Abstract
We analyze here the relation between alternative splicing and gene duplication in light of recent genomic data, with a focus on the human genome. We show that the previously reported negative correlation between level of alternative splicing and family size no longer holds true. We clarify this pattern and show that it is sufficiently explained by two factors. First, genes progressively gain new splice variants with time. The gain is consistent with a selectively relaxed regime, until purifying selection slows it down as aging genes accumulate a large number of variants. Second, we show that duplication does not lead to a loss of splice forms, but rather that genes with low levels of alternative splicing tend to duplicate more frequently. This leads us to reconsider the role of alternative splicing in duplicate retention.
Alternative splicing and gene duplication are thought to have major roles in the evolution of genomes. Both phenomena are widespread (Lynch and Conery 2000; Wang et al. 2008), and both provide an increased diversity of protein sequence, structure, and function (Graveley 2001; Chothia et al. 2003; Koonin and Wolf 2010). A negative correlation between these two processes has been reported: Genes belonging to large families were found to have fewer alternative splice forms than singletons or genes belonging to small families (Kopelman et al. 2005; Su et al. 2006; Talavera et al. 2007). This observation suggests a “function-sharing model,” in which the two mechanisms could be alternative ways to generate new protein forms and would be used in evolution in an interchangeable manner.
However, this interpretation is subject to controversy. First, there are fundamental differences between the generation of new duplicates or of new splice forms, as well as in their effects on protein sequence and structure (Talavera et al. 2007). Second, a mechanistic scenario accounting for this negative correlation is still missing. Su et al. (2006) hypothesized that this pattern could be explained by a loss of splice forms after duplication, but this appears to stand in contradiction with the observation that older duplicate genes experience more alternative splicing than recent ones (Kopelman et al. 2005; Su et al. 2006). Here, we provide new insight into this question and show how the age of a gene and its evolutionary history influence its level of alternative splicing.
Results
A complex relation between duplication and alternative splicing
Using the approach that identified the negative correlation, with more recent data, a parabola curve is observed between the proportion of genes undergoing alternative splicing and gene family size (Fig. 1A; Supplemental Fig. S1). The parabola has a significantly better fit than a simple linear model (P = 0.01; Supplemental Fig. S2). Notably, singletons have significantly fewer splice forms than genes of family size two (Fisher's exact test, P = 3.8 × 10−14). While there is a decrease in splicing in larger gene families, only those with 10 or more members have lower alternative splicing than singletons. This pattern is not specific to human: it holds in mouse and in zebrafish (although the proportion of genes with known alternative splicing is lower in zebrafish than in human and mouse due to a lower EST coverage; Supplemental Figs. S3, S4).
Figure 1.
Relation between gene family size and production of alternatively spliced variants in human genes. (A) Fraction of genes containing more than one splice variant (i.e., with alternative splicing). (B) Mean number of alternative spliced variants. Binning of family size was made as in Su et al. (2006), but the relation is independent of binning: Similar results were obtained when binning was made as in Kopelman et al. (2005) (Supplemental Fig. S1) or when no binning was made (Supplemental Fig. S2). The number of genes in each bin is indicated at the bottom of the bars.
These observations are not a simple consequence of methodological choices: Using the mean number of splice forms per gene, as in Su et al. (2006), yields similar results (Fig. 1B), as does a different gene family prediction method (Ensembl protein families; Supplemental Fig. S5) or a different estimate of the number of splice forms per gene (UCSC Genome Browser) (Supplemental Fig. S6). Estimates of the number of splice forms could be biased by expressed sequence tag (EST) coverage of highly expressed genes, but controlling for EST counts does not change the results (Supplemental Fig. S7), probably thanks to the deep coverage of human ESTs (Brett et al. 2002; Kopelman et al. 2005; Su et al. 2006). Similarly, controlling for the number of constitutive exons in genes, the selective pressure acting on protein sequences (dN/dS ratio), or maximum transcript length, did not alter the results (Supplemental Figs. S8–S10). And neither does removing very recent duplication events, which might include genome assembly errors (Supplemental Fig. S11).
In light of our results, we note that singletons did not consistently show the highest level of alternative splicing in previous studies (Kopelman et al. 2005; Su et al. 2006; Jin et al. 2008). Thus, these results question the hypothesis of equivalence between duplication and alternative splicing, since genes that never or rarely duplicated do not undergo alternative splicing more often.
Age matters
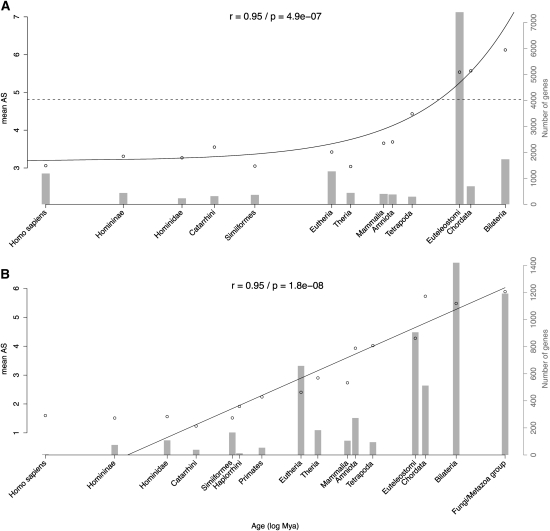
It has been suggested that older duplicates undergo more alternative splicing than recent duplicates (Kopelman et al. 2005; Su et al. 2006; Shabalina et al. 2010). We dated duplication events using tree reconciliation from Ensembl Compara (Vilella et al. 2009), which allows higher resolution than previously used methods. We find a highly significant linear relation between the age of the most recent duplication and the mean number of splice variants (Fig. 2A; note the logarithmic scale on the x-axis; see Supplemental Fig. S12 for a linear-scale representation). The same trend is seen using the proportion of genes undergoing alternative splicing (Supplemental Fig. S13). This linear model explains a remarkably high portion of the variance of the data set (R2 = 90%).
Figure 2.
Relation between gene age and production of alternatively spliced variants in human. (A) Mean number of alternative spliced variants is plotted against the age of the last gene duplication. The dashed line represents the mean level of alternative splicing of genes that did not experience duplication (singletons). For better visualization, only age categories with more than 30 genes are shown. The gray background histogram represents the number of genes in each category of age (right y-axis). Similar results were obtained when using the fraction of genes undergoing alternative splicing (Supplemental Fig. S13). The x-axis is shown in log-scale. Estimates of divergence times in million years were obtained from the TimeTree database (see Methods). Only the taxonomic name of the lineage is displayed. A weighted linear regression was adjusted on the data, with real scale and log-transformed time. The best fitting model was kept, and the regression line, P-value, and r coefficients are displayed. (B) Mean number of alternative spliced variants is plotted against age of singletons. Similar results were obtained when using the fraction of genes undergoing alternative splicing (Supplemental Fig. S17).
These results could have been affected by the time points used in this analysis, imposed by the divergence times of sequenced species in Ensembl. We show that this is not the case since a nonparametric Spearman's rank correlation is also very significant (ρ = 0.80, P = 3 × 10−4). Additionally, the bony vertebrate time point (Eutelostomi) displays a high number of duplicates, most probably generated by two rounds of whole-genome duplication (Putnam et al. 2008). Although this data point fits the general trend, the large number of genes in this category could strongly influence the weighted regression. Removing it from the data set did not affect the results (r = 0.98, P = 9.8 × 10−9; data not shown). Since the other duplicates were probably generated by other mechanisms than whole genome duplication, this implies that there is no strong effect of the type of duplication.
It is probable that old duplicates belong to different functional gene classes from young duplicates. This can be a confounding effect if these different functional categories differ regarding alternative splicing. To control for this effect, we divided our data set into top-level Gene Ontology Molecular Function categories, and we analyzed if the same trend was seen in each category. In all cases, the trend was similar to Figure 2A (Supplemental Fig. S14). For example, catalytic activity, which has many old duplicates, and structural molecule activity, which has many recent duplicates, show very similar patterns. In only one case was it not significant (Supplemental Fig. S14D, transporter activity, P = 0.21). While this may reflect a specificity of this class of genes, it is most probably due to a lack of statistical power: Very few genes have duplicated outside whole genome duplication in this category. Globally, it seems unlikely that the trend seen in Figure 2A is due to a functional bias.
The methodology used here and in previous studies (Kopelman et al. 2005; Su et al. 2006) does not estimate the age of each duplicate copy strictly speaking, but rather the time since the most recent event of duplication. If duplication is asymmetric, as in the case of retrogenes, the new copy will be as young as this recent duplication event, while the parent copy will be older. It should be noted that asymmetric events have been estimated to generate a minority of mammalian paralogs (Cusack and Wolfe 2007) and that retrogenes are expected to have no introns, thus very little if any alternative splicing. To evaluate the putative impact of asymmetric duplication on our results, we measured the difference in isoform number between pairs of paralogs (Supplemental Fig. S15). If asymmetry has an important impact, we expect young duplicates to differ strongly in the number of isoforms. But we observe the smallest difference between young duplicates, which indicates a weak effect of asymmetry at birth. Thus the average measure used here and in previous studies (Kopelman et al. 2005; Su et al. 2006) appears to summarize adequately the effect of duplication age on alternative splicing.
The results are consistent with the hypothesis that duplication is followed by a progressive acquisition of new splice forms. The linear regression (Fig. 2A) estimates a rate of 2.6 × 10−3 new splice forms per gene per million years, or one new splice form per gene every 385 million years. Interestingly, this rate is very close to the rate of exon gain estimated in the mouse lineage (2.7 × 10−3) (Wang et al. 2005). A plateau seems nevertheless to be reached when >80% of the genes undergo alternative splicing (Supplemental Fig. S13), with a mean number of splice forms around six (Fig. 2A). Recent duplicates show half this level, with a mean around 3 splice forms per gene. Of note, a large spread of the distribution of number of alternative splice variants is observed for all ages. This plateau should not be understood as an absolute limit, but rather denotes that the splice form acquisition process slows down for old genes (see Supplemental Fig. S16 for a comparison of the distribution of number of splice forms for young genes vs. old genes).
The increase in number of splice forms with evolutionary time is not specific to duplicates. We have dated singleton genes by the oldest node of their GeneTree (see Methods). We find a similar trend regarding mean level of alternative splicing, but it is best modeled as logarithmic (Fig. 2B; Supplemental Figs. S17, S18) (r = 0.95, P = 1.8 × 10−8). This model is consistent with an age-dependent rate of acquisition; older singletons acquire new splice forms at a slower pace. The initial state of young genes is logically low (around 1.5 splice forms per gene), with only few of them undergoing splicing (∼20%). It takes ∼40 million yr before new genes start to acquire alternative splice forms, while the process seems more rapid with duplicates. Of note, the potential bias induced by fast-evolving genes, whose age tends to be underestimated (Elhaik et al. 2006; Alba and Castresana 2007), goes against the trend observed here, making our model a conservative estimate. Besides, binning genes evolving at different rates (dN/dS) yields the same pattern (Supplemental Fig. S19). The acquisition of splice forms is thus not restricted to duplicate genes. Moreover, the rate of acquisition seems to be higher in singletons: They reach a similar plateau (mean level of around six splice forms per singleton, and 80% of genes undergoing alternative splicing) in a shorter time compared to duplicates, whereas their initial state was lower. For the relatively old genes that experienced the whole-genome duplication events ancestral to vertebrates, we see no significant difference in the level of alternative splicing between genes that were kept in duplicate or not (Kruskal-Wallis test, P = 0.24; Supplemental Fig. S20). This indicates that when the plateau is reached, different evolutionary histories are no longer reflected in the level of alternative splicing.
The trend of gain is not due to nonfunctional splice forms
It is debated if the annotated alternative splice forms are mostly functional, or if they are mostly “transcriptional noise” (Keren et al. 2010). To control for the impact of nonfunctional transcripts, we repeated the analysis with several strict quality criteria concerning splicing annotation. We first conserved only transcripts of our data set that have a protein product in Ensembl: the results were unchanged (Supplemental Fig. S21).
Next, we extracted manually annotated splicing events from SWISS-PROT (The UniProt Consortium 2010), adding the criterion that all splicing events should be confirmed by at least two observations of a given form in the literature. A significant correlation is found with the proportion of genes undergoing alternative splicing (Supplemental Fig. S22). The correlation between age of duplication and mean number of splice forms is also positive, but not quite significant (P = 0.08), probably because of the small quantity of splice events annotated with such stringent criteria.
Finally, we extracted from the H-DBAS database (Takeda et al. 2010) all human transcript forms that could be mapped to a mouse cDNA. These evolutionarily conserved splice forms are likely to be functional (Yeo et al. 2005). A significant trend is seen with the proportion of genes having more than one conserved splice form (Supplemental Fig. S23). It is marginally significant (P = 0.018) for the mean number of conserved splice forms, again because of less data.
We can thus conclude that our results are not due to spurious annotations in genomic data.
Which genes preferentially acquire new splice forms?
To gain better insight into the process of progressive acquisition of splice forms, we divided our data set of singletons into different subcategories. After fitting linear models, comparing the regression slopes shows that the acquisition of splice forms is faster for genes with fewer constitutive exons (Supplemental Fig. S24A), but slower for genes with shorter transcript length (although the trend is weak) (Supplemental Fig. S24B). This apparent contradiction might be due to the interplay between mechanistic causes (more opportunities of variation in longer transcripts) and selective constraints (less purifying selection on genes with less constitutive exons). The genes under strongest purifying selection on the protein sequence have the lowest rate of splice form acquisition (Supplemental Fig. S24C). Considering EST counts, no rate difference is seen between rates of acquisition for genes belonging to different bins, although the absolute number of forms detected is lower for genes with fewer ESTs (Supplemental Fig. S24D).
Finally, rates differ between splicing patterns. Exon/intron isoform events have the highest rate of acquisition with age (Supplemental Fig. S24E), followed by cassette exons and intron retention events. Mutually exclusive exons have the slowest dynamics, probably because they involve a complex combinatorial pattern. Thus the gain of new isoforms might not be occurring mainly through exon duplications, contrary to some suggestions in the literature (for review, see Keren et al. 2010). This ranking, although consistent with the reported proportions of the different events in the genome (Kim et al. 2007; Wang et al. 2008), is to be interpreted with caution, as the computational detection of these events might suffer from potential biases and methodological limits; experimental validation would be needed to draw more secure conclusions.
The biases induced by duplication
To explain that genes belonging to large families tend to show lower levels of alternative splicing, it has been suggested that a rapid loss of alternative splicing might occur after gene duplication (Su et al. 2006). First, this interpretation is questioned by the observation that very recently duplicated genes show a low level of alternative splicing (Fig. 2A), including genes whose last duplication was after the split between human and chimpanzee (∼6 million yr ago). Second, this conclusion was reached after comparison of levels of alternative splicing for human genes that duplicated since the divergence with mouse, versus genes that did not. This methodology does not account for preduplication biases (Studer and Robinson-Rechavi 2009).
To correct for such biases, we can approximate the ancestral state before gene duplication by the level of alternative splicing of an orthologous gene that diverged before the duplication. We compared the number of isoforms between mouse genes, either orthologous to a single human gene (one-to-one orthologs), or orthologous to several human genes that duplicated after the human–mouse split (one-to-many orthologs). The number of splice forms is significantly different between the two groups (Fig. 3A; Wilcoxon test P = 0.00081), one-to-one orthologs having more splice forms than one-to-many (61% of the genes undergo alternative splicing vs. 53%). This shows that genes with less alternative splicing tend to duplicate more.
Figure 3.
Relation between duplication and alternative splicing. (A) Histograms comparing the number of splice forms between mouse one-to-one orthologs of human genes (no duplication in the human lineage; black) and mouse one-to-many orthologs of human genes (at least one duplication in the human lineage; gray). (B) Histograms comparing the ratio of the number of splice forms between human genes and their mouse one-to-one orthologs (if no duplication occurred in the human lineage; black) or their mouse one-to-many orthologs (if at least one duplication occurred in the human lineage; gray). For the latter, the mean number of splice forms in human genes was used (see Methods).
It is still possible that there is a loss of alternative splicing after duplication. To control for the preduplication bias, we used the ratio of the level of alternative splicing in a human gene to the level in its mouse ortholog. We compared the distribution of this ratio depending on whether the human gene duplicated or not since the divergence with mouse. There is no significant shift of the distributions of ratios between the two groups, indicating that no systematic loss is experienced by duplicate genes (Fig. 3B; Wilcoxon test, P = 0.25). Thus, the lower number of splice variants of duplicates (Su et al. 2006) appears to reflect a preduplication bias.
These results are averages at the genome scale. Of note, because of the large variance in the distribution of the number of splice forms, we still find that in 27% of the cases, duplicates experienced a loss of splice forms. But this is significantly less than the proportion of genes that experienced a gain of splice forms after duplication (49%; Fisher's exact test, P = 0.00042), and not significantly different from the proportion of loss for genes that experienced no duplication in the human lineage (32%; Fisher's exact test, P = 0.12).
Finally, if the genes that duplicate more have lower levels of alternative splicing, the trend reported for duplicates (Fig. 2A) might be biased. To correct for this, we examined the ages of a coherent group of duplicate genes that experienced their last duplication at the same time, with age calculated as for singletons (origin of the gene family). We observe a corrected trend, best modeled with a logarithmic rather than a linear model (Supplemental Fig. S25), very similar to singletons. Thus the dynamics of splice form acquisition might not be different between duplicates and singletons, once biased duplication is taken into account.
Discussion
Our results show that the relation between alternative splicing and gene duplication is more complex than previously reported, and that the age of a gene is a crucial factor to consider in this analysis. A progressive acquisition of splice forms is detected with evolutionary time, and this trend is shared by duplicates and singletons. The difference seen in the dynamics of acquisition in the two groups (Fig. 2) is potentially spurious, explained by a higher probability of duplication of genes with lower levels of alternative splicing. Overall, the mean trend on all genes seems to be best modeled as logarithmic, with a rate of splice form acquisition decreasing progressively over time (Fig. 2B; Supplemental Fig. S25). For old genes, a plateau is reached, both for duplicates and singletons, after which genes do not gain any more splice forms on average, or at a much slower rate. This slowdown might be due to purifying selection on old genes with many isoforms. Indeed, stronger purifying selection has been reported on old genes (Supplemental Fig. S26; Alba and Castresana 2005, 2007; Wolf et al. 2009). This suggests that the acquisition of splice forms in younger genes might be under relaxed selection and that purifying selection gets increasingly efficient at preventing the acquisition of new splice variants as genes get older.
This is consistent with the slower accumulation of those alternative splice forms that are most likely to be functional (Supplemental Figs. S21–S23). The idea that young genes accumulate splice forms nearly neutrally is also supported by evidence that the transition from constitutive to alternative splicing might be a neutral process (Lev-Maor et al. 2007), and more generally by the observation that alternative splicing is a noisy phenomenon (Artamonova and Gelfand 2007; Irimia et al. 2008). Near neutrality is consistent with the observation of higher levels of alternative splicing in vertebrates than in species with larger effective population sizes, such as yeast, nematodes, or fruit fly (Artamonova and Gelfand 2007; Irimia et al. 2008; Keren et al. 2010). In the latter, selection might be more efficient at slowing down the process of new splice forms acquisition. Indeed, in Drosophila melanogaster, the acquisition of new splice forms is slower and a plateau is reached more rapidly, compared to vertebrates (Supplemental Fig. S27; Su et al. 2006).
Considering that genes acquire new splice variants with evolutionary time, it is possible to reinterpret the relation between family size and alternative splicing (Fig. 1). The previously reported negative correlation does not hold at the genome level, mainly because singletons and genes from small families do not show the highest level of alternative splicing. It is likely that these constitute a very heterogeneous group, composed both of recent genes that had time to accumulate neither splice variants nor duplicates, and of old genes, with many splice variants, which never duplicated. Consistently, if we control for this effect by considering groups of genes of similar ages for the analysis, we generally recover a weak negative correlation between family size and level of alternative splicing (linear regression, from r = 0.79 and P = 7.5 × 10−7 for “Chordata” to r = 0.47 and P = 0.01 for the “Fungi/Metazoa” group; among age classes with ≥1000 genes, only “Eutheria” is not significant; fitting a parabola is never significant). Thus the global relation between level of alternative splicing and family size is confounded by the age of genes.
Finally, a preduplication bias is sufficient to explain the lower number of splice forms of recent duplicates compared to singletons. Genes with a low level of alternative splicing are likely to duplicate frequently, and genes with a higher level of splicing duplicate less frequently. We do not detect any evidence for the loss of isoforms suggested to result from duplication (Kopelman et al. 2005; Su et al. 2006), invalidating subfunctionalization through partitioning of splice variants as a major mechanism for duplicate gene retention. Of note, retention through splice variant neofunctionalization is not favored by our observations, since isoforms are gained at a rather slow pace, probably insufficient to explain duplicate retention on the short term (Innan and Kondrashov 2010).
In conclusion, it appears that the effects of gene duplication on alternative splicing might be quite limited. It is rather the level of alternative splicing of a gene that appears to influence its potential to duplicate.
Methods
Identification of duplicate genes and families
Gene families were obtained from the Ensembl database release 57 (Hubbard et al. 2009). We used the Perl API and BioMart (Smedley et al. 2009) to query the Ensembl Compara GeneTrees (Vilella et al. 2009) and scan for specific gene topologies. A precise description of the methods used for these trees can be found at http://www.ensembl.org/info/docs/compara/homology_method.html. We selected sets of genes with or without duplications on specific branches of the bilaterian phylogenetic tree. Duplications annotated as “dubious” by Ensembl Compara were not considered. The parsing of GeneTrees allowed us to retrieve the age of the most recent retained duplication for every gene, including the genes that duplicated at the time of the 2R whole-genome duplication but not since, and to calculate family size and dN/dS ratios. We dated singleton genes by their first appearance in the phylogeny; this consisted of retrieving the age of the oldest node of their GeneTree in Ensembl. To study the asymmetry of the number of isoforms after duplication (Supplemental Fig. S15), we restricted the analysis to duplicate pairs for which no later duplication occurred, so that the number of isoforms of the two duplicates could be compared easily. These data are available as supporting information.
To identify one-to-one and one-to-many cases of orthology between human and mouse genes, we used the Ensembl Perl API to explore trees and detect duplication nodes since the human–mouse split node. For genes that are singletons since that time, we paid special attention that they did not undergo a scenario of duplication followed by a subsequent loss.
The pairwise dN and dS between Homo sapiens and Mus musculus is calculated with codeml from the PAML package in the Ensembl pipeline (model = 0, NSsites = 0) (Yang 1997). Ensembl considers that dS values are saturated when they reach a threshold that is 2 × median(dS). See http://www.ensembl.org/info/docs/compara/homology_method.html for further details.
Independent estimates of family sizes were calculated using Ensembl protein families. These family predictions are based on the Tribe MCL clustering method, including all protein isoforms of every coding gene that Ensembl predicts, but also all fungi/metaozoa proteins present in SWISS-PROT and TrEMBL. See http://www.ensembl.org/info/docs/compara/family.html for further details. For analyses using groups of family sizes, we used the same binning as Su et al. (2006). But all results hold when using another binning, for example, similar to Kopelman et al. (2005).
Number of isoforms
For human (H. sapiens), mouse (M. musculus), zebrafish (Danio rerio), and fruit fly (D. melanogaster), we retrieved the number of transcripts for all protein-coding genes from Ensembl 57 (Hubbard et al. 2009). An independent estimation of the number of isoforms per gene was obtained for human from the UCSC Genome Browser (February 2009 assembly, GRCh37/hg19) (Rhead et al. 2010). We used the “knownIsoforms” table, which displays the clustering of UCSC transcripts (“knownGenes”). The mapping to Ensembl genes was then made using the “knownToEnsembl” mapping of UCSC known genes to Ensembl transcripts. Clusters that could be mapped to a unique Ensembl gene were kept. Transcript counts could be retrieved for 19,914 protein-coding Ensembl genes.
To retrieve alternative splice forms manually annotated in SWISS-PROT (The UniProt Consortium 2010), we downloaded the file of all human annotated entries from the Uniprot website (release 2010_04) (http://www.uniprot.org/uniprot/?query=organism%3a%22Homo+sapiens+[9606]%22+AND+reviewed%3ayes&sort=score&format=*). A Perl script was used to extract the number of splice forms (CC lines, “ALTERNATIVE PRODUCTS:”, “Event=Alternative splicing”). Splice forms with no experimental validation were discarded (“Note=No experimental confirmation available”). Only accessions with a one-to-one mapping to Ensembl protein-coding genes were kept for the analysis. Systematic annotation of alternative splicing is currently not performed in SWISS-PROT, but all intron–exon junctions of informed events are manually checked (Marie-Claude Blatter, pers. comm.). This data set is thus likely to contain mainly functional splicing events referenced in the literature.
To retrieve splice forms conserved with mouse, we used the H-DBAS database release 4 (Takeda et al. 2010). A scheme explaining the methodology used for assessing human–mouse conservation can be found at http://jbirc.jbic.or.jp/h-dbas/document/scheme_genomic_comparison.html.
From the file http://h-invitational.jp/download/h-dbas/H-DBAS_version4.tar.gz, we extracted the HIX loci and their HIT clusters that showed conservation with mouse (CDNA rows, column 26: Genomic conservation with mouse cDNA). Only HIX IDs with a one-to-one mapping to Ensembl protein-coding genes were kept for the analysis.
The measures of alternative splicing used throughout the manuscript are the proportion of genes undergoing alternative splicing and the mean number of splice forms per gene. These measures are traditionally used in the literature (e.g., Kopelman et al. 2005; Su et al. 2006; Talavera et al. 2007).
The comparisons of alternative splicing levels in human and mouse were made using data from Ensembl 57. The use of ratios of the number of splice forms in the two species can account for differences in genome annotation quality. For genes that experienced duplications since the human–mouse split, we used the ratio of the mean number of splice forms in human genes divided by the number of splice forms of the mouse ortholog.
Linear regressions
For the relation between alternative splicing and family size, a weighted linear regression between alternative splicing measure and family size was fit to the data, and an F-test was used to assess if the slope was significantly different from zero. Weights were the total number of genes for each increment of family size. We adjusted a parabola (polynomial model of order 2) in the same way and used an ANOVA to estimate if the increase in fit to the data (r) between the linear and parabola models was significant.
For the relation between gene age and production of alternatively spliced variants, a weighted linear regression was adjusted on the data, with real scale and log-transformed time. Weights were the total number of genes for each age category. The best fitting model (best r value) was kept and displayed.
Gene Ontology
Mapping of genes to Gene Ontology (GO) functional categories was downloaded from Ensembl release 57 (Hubbard et al. 2009). We propagated the mapping to the top-level categories of the GO slim ontology (a simplified version of the GO ontology), and we only retained categories mapped to more than 500 genes, namely, GO:0005488 (binding), GO:0003824 (catalytic activity), GO:0004871 (signal transducer activity), GO:0005198 (structural molecule activity), GO:0005215 (transporter activity), GO:0030234 (enzyme regulator activity), and GO:0030528 (transcription regulator activity).
EST counts
EST data were retrieved from Bgee release 7 (dataBase for Gene Expression Evolution, http://bgee.unil.ch/), a database comparing transcriptome data between species (Bastian et al. 2008), including EST libraries from UniGene (Pontius et al. 2003). The mapping of UniGene clusters on Ensembl genes is taken from Ensembl 57 (Hubbard et al. 2009), where a percentage of identity of 90% is set as the minimum threshold to link an Ensembl gene with a UniGene cluster. Only EST libraries obtained under nonpathological conditions, with no treatment (“normal” gene expression), are included in Bgee.
Transcript and splice form features
Transcript length and number of constitutive exons and alternative splicing events were retrieved from Ensembl 57 via BioMart (Smedley et al. 2009). Alternative splicing events are classified with different codes (A5SS, alternative 5′ splice site; A3SS, alternative 3′ splice site; II, intron isoform; EI, exon isoform; CE, cassette exon; IR, intron retention; MXE, mutually exclusive exons). For the analysis, A5SS, A3SS, EI, and II events were grouped together because they represent similar events and the overlap between these categories was high. Other alternative splice forms (e.g., alternative first exon, alternative last exon, alternative termination, alternative initiation) are not predicted in Ensembl because they require experimental evidence (see, e.g., Wang et al. 2008).
Taxonomy
Molecular estimates of the age of taxonomic groups was obtained from the database TimeTree (Hedges et al. 2006; http://www.timetree.org, April 2010). When available the “TimeTree expert” result was used. Otherwise, the weighted average (nuclear + mitochondrial) was used.
Tools
R was used for statistical analyses and plotting (http://www.R-project.org/; R Development Core Team 2007).
Acknowledgments
We thank members of the Robinson-Rechavi lab for helpful discussions and Marie-Claude Blatter for help with SWISS-PROT annotation of alternative splicing. We acknowledge funding from Etat de Vaud and Swiss National Science Foundation grant 116798.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.113803.110.
References
- Alba MM, Castresana J 2005. Inverse relationship between evolutionary rate and age of mammalian genes. Mol Biol Evol 22: 598–606 [DOI] [PubMed] [Google Scholar]
- Alba MM, Castresana J 2007. On homology searches by protein Blast and the characterization of the age of genes. BMC Evol Biol 7: 53 doi: 10.1186/1471-2148-7-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artamonova II, Gelfand MS 2007. Comparative genomics and evolution of alternative splicing: The pessimists' science. Chem Rev 107: 3407–3430 [DOI] [PubMed] [Google Scholar]
- Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M 2008. Bgee: Integrating and comparing heterogeneous transcriptome data among species. In DILS: Data Integration in the Life Sciences 5th International Workshop, DILS 2008, Evry, France, June 25–27, 2008. Proceedings (ed. Bairoch A et al. ), Vol. 5109, pp. 124–131 Springer, New York [Google Scholar]
- Brett D, Pospisil H, Valcarcel J, Reich J, Bork P 2002. Alternative splicing and genome complexity. Nat Genet 30: 29–30 [DOI] [PubMed] [Google Scholar]
- Chothia C, Gough J, Vogel C, Teichmann SA 2003. Evolution of the protein repertoire. Science 300: 1701–1703 [DOI] [PubMed] [Google Scholar]
- Cusack BP, Wolfe KH 2007. Not born equal: Increased rate asymmetry in relocated and retrotransposed rodent gene duplicates. Mol Biol Evol 24: 679–686 [DOI] [PubMed] [Google Scholar]
- Elhaik E, Sabath N, Graur D 2006. The “inverse relationship between evolutionary rate and age of mammalian genes” is an artifact of increased genetic distance with rate of evolution and time of divergence. Mol Biol Evol 23: 1–3 [DOI] [PubMed] [Google Scholar]
- Graveley BR 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S 2006. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. 2009. Ensembl 2009. Nucleic Acids Res 37: D690–D697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F 2010. The evolution of gene duplications: Classifying and distinguishing between models. Nat Rev Genet 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Garcia-Fernandez J, Vinther J, Roy SW 2008. Widespread evolutionary conservation of alternatively spliced exons in Caenorhabditis. Mol Biol Evol 25: 375–382 [DOI] [PubMed] [Google Scholar]
- Jin L, Kryukov K, Clemente JC, Komiyama T, Suzuki Y, Imanishi T, Ikeo K, Gojobori T 2008. The evolutionary relationship between gene duplication and alternative splicing. Gene 427: 19–31 [DOI] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G 2010. Alternative splicing and evolution: Diversification, exon definition and function. Nat Rev Genet 11: 345–355 [DOI] [PubMed] [Google Scholar]
- Kim E, Magen A, Ast G 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35: 125–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI 2010. Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet 11: 487–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelman NM, Lancet D, Yanai I 2005. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 37: 588–589 [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Goren A, Sela N, Kim E, Keren H, Doron-Faigenboim A, Leibman-Barak S, Pupko T, Ast G 2007. The “alternative” choice of constitutive exons throughout evolution. PLoS Genet 3: e203 doi: 10.1371/journal.pgen.0030203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Pontius JU, Wagner L, Schuler GD 2003. UniGene: A unified view of the transcriptome. In The NCBI handbook (ed. McEntyre J, Ostell J), pp. 1–12 National Library of Medicine, US, NCBI, Bethesda, MD [Google Scholar]
- Putnam NH, Butts T, Ferrier DEK, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu J-K, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453: 1064–1071 [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2007. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Ogurtsov AY, Spiridonov AN, Novichkov PS, Spiridonov NA, Koonin EV 2010. Distinct patterns of expression and evolution of intronless and intron-containing mammalian genes. Mol Biol Evol 27: 1745–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A 2009. BioMart—biological queries made easy. BMC Genomics 10: 22 doi: 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M 2009. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet 25: 210–216 [DOI] [PubMed] [Google Scholar]
- Su Z, Wang J, Yu J, Huang X, Gu X 2006. Evolution of alternative splicing after gene duplication. Genome Res 16: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda J, Suzuki Y, Sakate R, Sato Y, Gojobori T, Imanishi T, Sugano S 2010. H-DBAS: human-transcriptome database for alternative splicing: Update 2010. Nucleic Acids Res 38: D86–D90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera D, Vogel C, Orozco M, Teichmann SA, de la Cruz X 2007. The (in)dependence of alternative splicing and gene duplication. PLoS Comput Biol 3: e33 doi: 10.1371/journal.pcbi.0030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium 2010. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res 38: D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E 2009. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zheng H, Yang S, Yu H, Li J, Jiang H, Su J, Yang L, Zhang J, McDermott J, et al. 2005. Origin and evolution of new exons in rodents. Genome Res 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf YI, Novichkov PS, Karev GP, Koonin EV, Lipman DJ 2009. The universal distribution of evolutionary rates of genes and distinct characteristics of eukaryotic genes of different apparent ages. Proc Natl Acad Sci 106: 7273–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]
- Yeo GW, Van Nostrand E, Holste D, Poggio T, Burge CB 2005. Identification and analysis of alternative splicing events conserved in human and mouse. Proc Natl Acad Sci 102: 2850–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]