 935-940 [DOI] [PubMed]
- 17.Martin DP, Williamson C, Posada D (2005) RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics 21: 260-262 [DOI] [PubMed]
- 18.Moury B, Morel C, Johansen E, Jacquemond M (2002) Evidence for diversifying selection in Potato virus Y and in the coat protein of other potyviruses. J Gen Virol 83: 2563-2573 [DOI] [PubMed]
- 19.Moury B (2010) A new lineage sheds light on the evolutionary history of Potato virus Y. Molecular Plant Pathol 11: 161-168 [DOI] [PMC free article] [PubMed]
- 20.Page RDM (1996) TreeView: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357-358 [DOI] [PubMed]
- 21.Rolland M, Glais L, Kerlan C, Jacquot E (2008) A multiple single nucleotide polymorphisms interrogation assay for reliable Potato virus Y group and variant characterization. J Virol Methods 147: 108-117 [DOI] [PubMed]
- 22.Romero A, Blanco-Urgoiti B, Soto MJ, Fereres A, Ponz F (2001) Characterization of typical pepper isolates of PVY reveals multiple pathotypes within a single genetic strain. Virus Res 79: 71-80 [DOI] [PubMed]
- 23.Rozendaal A (1938) Some remarks on a virus disease of the potato variety Duke of York and its connection with the stipple streak disease. Landbouwkundig tijdschrift 619: 1063-1088
- 24.Schubert J, Fomitcheva V, Sztangret-Wišniewska J (2007) Differentiation of Potato virus Y strains using improved sets of diagnostic PCR-primers. J Virol Methods 140: 66-74 [DOI] [PubMed]
- 25.Singh RP, Valkonen JPT, Gray SM, Boonham N, Jones RAC, Kerlan C, Schubert J (2008) Discussion paper: The naming of Potato virus Y strains infecting potato. Arch Virol 153: 1-13 [DOI] [PubMed]
- 26.Smith KM (1931) Composite nature of certain potato viruses of the mosaic group. Nature 127: 702
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882 [DOI] [PMC free article] [PubMed]
- 28.Van der Vlugt RAA, Verbeek M, Cuperus C, Piron PGM, De Haan E, Van de Bovenkamp GW (2008) Strains of Potato virus Y in Dutch seed potato culture. Potato Res 51: 191–192.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 2 of the “Final report about the PVY-Ringtest 1991 in the Framework of COST-88”. List of PVY-isolates used as antigens during the ring test. (TIFF 28963 kb)


