Abstract
The rate of RNA polymerase II (Pol II) elongation can influence splice site selection in nascent transcripts, yet the extent and physiological relevance of this kinetic coupling between transcription and alternative splicing (AS) is not well understood. We performed experiments to perturb Pol II elongation and then globally compared AS patterns with genome-wide Pol II occupancy. RNA binding and RNA processing functions were significantly enriched among the genes with Pol II elongation inhibition-dependent changes in AS. Under conditions that interfere with Pol II elongation, including cell stress, increased Pol II occupancy was detected in the intronic regions flanking the alternative exons in these genes, and these exons generally became more included. A disproportionately high fraction of these exons introduced premature termination codons that elicited nonsense-mediated mRNA decay (NMD), thereby further reducing transcript levels. Our results provide evidence that kinetic coupling between transcription, AS, and NMD affords a rapid mechanism by which cells can respond to changes in growth conditions, including cell stress, to coordinate the levels of RNA processing factors with mRNA levels.
Different steps in the synthesis, processing, and turnover of transcripts produced by RNA polymerase II (Pol II) are highly interconnected and can influence each other (Maniatis and Reed 2002; Bentley 2005; Moore and Proudfoot 2009). Microscopy-based evidence has suggested that the majority of precursor (pre-)mRNA processing events occur as transcripts are synthesized by Pol II (Beyer and Osheim 1988; for reviews, see Carmo-Fonseca and Carvalho 2007; Zhong et al. 2009), and additional studies have revealed physical and functional interactions between the Pol II machinery and factors involved in 5′-end cap formation, splicing, and 3′-end processing (Bentley 2005; Kornblihtt 2007; Allemand et al. 2008). An important consequence of the communication between Pol II and pre-mRNA processing factors is efficient and coordinated pre-mRNA processing steps.
It has been estimated that >95% of human multi-exon genes produce alternatively spliced transcripts (Pan et al. 2008; Wang et al. 2008). However, the extent to which transcriptional regulation directly impacts alternative splicing (AS) and the physiological roles of coupled regulation between these processes are not well understood. Nevertheless, numerous studies employing model pre-mRNA reporters and a smaller number of studies investigating endogenous transcripts have identified exons that display altered regulation when transcriptional activity is modulated (Allemand et al. 2008; Sanchez et al. 2008; Barboric et al. 2009; Schor et al. 2009; for reviews, see Kornblihtt et al. 2004; Kornblihtt 2007). Based on the results from these studies, two distinct but nonmutually exclusive models have been proposed to account for these effects: (1) the “recruitment model” and (2) the “kinetic model.”
In the recruitment model, one or more AS regulators physically interact with factors that control transcription to affect subsequent splicing decisions (Kornblihtt et al. 2004; Kornblihtt 2007). Examples include Ser/Arg-repeat (SR) splicing factors or other splicing regulators associated with transcriptional activators, coactivators, Pol II, or chromatin (Ge et al. 1998; Monsalve et al. 2000; Rosonina et al. 2005; de la Mata and Kornblihtt 2006; Das et al. 2007; Loomis et al. 2009; Luco et al. 2010). Such associations may also act in a bidirectional manner, since splicing factors bound to complexes on nascent pre-mRNA are also thought to modulate transcriptional activity (Fong and Zhou 2001; Lin et al. 2008).
The kinetic model, which is supported by numerous experiments from the Kornblihtt group and others, posits that factors affecting Pol II elongation rate regulate AS by controlling the accessibility of cis-competing splice sites in nascent RNA (de la Mata et al. 2003). In the most extensively studied examples, alternative exons flanked by one or more suboptimal splice site are in competition with stronger splice sites belonging to upstream and downstream exons (Kornblihtt et al. 2004; Kornblihtt 2007). If Pol II stalls within the regulated exon or neighboring intron sequences, there is more time for factors to recognize the suboptimal splicing signals, which therefore results in increased alternative exon inclusion. If Pol II elongation is more rapid, the stronger distal splice sites are preferentially utilized, resulting in increased alternative exon skipping. Reduced Pol II elongation can also result in increased exon skipping, for example, by facilitating the binding of negative-acting regulators to pre-mRNA silencer sequences that are proximal to alternative exons (Pagani et al. 2003).
In this study, we have globally analyzed the extent to which changes in Pol II elongation can affect the regulation of endogenous AS events, and we have also investigated the possible physiological relevance of Pol II-coupled splicing regulation. By comparing directly the results from profiling AS using a custom microarray, with results from profiling steady-state Pol II occupancy using chromatin immunoprecipitation coupled to high-throughput DNA sequencing (ChIP-seq), we provide evidence that altered rates of Pol II elongation or occupancy preferentially affect exons in genes that encode RNA binding proteins, including many splicing and RNA processing factors. Upon reduced Pol II elongation, these alternative exons predominantly display increased inclusion levels and many of them introduce premature termination codons (PTCs) into the spliced mRNA that elicit nonsense-mediated mRNA decay (NMD). Our data and analyses further suggest a role for Pol II-coupled AS regulation of transcripts from RNA processing factor genes as an evolutionarily conserved response to cellular stress.
Results
Global detection of AS events affected by inhibition of RNA polymerase II elongation
To identify AS events influenced by changes in Pol II elongation, we performed AS microarray profiling (Pan et al. 2004; Shai et al. 2006) of poly(A)+ RNA from stimulated Jurkat T lymphoma cells treated with two drugs that inhibit Pol II elongation: 5,6-dichloro-1-β-D-ribofuranosyl-benzimidazole (DRB) and camptothecin (Fig. 1A). DRB inhibits the kinase activity of the CDK9 subunit of the positive transcription elongation factor P-TEFb (Marshall et al. 1996; Sims et al. 2004; Saunders et al. 2006), and camptothecin creates topoisomerase I-DNA adducts which physically interfere with Pol II elongation (Kessel 1971; Darzacq et al. 2007). Treatment using these two drugs therefore provides a means to assess the consequences of mechanistically distinct modes of Pol II elongation inhibition on AS and mRNA levels.
Figure 1.
Global profiling of AS and mRNA levels affected by DRB and camptothecin inhibition of RNA polymerase II elongation. (A) Overview of the experimental and data analysis strategy. Jurkat T cells were mock-treated (with DMSO) or treated with three different concentrations of DRB (10, 25, and 50 μg/mL in DMSO) or camptothecin (4, 8, and 16 ng/mL in DMSO) for 24 h, and then stimulated with PMA for 6 h. Cy3- and Cy5-labeled cDNA prepared from poly(A)+ RNA isolated from the treated cells was hybridized to a custom microarray containing sets of exon and junction-specific probes (bold lines) representing 5782 human cassette-type AS events that are conserved in mouse transcripts. A, alternative exon; C1 and C2, 5′ and 3′ flanking constitutive exons. (B) Analysis of the microarray data reveals genes with changes in AS and mRNA levels. The Venn diagrams indicate the numbers of genes that change in each direction (color-coded according to increased/decreased mRNA level and increased/decreased splicing level) upon DRB and camptothecin (Camp) treatments. Numbers of genes with significantly overlapping changes at the mRNA and splicing levels are indicated in red (P-value < 7.0 × 10−5). Only genes with coincident increased exon inclusion and decreased mRNA levels are significantly enriched in Gene Ontology (GO) categories (red text; FDR < 0.005).
Following treatment with partially inhibitory concentrations of each drug (Lam et al. 2001; Gomes et al. 2006), the cells were stimulated with phorbol 12-myristate 13-acetate (PMA) to induce the production of new transcripts, so as to facilitate the detection of splicing and mRNA level changes against a background of pre-existing mRNA. Confirming the efficacy of this strategy, most of the DRB-induced splicing and transcript level changes analyzed could be detected without PMA treatment, but stimulation with PMA following DRB treatment resulted in the detection of more pronounced transcript and splicing level changes for the same set of genes (Supplemental Fig. S1).
Fluorescent-labeled cDNA prepared from the drug-treated cells was hybridized to a custom microarray (Calarco et al. 2007) containing sets of exon and splice junction probes representing 5782 human cassette AS events that are conserved between human and mouse. Analysis of the data (see Supplemental Methods) revealed genes with both steady-state mRNA and AS level changes over two or more of the three concentrations of each drug analyzed. To assess the accuracy of the microarray-based measurements, 55 genes with predicted DRB-dependent AS level changes, concomitant with increased or decreased steady-state mRNA levels, were randomly selected for analysis by semiquantitative RT-PCR assays. AS and mRNA level changes for 50 (85%) of these genes were validated (Figs. 2A; Supplemental Figs. S2, S3A; data not shown). Conversely, of 19 analyzed genes that did not display microarray-predicted AS or mRNA level changes, only two were found to display transcript level changes (Supplemental Fig. S3B). Similarly, predicted camptothecin-dependent AS and mRNA level changes for 22/30 (73%) analyzed genes were validated by RT-PCR assays (Fig. 2B; Supplemental Fig. S4). All of the genes with microarray-detected DRB- and/or camptothecin-dependent AS and mRNA level changes are listed in Supplemental Tables S1 and S2.
Figure 2.
Validation of microarray-detected changes in AS and mRNA levels following Pol II elongation inhibition. (A,B) Primer pairs specific for constitutive exons flanking each alternative exon (see Fig. 1A) were used to amplify exon-included and exon-skipped spliced variants, which are indicated by red and yellow circles, respectively. Each RT-PCR analysis was performed in triplicate on total RNA isolated from mock-treated control (“C”) cells, or cells treated with the three different concentrations of DRB (A) or camptothecin (B) (see Fig. 1A). The common color scale represents the percent of transcripts with exon inclusion (%In) and mRNA levels (Trans), as predicted from the microarray data and quantified from the RT-PCR data. Transcript levels on the arcsinh scale were represented as the level in each treatment over the median level for all treatments for the same gene. Numerical values below each panel represent average measurements from the three independent RT-PCR experiments. RT-PCR assays monitoring levels of 5S rRNA and U1 snRNA were used to control for loading and recovery. Additional RT-PCR validation experiments for the same treatments, together with numerical quantification of the data, are shown in Supplemental Figures S2, S3, and S4.
Collectively, the validation experiments confirm that our microarray profiling data and analysis reliably detected genes with altered splicing and mRNA levels in response to treatments that inhibit Pol II elongation.
Genes with DRB- and camptothecin-dependent changes in AS and mRNA levels are enriched in RNA binding and RNA processing functions
As expected for drugs that inhibit Pol II elongation, the majority of the microarray-profiled genes displaying DRB- and camptothecin-induced changes had reduced steady-state mRNA levels. Of 979 genes with at least a 1.5-fold DRB-induced change in steady-state mRNA level, 781 (80%) displayed decreased levels. Similarly, of 880 profiled genes displaying a 1.5-fold or greater change in mRNA level following camptothecin treatment, 787 (89%) displayed reduced levels. Significant proportions of the sets of genes with decreased mRNA levels also displayed changes in AS levels. For example, in DRB treated cells, of the 781 genes displaying ≥1.5-fold reduced mRNA levels, 146 also showed a ≥10% increase in exon inclusion level (Fig. 1B). This represented a statistically significant overlap between genes with reductions in mRNA levels and increased exon inclusion levels (P-value 3.71 × 10−9, hypergeometric distribution). Notably, no other groups of genes with a combination of mRNA and splicing level changes in response to DRB treatment displayed statistically significant overlaps (Fig. 1B).
Most of the DRB-dependent changes resulting in coupled decreases in steady-state mRNA levels and increases in exon inclusion levels likely occurred as a direct consequence of inhibition of P-TEFb, since Western blotting experiments did not reveal an appreciable change in the levels of either the CDK9 or cyclin T1 subunits of P-TEFb as a consequence of DRB treatment (Supplemental Fig. S5A). Also, as will be described later, ChIP-seq experiments revealed specific and statistically significant increases in Pol II density within intronic regions proximal to regulated exons in this group of genes (Fig. 5, see below).
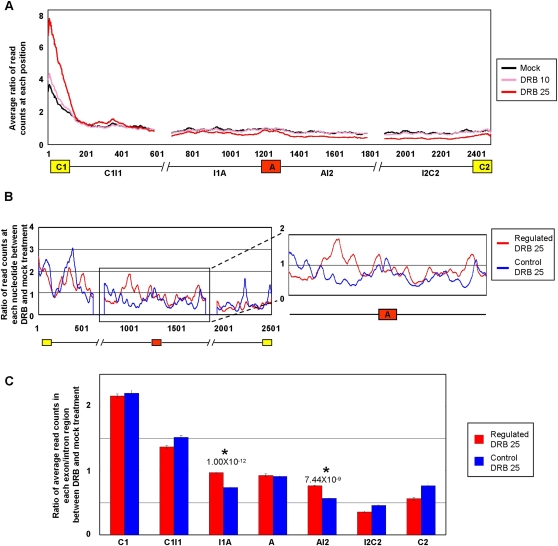
Figure 5.
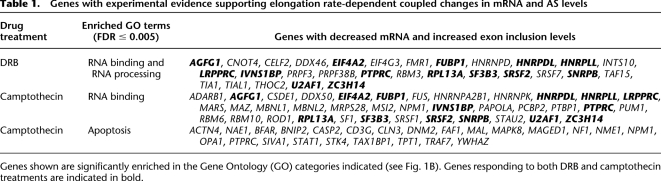
Increased Pol II occupancy over intron sequences flanking alternative exons that display Pol II elongation rate-dependent regulation of AS. Jurkat cells were treated with or without DRB as described in Figure 1, cross-linked with formaldehyde, and Pol II cross-linked to sheared chromatin fragments was immunoprecipitated with a monoclonal antibody (4H8) specific for the Pol II CTD. Illumina short (44 nt) sequence reads were generated from the immunoprecipitated chromatin fragments and these were mapped to the exons and flanking 500 nt intron sequences, corresponding to the C1–A–C2 regions analyzed by microarray profiling in Figure 1. C1, first/upstream constitutive exon; C2, second/downstream constitutive exon; A, alternative exon; I1, first/upstream intron; I2, second/downstream intron. (A) Plots showing merged distributions of Pol II over the profiled exons and intron sequences of all microarray profiled genes. The Pol II distributions are shown as averages of ratios of read counts at each position over a value representing the total number of reads mapped to all exons and introns in the gene set over the total length of the exons and introns in that set. Lengths of exons were normalized by binning into 100 equal-size segments to facilitate direct comparison. Plots are shown for mock (control) treatment and treatments with 10 and 25 μg/mL DRB. (B) Pol II occupancy for microarray-profiled genes with DRB (25 μg/mL)-dependent increases in exon inclusion and decreases in mRNA levels, designated as “Regulated” genes (red trace). A similar number of genes with comparable mRNA levels but that do not detectably respond to DRB (25 μg/mL) are shown for comparison, designated as “Control” genes (blue trace). Additional control traces are shown in Supplemental Figure S11. The traces show the average ratios of read counts at each position over a sliding window of 20 nt in the DRB-treated versus mock-treated samples (see Methods for details). (C) Bar graph quantifying average ratios of Pol II occupancy within each indicated region, as determined from the data in panel B. Regions displaying significant increases in Pol II occupancy in the DRB-treated relative to mock treated samples, between the Regulated and Control sets of genes, are indicated. Error bars indicate standard error and P-values were calculated using Wilcoxon signed-rank test.
Upon camptothecin treatment, genes with decreased mRNA levels and increased exon inclusion levels also displayed the most extensive and statistically significant overlap (P-value 2.45 × 10−14, hypergeometric distribution), although significant overlap was also observed between camptothecin-responsive genes displaying decreased mRNA levels and decreased exon inclusion levels (P-value 6.57 × 10−5, hypergeometric distribution; see below; Fig. 1B). When comparing genes affected by both DRB and camptothecin treatments, only the groups of genes that displayed the combination of reduced steady-state mRNA levels and increased exon inclusion levels significantly overlapped, with 34 genes responding in a similar manner to both drugs (P-value = 0.007, hypergeometric distribution; Fig. 1B; Table 1; see below). These results provide evidence that a subset of transcription-coupled AS events are similarly regulated as a consequence of treatments that reduce Pol II elongation by distinct mechanisms. Moreover, the results are consistent with previous studies showing that inhibition of Pol II elongation can lead to increased exon skipping as well as increased exon inclusion (de la Mata et al. 2003; Pagani et al. 2003).
Table 1.
Genes with experimental evidence supporting elongation rate-dependent coupled changes in mRNA and AS levels
Genes shown are significantly enriched in the Gene Ontology (GO) categories indicated (see Fig. 1B). Genes responding to both DRB and camptothecin treatments are indicated in bold.
To assess whether any of the groups of DRB- and camptothecin-responsive genes may be regulated in a functionally coordinated manner in response to Pol II inhibition, each group was tested for significant enrichment of specific Gene Ontology (GO) terms (see Supplemental Methods). Notably, the groups of genes with DRB- or camptothecin-induced decreased mRNA levels and increased exon inclusion levels displayed significant GO-category enrichment (false discovery rate [FDR] < 0.005). Consistent with the observation that these sets of genes significantly overlap one another (Fig. 1B), both were significantly enriched in RNA binding and RNA processing functions (Fig. 1B; Table 1). Additionally, consistent with recent results implicating elongation-dependent splicing regulation in the control of apoptosis genes (Munoz et al. 2009), the group of genes with coincident decreased mRNA and increased exon inclusion levels in response to camptothecin treatment was also enriched in apoptosis-associated functions. While we do not exclude the possibility that genes belonging to the other regulated groups (Figs. 1B; Supplemental Figs. S3, S4) also represent potentially interesting examples of kinetic coupling, the smaller numbers of these groups and their lack of significant enrichment of GO categories could reflect that many of them displayed changes arising from possible indirect effects (see below), or that they included bona fide examples of kinetic coupling represented by a more diverse spectrum of gene functional categories.
The observation that DRB and camptothecin treatments inhibit Pol II elongation by entirely different mechanisms, yet result in significantly overlapping sets of functionally related genes with coincident changes in AS and mRNA expression levels, strongly suggests that these drugs impact the kinetic coupling between Pol II elongation and splicing regulation. In the experiments that follow, we have focused our attention on representative members of this group that encode factors involved in RNA processing and other aspects of RNA metabolism.
Pol II mutations that disrupt transcription elongation affect AS and mRNA levels of RNA binding and processing factor genes
To further determine whether the DRB and camptothecin-dependent changes in mRNA and AS levels observed above are a consequence of inhibition of Pol II elongation activity, we next asked whether similar effects are observed when transcription is driven by Pol IIs harboring mutations that reduce elongation activity.
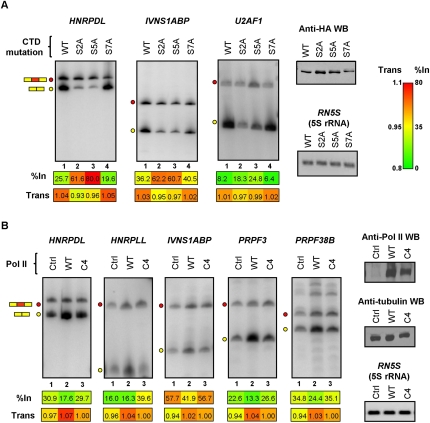
Raji (B cell lymphoma) lines were initially employed that can be induced with tetracycline to express alpha-amanitin-resistant forms of either wild-type Pol II or Pol IIs harboring alanine substitutions at each of the serine residues (Ser 2, Ser 5, or Ser 7) within the CTD heptapeptide repeats (Meininghaus et al. 2000; Chapman et al. 2005, 2007). Phosphorylation of Ser 5 within the CTD heptapeptide repeats is associated with transcription initiation and recruitment of the 5′-end capping machinery (Ho and Shuman 1999), which in turn stimulates transcription elongation (Schroeder et al. 2000). Phosphorylation of Ser 2 residues in the CTD is required for transcription elongation and the recruitment of RNA processing factors (Komarnitsky et al. 2000; Cho et al. 2001; Ahn et al. 2004). In contrast, phosphorylation of Ser 7 is specifically associated with snRNA expression (Chapman et al. 2007; Egloff et al. 2007). We therefore expected that mutation of Ser 2 and Ser 5 but not mutation of Ser 7 residues within the CTD heptapeptide repeats would specifically affect mRNA and AS levels of at least some of the same genes shown above to be affected by DRB and/or camptothecin treatment.
Following tetracycline induction, and then alpha-amanitin treatment to destroy endogenous Pol II (Nguyen et al. 1996), the Pol II-expressing cell lines were stimulated with PMA, as before to induce new gene expression. In cells expressing polymerases with mutations at Ser 2 or Ser 5 positions within the CTD repeats, 12 of 15 of the RNA binding and processing factor genes analyzed in Figure 2 and Supplemental Figures S2, S4 displayed coincident increases in alternative exon inclusion levels and decreases in steady-state mRNA levels (Figs. 3A; Supplemental Fig. S6A). In contrast, none of these genes displayed significant changes in AS or mRNA levels when transcribed by Pol II harboring a CTD with S7A substitutions. Moreover, the 17 genes shown previously (Supplemental Fig. S3B) not to display detectable splicing or mRNA level changes in response to DRB treatment also did not display changes when transcribed by mutant Pol IIs with S2A and S7A substitutions, and only one of these genes showed a change when transcribed by Pol II with S5A substitutions (Supplemental Fig. S6B). Therefore, only mutations of CTD serine residues known to impact transcription elongation affect AS and mRNA levels of genes that are similarly affected by small molecule inhibitors of Pol II elongation.
Figure 3.
Mutations of Pol II that affect transcription elongation consistently affect splicing and mRNA levels of RNA binding protein genes. (A) RT-PCR assays performed on total RNA isolated from Raji B cell lines expressing tetracycline-inducible, alpha-amanitin-resistant, and HA-tagged RNA Pol IIs harboring alanine-substituting mutations at every Ser 2, Ser 5, or Ser 7 position within the CTD heptapeptide repeats (see Results). The cell lines were treated with tetracycline to induce expression of the alpha-amanitin-resistant polymerases for 24 h, then endogenous Pol II was destroyed by treatment with alpha-amanitin for 48 h. Lysates from equal numbers of cells for each cell line were analyzed for Pol II levels by Western blotting using anti-HA antibody (Anti-HA WB) and total RNA was analyzed by RT-PCR assays as described in Figure 2. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S6. (B) Control (Ctrl) and expression plasmids for alpha-amanitin-resistant wild-type (WT) or C4 slow mutant (C4) polymerases were transiently expressed in 293T cells as described in Methods. RNA harvested from the transfected cells was analyzed by RT-PCR as described above. Expression levels of the exogenous Pol IIs were confirmed by Western blotting using an antibody (N-20), which recognizes the N-terminal domain of the largest subunit of Pol II. Tubulin detection was used as loading control for the Western blot, and 5S rRNA detection was used as loading control for RT-PCR assays. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S7.
We next investigated effects on AS and mRNA levels when the DRB/camptothecin-responsive genes described above are transcribed by a “slow” (C4) mutant Pol II which harbors an amino acid substitution in the catalytic domain of the Pol II large subunit that causes reduced rates of Pol II elongation (Coulter and Greenleaf 1985; Chen et al. 1993; de la Mata et al. 2003). Plasmids expressing alpha-amanitin-resistant wild-type and C4 slow Pol IIs were transfected into 293T cells. The cells were then cultured in the presence of alpha-amanitin to destroy endogenous Pol II, and harvested for RNA and protein 48 h later. Western blotting of the cell lysates with anti-Pol II antibody confirmed that endogenous Pol II was efficiently depleted whereas the alpha-amanitin-resistant wild-type and C4 mutant Pol IIs were expressed at comparable levels (Fig. 3B).
RT-PCR analysis showed that most (12/19) of the assayed genes displayed coincident reductions in mRNA levels and increased alternative exon inclusion levels when transcription was carried out by the C4 slow Pol II, as compared to wild-type Pol II (Fig. 3B; Supplemental Fig. S7A). In contrast, all (10/10) assayed genes that were not responsive to DRB/camptothecin treatment did not show a change when the C4 slow Pol II was expressed (Supplemental Fig. S7B). The C4 slow Pol II-dependent splicing and mRNA level changes detected in Figure 3B and Supplemental Figure S7A were less pronounced than those observed in the earlier experiments, and not all of the genes responding to DRB/camptothecin treatment, or to transcription by Pol II CTD mutants, displayed detectable changes when transcribed by this polymerase. These differences are due, at least in part, to the reduced capacity of detection of mRNA and AS changes in transcripts produced from the alpha-amanitin-resistant Pol IIs against the substantial background of endogenous Pol II-produced RNA remaining after alpha-amanitin treatment (i.e., cf. lanes 1 with lanes 2 and 3 in Fig. 3B and Supplemental Fig. S7A). Moreover, this reduced capacity of detection was exacerbated by the lack of induction of new transcription as was performed in the experiments employing PMA induction of Jurkat and Raji cell lines. Finally, it was also possible that some of the differences might be due to cell type-specific effects.
Despite the experimental differences outlined above, more than half of the assayed genes with DRB/camptothecin-dependent changes in both mRNA and AS levels displayed similar changes when transcribed by the C4 slow Pol II. This result therefore strongly supports the conclusion that most of these coupled effects are the consequence of reduced Pol II elongation.
Pol II elongation-dependent AS events can elicit NMD
Recent studies have shown that many genes encoding regulatory and basal splicing factors contain AS events that can introduce PTCs which elicit NMD (Lareau et al. 2007; Ni et al. 2007; Saltzman et al. 2008). Interestingly, several of the Pol II elongation perturbation-sensitive AS events identified in the present study overlapped those defined as PTC-introducing AS events in these previous studies. This raised the intriguing possibility that changes in Pol II elongation could in part affect transcript levels by activating mRNA turnover. In fact, among the DRB/camptothecin-sensitive AS events in genes encoding RNA binding and processing factors 21/73 (29%) are predicted to introduce a PTC when included in spliced mRNA. This proportion differs significantly from the Pol II elongation-sensitive exons from genes belonging to other functional categories, of which 33/441 (7%) have the potential to introduce a PTC upon inclusion in spliced mRNA (P-value 2.83 × 10−15, χ2 test).
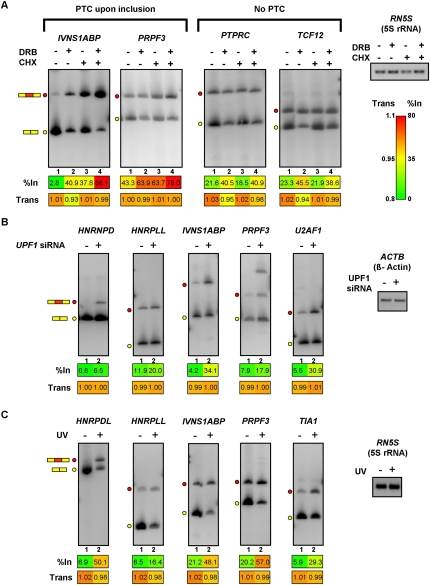
We next asked whether inhibition of the NMD pathway by translational block in Jurkat cells exacerbates the effects of DRB treatment, to result in further increased levels of the PTC-containing splice isoforms. To block NMD, Jurkat cells were treated with the translation inhibitor cycloheximide (CHX). As shown in Figure 4A and Supplemental Figure S8A, 12/13 of the annotated PTC-introducing AS events displayed increased levels of their PTC-containing isoforms following CHX treatment. In all cases, overall transcript levels were reduced when cells were treated with both CHX and DRB, than when treated with CHX alone (compare individual with combined treatments in Fig 4A [IVNS1ABP and PRPF3] and in Supplemental Fig. S8A). This result is consistent with additive effects on mRNA levels arising from NMD and the inhibition of Pol II elongation.
Figure 4.
Roles for NMD and cell stress in the control of splice variant levels regulated by Pol II elongation rate. (A) Cycloheximide (CHX) treatment of PMA-stimulated Jurkat cells promotes increased levels of PTC-containing splice variants that also respond to DRB and camptothecin treatments. Jurkat cells were treated with DRB for 24 h, followed by PMA stimulation. Cycloheximide was added an hour after the addition of PMA, and RNA was isolated 5 h later. RT-PCR analysis of the RNA samples was performed as described in Figure 2. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S8. (B) Knockdown of the essential NMD factor UPF1 in HeLa cells results in specific increases in exon-included, PTC-containing splice variants that are responsive to treatments that inhibit Pol II. UPF1 was knocked down in HeLa cells by transfection of a specific siRNA (Kim et al. 2005) for 48 h, prior to harvesting total RNA for analysis by RT-PCR assays, which were performed as described in Figure 2. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S9. (C) UV irradiation of Jurkat cells results in increased levels of exon inclusion and decreased levels of mRNA for RNA processing factor genes that respond to Pol II elongation perturbation. Jurkat cells were exposed to 120 J/m2 UV and allowed to recover for 6 h prior to harvesting total RNA for RT-PCR assays, which were performed as described in Figure 2. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S10. Loading/recovery controls in A–C are as shown in Figures 2 and 3.
The increased levels of PTC-isoforms observed upon treatment with CHX were likely due to the specific inhibition of NMD, since splice variants from nine out of ten genes containing alternative exons that do not have the potential to introduce a PTC did not display a change in level following treatment with CHX (PTPRC and TCF12 in Fig. 4A; see also Supplemental Fig. S8B). Moreover, confirming that the majority of the DRB- and camptothecin-sensitive splicing events with predicted PTC-introducing AS events were bona fide targets of the NMD pathway, all 11 of the PTC-containing splice isoforms that responded to CHX treatment also increased in level following the specific knockdown of the core NMD factor UPF1 in HeLa cells (Fig. 4B; Supplemental Fig. S9).
Collectively, the results described above support the conclusion that NMD contributes in an additive manner to the reduced steady-state levels of transcripts with PTC-introducing exons that are controlled by changes in the rate Pol II elongation.
Effects of cell stress on Pol II elongation-dependent AS of RNA processing factor transcripts
Exposure of cells to ultraviolet (UV) light can affect the AS levels of transcripts encoding factors involved in p53-independent apoptosis, through a mechanism linked to the inhibition of Pol II elongation via hyperphosphorylation of the CTD (Munoz et al. 2009). These authors focused on changes in splicing of transcripts from a set of genes having functions predominantly associated with cell cycle and cancer, including many genes with functions in apoptosis. To determine whether UV irradiation affects the AS of transcripts from genes encoding RNA binding and processing factors, we exposed Jurkat cells to UV irradiation, and RNA was collected for analysis 6 h later.
Most (nine of 13) of the Pol II elongation perturbation-responsive transcripts described above displayed increased exon inclusion levels and decreased mRNA levels (Fig. 4C; Supplemental Fig. S10A). These effects were specific since only two of 17 tested genes that did not respond to DRB treatment displayed a change in splicing or mRNA level following UV treatment (Supplemental Fig. S10B). Thus, in addition to affecting the regulation of AS of genes involved in apoptosis (Munoz et al. 2009), UV exposure alters the mRNA and AS levels of RNA binding and processing factor genes that are sensitive to conditions impacting Pol II elongation. These findings suggest that changes in the expression of RNA binding and processing factor genes through elongation-dependent AS regulation may constitute an important response to UV-induced cell stress.
Increased Pol II occupancy is associated with AS events regulated by Pol II elongation rate
The kinetic coupling model posits that reduced rates of elongation, whether through reduced Pol II processivity or increased pause time, facilitate the recognition of suboptimal splice sites adjacent to regulated alternative exons (see introduction). We therefore asked whether the group of genes identified above as having coupled decreases in mRNA levels and increases in alternative exon inclusion levels (referred to below as the “regulated” gene set) harbors significant differences in Pol II occupancy proximal to these regulated exons, relative to the equivalent regions proximal to alternative exons in genes that do not respond to perturbations of Pol II elongation (referred to below as the “control” gene set). A full list of the regulated and control gene sets used in the analysis described below is provided in Supplemental Table S3.
To monitor Pol II distributions, ChIP-seq was performed using an anti-Pol II antibody (4H8) and cross-linked chromatin preparations from Jurkat cells, treated with or without DRB prior to PMA stimulation, as described above for the AS microarray profiling experiments (Fig. 5; Supplemental Fig. S11). In order to directly compare Pol II densities between the different conditions and between the regulated and control gene sets, Pol II ChIP-seq signals at each nucleotide position within the alternative and constitutive exons, and flanking intron sequences, were determined as ratios of the number of read counts at each position relative to the mean number of counts over the surveyed regions. The ratios of Pol II signals across the profiled regions in the DRB-treated versus mock-treated samples were then compared for the regulated and control gene sets (see Fig. 5 legend and Methods for further details).
To first assess the specificity of the Jurkat cell ChIP-seq data, we analyzed Pol II signals across the alternative exons and neighboring intron and exon regions corresponding to all unique cassette AS events profiled on our microarray. When analyzing averaged data from the different gene regions, Pol II signals were highest around the 5′ constitutive exons, and these signals increased markedly with increased DRB concentration (compare traces generated from the 10 μg/mL and 25 μg/mL DRB treatments in Fig. 5A). Given that mapping information indicated that at least 15% of the 5′ exons are first or alternative first exons and are therefore promoter proximal, this pattern is consistent with previous results showing a steady-state occupancy of Pol II over promoters and 5′-end regions of genes, and the effect of DRB in suppressing the release of Pol II at these regions (Core and Lis 2008). Also consistent with previous reports indicating DRB-induced changes in Pol II density downstream from promoter/5′-end regions (Core et al. 2008; Baugh et al. 2009; Nechaev et al. 2010), we observed a gradual reduction of Pol II signal 5′ to 3′ across the profiled regions (Fig. 5A). This reduction was more pronounced in the presence than in the absence of DRB. It was also more pronounced over specific profiled alternative exons relative to neighboring intron sequences (see below).
The reduction in Pol II signal in regions downstream from the first constitutive exon was not due to the inability of the 4H8 antibody to recognize hyperphosphorylated Pol II, the form of Pol II most often correlated with elongating Pol II, since Western blotting experiments showed that this antibody preferentially recognizes a hyperphosphorylated form rather than a hypophosphorylated form of Pol II (Supplemental Fig. S5B). Thus, our Pol II ChIP-seq data supports the conclusion that, whereas the strongest inhibitory effect of DRB is on elongation at the 5′ ends of responsive genes (Chodosh et al. 1989), it can also reduce Pol II elongation within downstream regions of genes, including those that display coupled changes in mRNA and AS levels.
Consistent with reduced Pol II elongation rates contributing directly to increased inclusion levels of the alternative exons found earlier to be regulated by DRB and the other modes of Pol II elongation inhibition, we observed statistically significant DRB-dependent increases in the occupancy of Pol II over the 500 nt of intronic sequences flanking the alternative exons in the regulated gene set (I1A and AI2 regions in Fig. 5B,C). Supporting the specificity of these DRB-dependent differences in Pol II occupancy, significant increases in Pol II occupancy were not observed in other regions of the regulated versus control genes, including the alternative exons. In fact, changes in Pol II occupancy were reduced in other gene regions in the regulated versus control sets of genes (Fig. 5C). Moreover, increases in Pol II occupancy over the I1A and AI2 regions were not detected in other sets of control genes analyzed, including those that have decreases in both AS and mRNA levels and those that have decreases in mRNA levels but no changes in splicing levels (Supplemental Fig. S11).
To further confirm whether increased Pol II occupancy within intronic regions flanking DRB-responsive exons is a prominent feature of RNA binding and processing factor genes, we next sorted the total set of 4232 microarray profiled genes (which are representative of diverse gene functional categories) based on the relative strength of the Pol II ChIP-seq signal within the 500 nt of intronic sequences flanking the profiled alternative exons, relative to the strength of the Pol II ChIP-seq signal in the neighboring downstream constitutive exons (as an intragenic control for Pol II occupancy). Consistent with the results described above, genes with the highest relative Pol II occupancy in these intronic regions were significantly enriched in RNA processing functions (FDR ≤ 0.049) (see Supplemental Information for details).
Significant differences in splice site strength are a general feature of AS events regulated by changes in Pol II elongation
We next assessed whether differences in the strengths of splice sites between the regulated set and control sets of genes analyzed above could mediate the effects of reduced Pol II elongation on AS regulation. To address this, we assigned strength scores (Yeo and Burge 2004) (see Supplemental Methods) to splice sites belonging to the alternative and flanking constitutive exons analyzed in each gene set (Supplemental Fig. S13A). We then compared the relative strengths of splice sites both within and between the regulated and control sets of genes.
Consistent with the kinetic model, the average strength of the 3′ splice sites of the alternative exons from the regulated genes are significantly weaker than the average strength of the 3′ splice sites of the downstream constitutive exons (7.31 vs. 9.41; P-value 5.87 × 10−7, Wilcoxon signed-rank test, Supplemental Fig. S13A, top panel). In contrast, the average strengths of the corresponding splice sites in the control gene sets are not significantly different (8.02 vs. 8.51; P-value = 0.21, Wilcoxon signed-rank test, Supplemental Fig. S13A, bottom panel). Similarly, the average strengths of the 5′ splice sites of the regulated alternative exons are also significantly weaker than those of the upstream constitutive exons (7.31 vs. 8.42; P-value = 2.80 × 10−3, Wilcoxon signed-rank test, Supplemental Fig. S13A, top panel), and, again, this trend was not observed when comparing the corresponding splice sites of the control genes (8.06 vs. 8.52; P-value = 0.50, Wilcoxon signed-rank test, Supplemental Fig. S13A, bottom panel).
When comparing between the regulated and control gene sets we observed that the elongation rate-responsive alternative exons are flanked by 5′ and 3′ splice sites (red lines) that are on average significantly weaker than the corresponding splice sites flanking the control alternative exons (blue lines) (see Supplemental Fig. S13A,C,D; P-value ≤ 0.040, Wilcoxon signed-rank test). Additionally, the downstream constitutive exons have significantly stronger 3′ splice sites than those of the constitutive exons downstream from alternative exons in the control genes (Supplemental Fig. S13A,E; P-value < 0.001, Wilcoxon signed-rank test). These findings therefore provide evidence that the responsiveness of alternative exons to changes in transcription elongation is generally associated with the exons having splice sites that are weaker on average than those found in control, nonresponsive alternative exons, as well as compared to those in the adjacent constitutive exons. What also appears to be generally important, however, is the presence of a relatively strong 3′ splice site adjacent to the downstream constitutive exons.
Taken together, the results indicate that increased levels of Pol II occupancy over intronic regions neighboring regulated alternative exons in RNA binding and processing factor genes, together with significant differences in splice site strength associated with these and the downstream constitutive exons, are important general features of Pol II elongation rate-dependent regulation of AS.
An evolutionarily conserved role for Pol II elongation-dependent regulation of splicing factor genes
Thus far, we have shown that changes in Pol II elongation impacts the regulation of RNA processing factor genes at both transcription and AS levels, and that this mechanism may be important in response to stress conditions such as UV exposure in mammalian cells. To assess whether such a mechanism may play a role in response to stress in a whole organism context, we next analyzed data from a recent study (Baugh et al. 2009) comparing mRNA expression profiles and Pol II distributions across genes in Caenorhabditis elegans when subjected to starvation, which causes larval stage 1 (L1) arrest, and during recovery from starvation. Baugh et al. (2009) observed that Pol II accumulates at the promoter regions of growth and developmental genes during starvation-induced L1 arrest, relative to recovery conditions (Baugh et al. 2009). Here, we asked whether there are significantly increased levels of Pol II occupancy within alternative exons and/or flanking intron regions in a manually curated set of RNA processing factor genes during starvation conditions, relative to Pol II occupancy in the equivalent regions in a set of randomly selected genes with comparable steady-state mRNA levels.
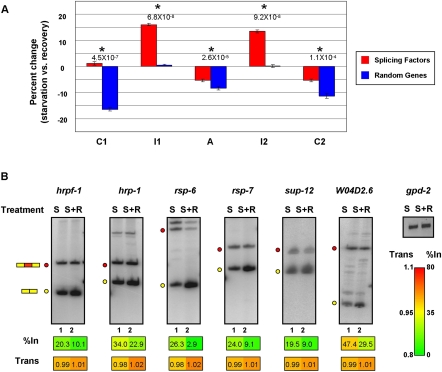
Similar to the results shown in Figure 5A, on average we observed an increase in Pol II occupancy over C. elegans exons relative to neighboring intron sequences (Supplemental Fig. S12A). However, consistent with the results shown in Figure 5B,C for Jurkat cells, we also observed a significant increase in Pol II occupancy over intronic regions flanking alternative exons in the set of RNA processing factor genes in C. elegans grown under starvation versus recovery conditions, relative to the change in Pol II occupancy between these conditions and over the corresponding regions in the randomly selected control genes. As shown in Figure 6A, the percent change in Pol II occupancy between starvation versus recovery conditions significantly increased only in the intronic regions flanking the alternative exons of the RNA processing factor genes (P-value ≤ 9.2 × 10−8, Wilcoxon signed-rank test). Moreover, while we observed an overall decrease in Pol II occupancy over exons during recovery versus starvation conditions, consistent with an observed release of Pol II from promoter regions during growth recovery (Baugh et al. 2009), this decrease was significantly less pronounced in the RNA processing factor genes than in the control genes (P-value ≤ 1.1 × 10−4, Wilcoxon signed-rank test; Fig. 6A).
Figure 6.
Transcription elongation-dependent regulation of splicing factor genes during starvation of C. elegans. (A) Analysis of the changes in average Pol II distribution (as measured by ChIP-seq) in C. elegans under starvation and growth recovery conditions, for C1–I1–A–I2–C2 regions from a set of splicing factor genes, and C1–I1–A–I2–C2 regions from an equivalent number of randomly selected genes that have comparable expression levels. Bar graphs show the percent change of Pol II signal within each individual region under starvation versus recovery conditions. Analyzed data are from Baugh et al. (2009); see Methods for details. Asterisks highlight statistically significant changes in Pol II occupancy between starvation and recovery conditions (P-values ≤ 1.1 × 10−4, Wilcoxon signed-rank test). (B) RT-PCR analysis of AS events in splicing factor genes corresponding to those displaying increased Pol II occupancy in flanking intron regions upon starvation conditions (A). RT-PCR assays were performed as described in Figure 2 on total RNA isolated from worms subjected to starvation and recovery conditions. RNA levels were normalized to gdp-2 mRNA. S, starved worms; S+R, starved worms reintroduced to food. Numerical values below each panel represent average measurements from three independent experiments. Additional RT-PCR experiments and numerical quantification of the data are shown in Supplemental Figure S12.
Finally, to assess whether the observed differences in Pol II occupancy between starvation and recovery conditions detected impact the regulation of AS, we performed RT-PCR assays on a subset of both the RNA processing factor and control genes in worms subjected to the same starvation and recovery conditions as described in Baugh et al. (2009). Six of eight analyzed RNA processing factor genes displayed increased exon inclusion and decreased transcript levels under starvation conditions relative to recovery conditions (Fig. 6B; Supplemental Fig. S12B). In contrast, only one of the seven genes in the control set had decreased mRNA and increased exon inclusion levels, and the remaining exons showed either no change or changes in either AS or mRNA levels, but not both (Supplemental Fig. S12C). In addition, as observed for mammalian exons, there was a significantly higher proportion of alternative exons in C. elegans RNA processing factor genes that could introduce a PTC upon inclusion than in the randomly selected set of control genes. Of the 38 alternative exons from the RNA processing factor genes, 12 (32%) could introduce a PTC upon inclusion, whereas three (9%) of the 35 exons from the randomly selected control genes had this capacity (P-value 4.82 × 10−19, χ2 test).
These results provide evidence that reduced Pol II elongation, coinciding with increased occupancy of Pol II over intron regions adjacent to alternative exons, represents an evolutionarily conserved mechanism to control splicing levels in response to stress conditions. This conserved mechanism appears to preferentially impact splicing levels of alternative exons in the RNA processing factor and other RNA binding protein genes, in part by activating NMD of the regulated transcripts.
Discussion
The results of this study collectively provide evidence that an important role for Pol II elongation-dependent changes in AS regulation is to modulate the levels of expression of genes with critical functions in RNA metabolism, including pre-mRNA splicing factors and other proteins with RNA binding activity. Most of the analyzed genes with coupled decreases in steady-state mRNA levels and increases in exon inclusion levels were similarly affected by a variety of conditions that impact Pol II elongation, including two drugs and three different types of Pol II mutations that inhibit elongation activity, whereas a smaller number of genes displayed condition-specific effects. The results suggest that many transcription-coupled AS events can be regulated by distinct mechanisms that affect Pol II elongation rate or Pol II occupancy, and that these effects can occur in different cell types.
Our observation that many exons in RNA processing factor genes sensitive to Pol II elongation perturbation are PTC-introducing and NMD-eliciting further suggests that cells may rapidly adjust the levels of many RNA processing factors in response to changes in growth conditions, including cell stress, that directly or indirectly impact Pol II elongation. The inclusion of PTC-containing exons contributes additively to decreased mRNA levels when transcription elongation activity is reduced. This mechanism likely acts in conjunction with previously described examples of auto- and cross-regulation of splicing factors and other RNA binding domain proteins, whereby the binding of these factors to their own transcripts or the transcripts of closely related gene family members promotes increased levels of PTC-containing splice variants, which are subjected to NMD (Sureau et al. 2001; Wollerton et al. 2004; Hase et al. 2006; Lareau et al. 2007; Rossbach et al. 2009; Saltzman et al. 2011).
Our results support the conclusion that Pol II elongation rate-dependent regulation of AS simultaneously controls a network of PTC-introducing exons and other exons in genes that have RNA processing and related functions. Pol II elongation-dependent control of AS, in conjunction with the aforementioned layers of regulation, therefore appears to serve as a mechanism to coordinate the cellular requirements of splicing and other RNA binding and regulatory factors in response to changes in transcript levels. These results are reminiscent of recent findings in yeast, in which specific stress conditions can result in the widespread accumulation of introns in transcripts encoding ribosomal protein genes, a subset of which introduces PTCs that trigger NMD (Pleiss et al. 2007; Sayani et al. 2008). Moreover, also consistent with our results, it has been reported that a PTC-introducing exon in the C. elegans splicing factor gene uaf-1, which is a homolog of the mammalian U2AF2, is more highly included under starvation conditions (Zorio et al. 1997).
The Pol II elongation-dependent effect on AS levels correlated with statistically significant increases in Pol II occupancy over intron sequences flanking the regulated alternative exons in RNA binding and processing factor genes. The possible mechanistic significance of this observation is emphasized by the finding that similar patterns of Pol II accumulation were detected in both human cells and in C. elegans when Pol II elongation activity was reduced. In both cases, the increased accumulation of Pol II over intron sequences specifically coincided with increased levels of inclusion of the adjacent alternative exons. These observations and our detection of highly consistent effects on coupled changes in mRNA and splicing levels when Pol II elongation is inhibited by very different mechanisms argue that it is unlikely that many of these regulated events are simply a consequence of indirect effects. Further supporting this conclusion is the observation that the strengths of the splice sites adjacent to the Pol II elongation-sensitive alternative exons are significantly different from those found adjacent to the downstream constitutive exons and also from those flanking alternative exons and downstream constitutive exons in the nonresponsive control genes. The combined observations therefore generally support a version of the kinetic model in which Pol II accumulation over the intronic regions flanking the regulated exons favors the recognition of suboptimal splice sites adjacent to the regulated exons, over recognition of the significantly stronger downstream 3′ splice site.
A recent study has shown that kinetic coupling of transcription elongation and AS is not simply a consequence of a “first come first serve” mode of splicing, with the upstream intron being spliced first when Pol II is paused within the alternative exon or downstream intron (Pandya-Jones and Black 2009; de la Mata et al. 2010). Instead, pausing of Pol II appears to facilitate the spliceosome to commit to exon inclusion without requiring ordered intron removal. Similarly, in the present study, Pol II pausing in either the upstream or downstream intronic regions could commit either intron to splicing by allowing time for the association of splicing components with stalled polymerase and nascent pre-mRNA to occur, thereby resulting in increased exon inclusion. The detection of significantly weaker splice sites associated with the Pol II elongation sensitive exons and strong downstream constitutive exon 3′ splice sites further supports this proposal. It is interesting to consider that the highly conserved intronic sequences, some of which encompass ultraconserved elements that surround the regulated exons (Lareau et al. 2007; Ni et al. 2007; Saltzman et al. 2008), could function to control splicing levels by acting as Pol II pause sites, in addition to functioning more directly to control splicing at the level of RNA recognition.
In summary, by profiling AS and steady-state mRNA levels together with Pol II occupancy under experimental and physiological conditions that impact Pol II elongation, we provide evidence for an in vivo role for the kinetic coupling of Pol II elongation and AS in the regulation of the expression of many RNA binding proteins including many splicing factors. This function appears to play an evolutionarily conserved role in response to stress conditions such that the levels of RNA binding proteins are coordinated with steady-state mRNA levels.
Methods
Cell culture, treatments, and RNA purification
The Trex-Jurkat (Invitrogen) cell line was cultured in RPMI-1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS). For DRB (Sigma) treatments or for control treatment (DMSO alone), 10, 25, 50 μg/mL DRB in DMSO was added to cells in fresh medium at a density of 1 × 106 cells. After 24 h, cells were stimulated by the addition of 20 ng/mL of PMA (Sigma) for 6 h. Camptothecin treatment was performed as described for the DRB treatment with 4, 8, and 16 ng/mL of camptothecin. For cycloheximide (Sigma) treatment, Jurkat cells were treated with 33 μg/mL DRB for 24 h, followed by addition of PMA (20 ng/mL). One hour after activation, 50 μg/mL CHX (in DMSO) was added and the cells were incubated for an additional 5 h. Raji cells were maintained and induced to express exogenous wild-type and various mutant Pol IIs as described previously (Meininghaus et al. 2000). HeLa cells and 293T cells were maintained in DMEM medium supplemented with 10% heat-inactivated FBS. For knockdown of UPF1, HeLa cells were transiently transfected with siRNAs as previously described (Kim et al. 2005). 293T cells were transfected with expression vectors for wild-type and the C4 slow Pol II for 18 h, followed by treatment of 5 μg/mL alpha-amanitin and were harvested 48 h later. For UV treatment, Jurkat cells were placed in PBS and irradiated with 120 J/m2 UVC using a UV Stratalinker 1800 (Stratagene). Cells were harvested after 6 h of recovery. C. elegans N2 strain was grown and synchronized as previously described (Baugh et al. 2009) and was starved for 12 h. The recovery sample was reintroduced to food for 1 h before harvesting. Total RNA was harvested using TRI Reagent (Sigma). For microarray profiling, poly(A)+ mRNA was purified using the NucleoTrap mRNA Midi Kit (Clontech) following the manufacturer's instructions.
Microarray design, hybridization, and data analysis
AS events represented on a customized human 244K microarray (Agilent) were identified as previously described (Pan et al. 2004). Microarray design was also as previously described (Pan et al. 2004). Hybridization of Cy3- and Cy5-labeled cDNA synthesized from poly(A)+ RNA from DRB, camptothecin, or DMSO-treated Jurkat cells was performed using a HS4800 Pro Hybridization Station (Tecan) at 42°C for 22 h. Microarray slides were scanned with an Agilent microarray scanner at 5 μm resolution and TIFF images were processed using Feature Extraction Software (Agilent). Processed data were then analyzed using the GenASAP algorithm (Shai et al. 2006). Further details are provided in Supplemental Methods.
RT-PCR assays
RT-PCR assays were carried out using the One-Step RT-PCR kit (QIAGEN) as described previously (Calarco et al. 2007); 15 ng of total RNA was used in each reaction. Primers specific for constitutive exons flanking alternative exons were used in each case. Results were quantified with the Typhoon Imager and the ImageQuant software (GE Healthcare). All primer sequences are available from the authors upon request.
Jurkat and C. elegans ChIP-seq data and analysis
Jurkat cells were treated as described above and ChIP-seq was performed as recently described (Schmidt et al. 2009). Immunoprecipitation was performed with the murine monoclonal antibody 4H8 (Abcam), which recognizes the C-terminal domain of Pol II. The immunoprecipitated material was end-repaired, A-tailed, ligated to sequencing adapters, amplified using 18 cycles of PCR, and size-selected (200–300 bp) material was subject to single-end sequencing using an Illumina Genome Analyzer, as per the manufacturers' recommendations. Details of ChIP-seq data analysis are provided in Supplemental Methods.
Acknowledgments
We thank Nik Matthews and James Hadfield at the CRI Genomic Core and the CRI Bioinformatics Core for ChIP-seq data, and Leo Lee for assistance with data analysis. We also thank Alberto Kornblihtt, Arneet Saltzman, Dirk Eick, Rob Chapman, Lynne Maquat, and Yoon-Ki Kim for providing reagents. Nuno Barbosa-Morais, John Calarco, Jonathan Ellis, Mathieu Gabut, Jim Ingles, and Arneet Saltzman kindly provided helpful suggestions and comments on the manuscript. The research was supported in part by operating grants from the Canadian Cancer Society/National Cancer Institute of Canada and from the Canadian Institute of Health Research (MOP-67011) to B.J.B., and from the Ontario Research Fund and Genome Canada (through the Ontario Genomics Institute) to B.J.B. and others.
Footnotes
[Supplemental material is available for this article. The microarray and sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE25495.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111070.110.
References
- Ahn SH, Kim M, Buratowski S 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Allemand E, Batsche E, Muchardt C 2008. Splicing, transcription, and chromatin: A menage a trois. Curr Opin Genet Dev 18: 145–151 [DOI] [PubMed] [Google Scholar]
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM 2009. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci 106: 7798–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW 2009. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 324: 92–94 [DOI] [PubMed] [Google Scholar]
- Bentley DL 2005. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN 1988. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2: 754–765 [DOI] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ 2007. Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev 21: 2963–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Carvalho C 2007. Nuclear organization and splicing control. Adv Exp Med Biol 623: 1–13 [DOI] [PubMed] [Google Scholar]
- Chapman RD, Conrad M, Eick D 2005. Role of the mammalian RNA polymerase II C-terminal domain (CTD) nonconsensus repeats in CTD stability and cell proliferation. Mol Cell Biol 25: 7665–7674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RD, Heidemann M, Albert TK, Mailhammer R, Flatley A, Meisterernst M, Kremmer E, Eick D 2007. Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318: 1780–1782 [DOI] [PubMed] [Google Scholar]
- Chen Y, Weeks J, Mortin MA, Greenleaf AL 1993. Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol 13: 4214–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev 15: 3319–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA 1989. 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem 264: 2250–2257 [PubMed] [Google Scholar]
- Core LJ, Lis JT 2008. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319: 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter DE, Greenleaf AL 1985. A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem 260: 13190–13198 [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH 2007. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol 14: 796–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Yu J, Zhang Z, Gygi MP, Krainer AR, Gygi SP, Reed R 2007. SR proteins function in coupling RNAP II transcription to pre-mRNA splicing. Mol Cell 26: 867–881 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Kornblihtt AR 2006. RNA polymerase II C-terminal domain mediates regulation of alternative splicing by SRp20. Nat Struct Mol Biol 13: 973–980 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR 2010. First come, first served revisited: Factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S 2007. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q 2001. Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933 [DOI] [PubMed] [Google Scholar]
- Ge H, Si Y, Wolffe AP 1998. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol Cell 2: 751–759 [DOI] [PubMed] [Google Scholar]
- Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 20: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Yalamanchili P, Visa N 2006. The Drosophila heterogeneous nuclear ribonucleoprotein M protein, HRP59, regulates alternative splicing and controls the production of its own mRNA. J Biol Chem 281: 39135–39141 [DOI] [PubMed] [Google Scholar]
- Ho CK, Shuman S 1999. Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Kessel D 1971. Some determinants of camptothecin responsiveness in leukemia L1210 cells. Cancer Res 31: 1883–1887 [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208 [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev 14: 2452–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR 2007. Coupling transcription and alternative splicing. Adv Exp Med Biol 623: 175–189 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G 2004. Multiple links between transcription and splicing. RNA 10: 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Pickeral OK, Peng AC, Rosenwald A, Hurt EM, Giltnane JM, Averett LM, Zhao H, Davis RE, Sathyamoorthy M, et al. 2001. Genomic-scale measurement of mRNA turnover and the mechanisms of action of the anti-cancer drug flavopiridol. Genome Biol 2, RESEARCH0041 doi: 10.1186/gb-2001-2-10-research0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE 2007. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature 446: 926–929 [DOI] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD 2008. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 15: 819–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D 2009. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol Cell 33: 450–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T 2010. Regulation of alternative splicing by histone modifications. Science 327: 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R 2002. An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem 271: 27176–27183 [DOI] [PubMed] [Google Scholar]
- Meininghaus M, Chapman RD, Horndasch M, Eick D 2000. Conditional expression of RNA polymerase II in mammalian cells. Deletion of the carboxyl-terminal domain of the large subunit affects early steps in transcription. J Biol Chem 275: 24375–24382 [DOI] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM 2000. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell 6: 307–316 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. 2009. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell 137: 708–720 [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K 2010. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science 327: 335–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VT, Giannoni F, Dubois MF, Seo SJ, Vigneron M, Kedinger C, Bensaude O 1996. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res 24: 2924–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O'Brien G, Shiue L, Clark TA, Blume JE, Ares M Jr 2007. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev 21: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F, Stuani C, Zuccato E, Kornblihtt AR, Baralle FE 2003. Promoter architecture modulates CFTR exon 9 skipping. J Biol Chem 278: 1511–1517 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell 16: 929–941 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C 2007. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell 27: 928–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Ip JY, Calarco JA, Bakowski MA, Emili A, McCracken S, Tucker P, Ingles CJ, Blencowe BJ 2005. Role for PSF in mediating transcriptional activator-dependent stimulation of pre-mRNA processing in vivo. Mol Cell Biol 25: 6734–6746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossbach O, Hung LH, Schreiner S, Grishina I, Heiner M, Hui J, Bindereif A 2009. Auto- and cross-regulation of the hnRNP L proteins by alternative splicing. Mol Cell Biol 29: 1442–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ 2008. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol 28: 4320–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, and Blencowe BJ 2011. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez G, Bittencourt D, Laud K, Barbier J, Delattre O, Auboeuf D, Dutertre M 2008. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci 105: 6004–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT 2006. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 7: 557–567 [DOI] [PubMed] [Google Scholar]
- Sayani S, Janis M, Lee CY, Toesca I, Chanfreau GF 2008. Widespread impact of nonsense-mediated mRNA decay on the yeast intronome. Mol Cell 31: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT 2009. ChIP-seq: Using high-throughput sequencing to discover protein-DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR 2009. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci 106: 4325–4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev 14: 2435–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shai O, Morris QD, Blencowe BJ, Frey BJ 2006. Inferring global levels of alternative splicing isoforms using a generative model of microarray data. Bioinformatics 22: 606–613 [DOI] [PubMed] [Google Scholar]
- Sims RJ III, Belotserkovskaya R, Reinberg D 2004. Elongation by RNA polymerase II: The short and long of it. Genes Dev 18: 2437–2468 [DOI] [PubMed] [Google Scholar]
- Sureau A, Gattoni R, Dooghe Y, Stevenin J, Soret J 2001. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J 20: 1785–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13: 91–100 [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394 [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell 35: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Lea K, Blumenthal T 1997. Cloning of Caenorhabditis U2AF65: An alternatively spliced RNA containing a novel exon. Mol Cell Biol 17: 946–953 [DOI] [PMC free article] [PubMed] [Google Scholar]