Abstract
In Mus spretus, the chloride channel 4 gene Clcn4-2 is X-linked and dosage compensated by X up-regulation and X inactivation, while in the closely related mouse species Mus musculus, Clcn4-2 has been translocated to chromosome 7. We sequenced Clcn4-2 in M. spretus and identified the breakpoints of the evolutionary translocation in the Mus lineage. Genetic and epigenetic differences were observed between the 5′ends of the autosomal and X-linked loci. Remarkably, Clcn4-2 introns have been truncated on chromosome 7 in M. musculus as compared with the X-linked loci from seven other eutherian mammals. Intron sequences specifically preserved in the X-linked loci were significantly enriched in AT-rich oligomers. Genome-wide analyses showed an overall enrichment in AT motifs unique to the eutherian X (except for genes that escape X inactivation), suggesting a role for these motifs in regulation of the X chromosome.
In mammals, X-linked genes are regulated by special epigenetic mechanisms, because females have two X chromosomes and males only have one, while autosomes are present in two copies. These regulatory mechanisms are (1) X up-regulation in both sexes to balance expression between X-linked and autosomal genes (Gupta et al. 2006; Nguyen and Disteche 2006) and (2) X inactivation in females (Lyon 1961). Ohno (1967) predicted that due to these unique regulatory mechanisms the gene content of the X chromosome would be highly conserved between mammalian species. The chloride channel 4 gene (hereafter called Clcn4 when considering multiple mammalian species) illustrates a rare exception to this conservation, since it is X-linked in most mammals including human, primates, dog, cow, and horse (CLCN4), as well as rat (Clcn4-2), but located on chromosome 7 in the laboratory mouse (Clcn4-2) (derived from a mixture of M. musculus musculus and M. musculus domesticus and thereafter referred to as M. musculus) (Palmer et al. 1995; Rugarli et al. 1995; Flicek et al. 2010).
Clcn4-2 is X-linked in the wild-derived mouse M. spretus, suggesting that it was translocated to an autosome in one branch of Mus (musculus) during evolution (Palmer et al. 1995; Rugarli et al. 1995). We have previously used F1 mice from crosses between Mus species to show that Clcn4-2 is subject to X inactivation (Rugarli et al. 1995) and that its expression is doubled on the active X from M. spretus compared with each autosomal allele from M. musculus (Adler et al. 1997). Thus, Clcn4-2 is subject to both types of dosage-compensation mechanisms, X up-regulation, and X inactivation. The different location of this gene in closely related mouse species provides a system to explore whether X-linked genes differ from autosomal genes in terms of DNA sequence and epigenetic modifications. Our hypothesis based on studies in other organisms is that specific sequence motifs may be enriched on the mammalian X to facilitate its regulation. For example, the Drosophila melanogaster X chromosome is enriched in specific motifs as entry points for the male-specific lethal complex that up-regulates X-linked genes in males (Alekseyenko et al. 2008). The Caenorhabditis elegans X is also enriched in specific motifs, in this case to recruit the complex that silences both X chromosomes in hermaphrodites (McDonel et al. 2006).
In this study we sequenced the M. spretus Clcn4-2 X-linked locus for comparison to the M. musculus autosomal locus. By defining the breakpoints of the translocation in the Mus lineage, we determined that the evolutionary rearrangement is complex. We established that the promoter regions and the chromatin structure of the autosomal and X-linked loci differed between M. musculus and M. spretus, consistent with increased expression on the X. Dramatic deletions of intronic sequences were observed in the autosomal gene from M. musculus compared with seven other eutherian species. Examination of intronic sequences conserved within the X-linked Clcn4 loci in human, rat, cow, dog, and M. spretus, but deleted in the autosomal gene, led to the identification of AT-rich motifs enriched on the entire X chromosome, where these motifs could play a role in its regulation.
Results
Sequence of M. spretus Clcn4-2 and definition of the evolutionary translocation breakpoints
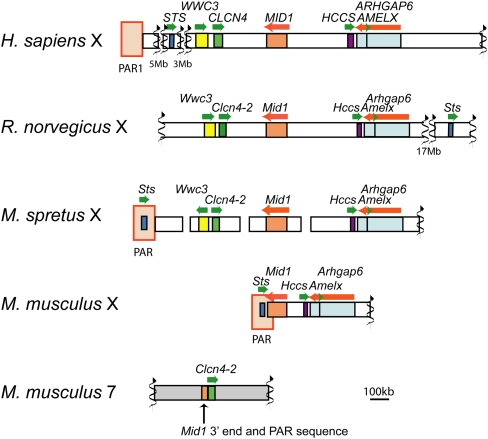
A BAC library constructed from M. spretus genomic DNA was screened by hybridization with labeled PCR products amplified from M. spretus DNA using primers based on conserved sequences between mouse, human, and rat loci (Rhead et al. 2010). High-density colony arrays were screened with probes for Clcn4-2 and for the flanking genes Wwc3 (present in human and predicted in rat, but absent in M. musculus) and Mid1. Eighteen positive BACs were identified, of which 13 were positive for Wwc3, three for Clcn4-2, and two for Mid1. Thus, Wwc3 is present in M. spretus, unlike the situation in M. musculus.
Two M. spretus BACs were sequenced to provide complete coverage of Clcn4-2. Ch35-246O15 (BAC31, 165,529 bp) contained the whole coding region except for the 5′UTR, while Ch35-316H16 (BAC29, 167,913 bp) contained the 5′UTR together with the complete sequence of the adjacent gene Wwc3 (Supplemental Fig. S1A). Each contig from the BAC sequence assembly was aligned against the rat genome to verify their position (Rhead et al. 2010). In M. spretus, Wwc3 and Clcn4-2 were arranged in opposite orientation compared with human, chimpanzee, orangutan, rhesus monkey, rat, cow, pig, and dog, suggesting an inversion (Fig. 1). BAC sequencing did not allow us to connect Mid1 to the Clcn4-2/Wwc3 cluster. However, fluorescence in situ hybridization (FISH) to metaphase chromosomes using a M. musculus-derived Mid1 probe (red) together with BAC29 (green) showed that the Wwc3-Clcn4-2 cluster is distal to Mid1 on the M. spretus X (Supplemental Fig. S1B).
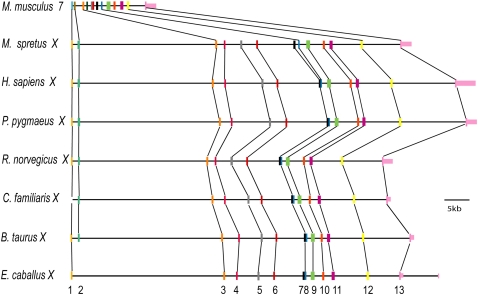
Figure 1.
Genomic landscape around Clcn4 in human, rat, M. spretus, and M. musculus. Clcn4-2 is the only gene translocated to autosome 7 in M. musculus, along with a small piece of Mid1 and of the PAR. Wwc3 is lost in M. musculus. The order of loci in M. spretus is based on previous mapping studies and our current data. The positions of the PAR and of additional genes, Sts, Hccs, Arhgap6, and Amelx, are shown for reference. Wwc3 in rat is predicted by N-SCAN (Rhead et al. 2010).
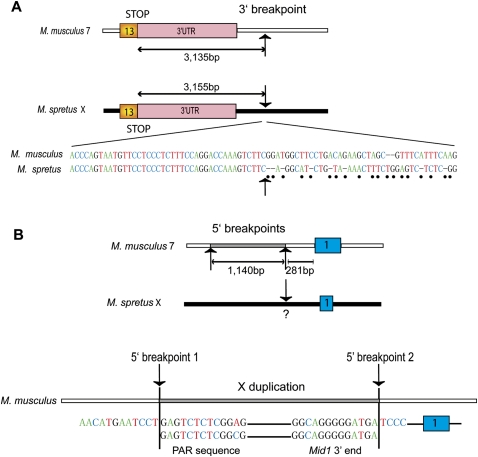
Clcn4-2 is the only intact gene involved in the evolutionary translocation to proximal chromosome 7 in M. musculus. To define the breakpoints, the M. spretus BAC library was screened with M. musculus probes from regions that flank Clcn4-2 on chromosome 7. This screen yielded multiple positive BACs due to the repeated nature of the region, which contains three families of multicopy and polymorphic genes consisting of vomeronasal, olfactory receptor, and zinc-finger genes (Rhead et al. 2010). Hence, chromosome 7 sequences could not reliably be obtained from M. spretus. Nonetheless, based on sequence alignments, the 3′end breakpoints of the translocation were clearly identified as the nucleotide positions where identity (97%) between the species terminated sharply (3135 and 3155 bp beyond the stop codons in M. musculus and M. spretus, respectively) (Fig. 2A).
Figure 2.
Breakpoints of the Clcn4-2 evolutionary translocation in the Mus lineage. (A) The 3′end breakpoints located about 3 kb from the stop codon in exon 13 in both species (arrows) are defined as the point in each sequence where M. musculus and M. spretus diverge (dots indicate nonconserved nucleotides). (B) The 5′end breakpoints are complex and involve duplication of part of Mid1 and PAR sequences (1040 bp). Breakpoint 1 marks the distal edge of the duplicated sequences and breakpoint 2, the proximal edge located 281-bp upstream of exon 1. The first evidence of sequence homology with M. spretus is within exon 1; however, a region upstream of exon 1 may have been translocated and subsequently diverged, hence the uncertainty of this breakpoint (?). Chromosome X is shown as a black horizontal bar, chromosome 7, as a white bar, and the duplicated X region within chromosome 7 as a gray bar. Schematics are not to scale.
A 13,080-bp M. spretus sequence directly upstream of Clcn4-2 5′end was conserved in rat and human (up to the breakpoint of the Wwc3 inversion in M. spretus), but not in M. musculus. Surprisingly, a 1140-bp duplicated fragment upstream of exon 1 on chromosome 7 displayed 93% identity to sequences corresponding to Mid1 3′end and to a portion of the pseudoautosomal region (PAR). This insertion of duplicated X-linked sequences not directly adjacent to Clcn4 in other mammals implies that there must be at least two breakpoints at the 5′end of Clcn4-2 in M. musculus. One breakpoint was tentatively defined as the point where the X duplicated sequence terminated and the chromosome-7-specific sequence begun (Fig. 2B). A second breakpoint must also be located 281-bp upstream of exon 1 to account for the insertion of duplicated sequences. We conclude that the evolutionary translocation to chromosome 7 is complex and involves both deletion and duplication.
Clcn4 gene structure, regulatory elements, and epigenetic modifications in mammals
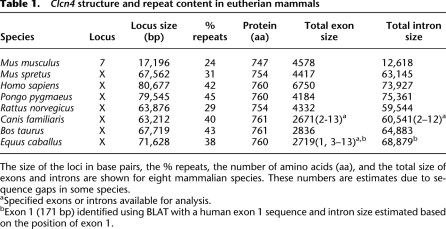
Clcn4 exon/intron structure was compared between eight mammalian species for which a complete or near-complete sequence of the gene was available. The exons were generally conserved between the X-linked loci (M. spretus, human, orangutan, rat, cow, dog, and horse) and the autosomal locus (M. musculus) (Table 1; Supplemental Table S1). The predicted protein size ranged between 747 and 761 amino acids. Exons 3–13 representing the coding sequence were highly conserved, with an overall 80%–97% identity between species (Supplemental Table S2). In contrast, exons 1 and 2 significantly differed both between the two Mus species (46% identity) and between M. spretus and other X-linked forms (42%–45% identity), except for rat (92% identity). In all species examined, the ATG translation start site was located at the beginning of exon 3, and the poly(A) signal, within exon 13 (Fig. 3A).
Table 1.
Clcn4 structure and repeat content in eutherian mammals
The size of the loci in base pairs, the % repeats, the number of amino acids (aa), and the total size of exons and introns are shown for eight mammalian species. These numbers are estimates due to sequence gaps in some species.
aSpecified exons or introns available for analysis.
bExon 1 (171 bp) identified using BLAT with a human exon 1 sequence and intron size estimated based on the position of exon 1.
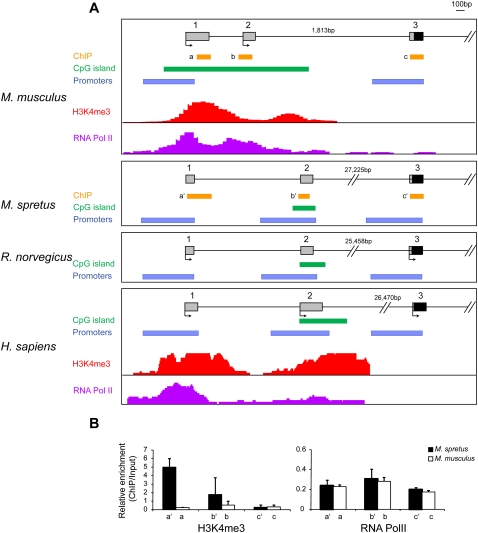
Figure 3.
Comparison of Clcn4 5′ end between eutherian mammals. (A) Map locations of putative promoters (P1, P2, and P3) identified by Genomatix analysis, of CpG islands, and of known regions enriched in H3K4me3 and PolII, in M. musculus, M. spretus, rat, and human. Exon numbers (1, 2, 3) are at top. Chromatin enrichment data was downloaded to the UCSC browser for M. musculus (Gupta et al. 2010) and for human (ENCODE) (Celniker et al. 2009). Arrows indicate starts of transcription based on reported cDNAs (Flicek et al. 2010). (B) Chromatin analysis of Clcn4-2 in the Patski cell line using ChIP followed by quantitative PCR to determine enrichment relative to input using primers specific for M. musculus and M. spretus loci. Relative enrichment is shown for H3K4me3 and RNA PolII using primers at exons 1 (a/a′), 2 (b/b′), and 3 (c/c′) whose position is indicated in A (Supplemental Table S6).
Three potential promoters (P1, P2, and P3) whose sequence was partially conserved between species were identified using Genomatix software (Fig. 3A; Supplemental Table S2). In M. spretus, M. musculus, rat, and human, P1 and P3 were found upstream of exons 1 and 3, respectively (Fig. 3A). Promoter P2 was identified upstream of exon 2 in M. spretus, rat, and human, but Genomatix analysis failed to detect P2 in M. musculus, even though transcripts starting in exon 2 have been reported (Fig. 3A; Rhead et al. 2010). A list of transcription factors that bind to Clcn4 promoter regions in all four species generated using Genomatix based on conserved sequence alignments include factors specific to genes expressed in brain (Supplemental Table S3). However, some factors present in M. spretus, human, and rat were absent in M. musculus, especially at promoter P2, which is poorly conserved. The 5′end of M. musculus Clcn4-2 is marked by a large CpG island overlapping exons 1 and 2, a characteristic feature of broad promoters with multiple start sites (Sandelin et al. 2007). In human, a small CpG island overlaps exon 2, and our own CpG island searches in M. spretus and in rat also showed a small CpG island at the corresponding location (Fig. 3A; Takai and Jones 2003). Enrichment in RNA polymerase II phosphorylated at serine 5 (PolII) and in histone H3 trimethylated at lysine 4 (H3K4me3), as well as DNase I hypersensitive sites has been detected in regions that overlap exons 1 and 2 and the CpG island in M. musculus (Fig. 3A; Flicek et al. 2010; Gupta et al. 2010). Similarly, the 5′end of human CLCN4 shows PolII occupancy, DNase I sensitivity, and is enriched in H3K4me3, suggesting that the chromatin is open to facilitate transcription initiation (Celniker et al. 2009).
To compare epigenetic modifications at the 5′end of M. musculus and M. spretus Clcn4-2 genes within the same cells, we used a cell line (Patski) derived from an F1 mouse from a cross between the species (Yang et al. 2010). H3K4me3 and PolII occupancy were determined by chromatin immunoprecipitation (ChIP), followed by quantitative PCR analyses using primers that distinguish loci based on DNA polymorphisms. While no significant difference in PolII occupancy was detected between loci, enrichment in H3K4me3 was observed at the spretus X-linked locus compared with the musculus autosomal locus at exons 1 and 2, but not exon 3 (Fig. 3B). Thus, the autosomal and X-linked Clcn4-2 genes differ in their chromatin structure in the cell line, potentially reflecting differences in expression levels and/or a different promoter usage.
Clcn4 introns conserved on the X are deleted on the M. musculus autosome
A major difference between the autosomal and X-linked forms of Clcn4 is a dramatic reduction in the total size of intron sequence on the autosome: All seven species of eutherian mammals with an X-linked form have large introns, totaling 60–75 kb, compared with 13 kb in M. musculus (Fig. 4; Table 1; Supplemental Table S1). Genomic comparisons using VISTA identified short M. musculus regions homologous to the M. spretus sequence retained at the edges of introns, thus preserving exon/intron boundaries (Supplemental Fig. S2). Even though intron 2 had a 25-kb deletion in M. musculus, promoter P3 upstream of exon 3 was retained (Fig. 3A). While most of the M. spretus introns were highly conserved in rat, their sequence partially diverged in human, except for five regions with at least 75% identity between M. musculus, M. spretus, rat, and human, which may represent regulatory elements essential for proper expression of the gene, regardless of its position on the X or on an autosome (Supplemental Fig. S2).
Figure 4.
Clcn4-2 introns are truncated on chromosome 7 in M. musculus compared to the X-linked Clcn4 loci in M. spretus, human, orangutan, rat, dog, cow, and horse (see also Supplemental Table S1). Exons are labeled 1–13.
In a more distant mammalian species (marsupial opossum) with an autosomal form of CLCN4, the introns were large (Supplemental Table S1). There was little evidence of intron sequence conservation with eutherian mammals, except for a few small regions partially conserved between M. spretus and opossum. Some of these regions (located within introns 2, 5, 11, and 12) were deleted in M. musculus, suggesting that the ancestral mammalian Clcn4 gene may have had large introns (Supplemental Fig S2). Thus, deletions of introns in M. musculus are recent events in a branch of Mus. No significant conservation was observed in chicken, except in protein-coding exons (Supplemental Fig S2).
The eutherian X chromosome is enriched in AT-rich motifs
The uniformly large size of introns in X-linked forms of Clcn4 compared with the autosomal gene suggested positive selection for retention of sequences specifically on the X (Fig. 4). To determine whether these sequences contain specific repeat elements or sequence motifs, RepeatMasker was used to catalog the repeat content of Clcn4 introns in human, rat, M. spretus, and M. musculus. Enrichment in the density of all repeats as well as of specific types of repeats (SINE, LINE, LTR) was observed in all X-linked loci compared with the autosomal locus (Supplemental Fig. S3). The autosomal opossum CLCN4 gene had a high LINE and total repeat content, a known characteristic of this marsupial genome (Mikkelsen et al. 2007).
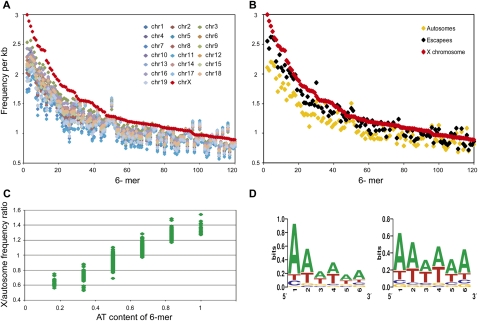
To identify unique sequence motifs, Clcn4 intron sequences specifically retained in the X-linked loci of five species (M. spretus, rat, human, cow, dog) but absent in the M. musculus autosomal locus were aligned using ExactPlus (Antonellis et al. 2006). We identified 108 sequence fragments ranging in size from 6 to 27 bp, which were perfectly conserved (100% identical) between the X-linked loci, but specifically lost at the autosomal locus. Searches for 6-mer motifs within these fragments (in both orientations) identified 427 6-mers (Supplemental Table S4). Surveys of the whole mouse genome using this list of 6-mers showed marked enrichment of a subset on the whole X compared with autosomes. This specific subset of 6-mers had 1.2–1.3 times greater frequency on the X, representing 117/427 (27%) and 91/427 (21%) 6-mers in the masked and unmasked genome, respectively (Fig. 5; Supplemental Table S4). Analysis of each individual mouse chromosome (1–19, X) showed that the X was unique in terms of enrichment in these motifs (Fig. 5A). Interestingly, genes known to escape X inactivation in mouse had an intermediate level of enrichment, higher than the autosomes, but lower than the rest of the X (Fig. 5B; Yang et al. 2010).
Figure 5.
The X chromosome is enriched in AT-rich motifs in mouse. (A) The mouse X chromosome (red symbols) is uniquely enriched in a subset of 6-mers as compared with each autosome. A total of 427 6-mers were initially defined as those present in introns of the X-linked Clcn4 loci in five species (M. spretus, rat, human, cow, dog), but absent in the autosomal locus in M. musculus (Supplemental Table S4). The frequencies per kilobase of genomic DNA for a subset 120 6-mers on the X and autosomes are shown for the repeat-masked mouse genome (6-mers were ordered by decreasing frequency per kilobase on the X). (B) The frequencies per kilobase for a subset of 120 6-mers on genes that escape X inactivation (black) show an intermediate enrichment, as compared with the X chromosome (red) and autosomes (yellow). Each point represents the frequency per kilobase for each 6-mer. (C) The relative X to autosome frequency per kilobase for each 6-mer increases with the AT content. Frequency ratios between the X and autosomes are shown as a function of the AT content of the 6-mers (expressed as a fraction of the six nucleotides). Analysis shown for the repeat-masked mouse genome (see Supplemental Figs. S4, S5 for other species). (D) AT-rich sequence motif logos for 6-mers enriched at least 1.2-fold on the X in the unmasked genome (left), and the repeat masked genome (right).
Remarkably, the subset of 6-mers specifically enriched on the mouse X had a uniformly high AT content with an A or T in 4–6/6 nucleotides in both the masked and unmasked mouse genome (Fig. 5C; Supplemental Table S4). Motif logos were generated for these 6-mers, based on a 1.2–1.3 greater frequency on the X versus autosomes (Fig. 5D). Genome-wide analyses in four other eutherian species (rat, human, dog, and cow) showed a similar enrichment in AT-motifs on the X, although this was most striking for the rodents (Supplemental Fig. S4). This is consistent with a low GC content of the X chromosome in eutherian mammals, especially in rodents, where the GC content is 39% for the X versus 42% for the autosomes (Rhead et al. 2010; Supplemental Table S5). Additional analyses of the human genome showed a lesser enrichment in AT-motifs on the short arm compared with the long arm (Supplemental Fig. S5). A similar, but less pronounced difference was observed between human genes that escape X inactivation (abundant on the short arm) and genes subject to X inactivation (Carrel and Willard 2005). In a reverse pattern to that observed in eutherian mammals, the opossum had a lower enrichment in AT motifs on the X consistent with a higher GC content (41%) compared with the autosomes (37%) in this marsupial species (Mikkelsen et al. 2007; Supplemental Fig. S4; Supplemental Table S5).
Discussion
A rare exception to Ohno's rule of conservation of the mammalian X chromosome is exemplified by Clcn4, a gene that is X-linked in most mammals, but autosomal in a subset of Mus species including the laboratory mouse M. musculus (Palmer et al. 1995; Rugarli et al. 1995). The evolutionary translocation event inserted Clcn4-2 as a single gene into a cluster of multicopy genes on chromosome 7 in M. musculus. In the current study, we established the genomic sequence of Clcn4-2 in M. spretus, a closely related mouse species with an X-linked form of the gene. We defined the breakpoints of the translocation, demonstrating that the 5′end breakpoints involve both loss of the adjacent gene Wwc3 and duplication of a small portion of Mid1 and PAR sequences in M. musculus. Surprisingly, comparisons between eight eutherian mammals revealed extensive truncations of intron sequences at the autosomal locus. Portions of the deleted intron sequences are conserved on the X in eutherian species that diverged much before the separation of the Mus species. Furthermore, a few small regions of conservation are apparently retained in a marsupial (opossum) where CLCN4 is autosomal. Thus, a parsimonious interpretation is that intronic sequences present in an ancestral mammal were subsequently lost on chromosome 7 in one Mus branch.
Our results indicate that the distal end of the M. spretus X chromosome significantly differs from that of M. musculus, even though the two species are separated by only 1–3 My of evolution. These findings support prior evidence of rapid evolution of the PAR whose boundary varies between species (Ellis and Goodfellow 1989; Graves et al. 1998). We determined that Mid1 is proximal to Clcn4-2 and Wwc3 in M. spretus, confirming that, unlike the situation in M. musculus where Mid1 straddles the pseudoautosomal boundary, this gene is outside of the PAR (Perry et al. 2001). Furthermore, we detected an inversion between Clcn4-2 and Wwc3 in M. spretus. The location of the pseudoautosomal boundary remains to be defined in M. spretus once the sequence of the Y chromosome is determined. Interestingly, truncation of introns has been documented in the pseudoautosomal 3′end portion of Mid1 in M. musculus compared with the X-linked version of Mid1 in M. spretus, which was interpreted in terms of an increase in GC content and high recombination within the PAR (Montoya-Burgos et al. 2003).
In M. spretus, Clcn4-2 is dosage compensated by X up-regulation to double its expression compared with the autosomal gene in M. musculus and by X inactivation to silence one allele in females (Rugarli et al. 1995; Adler et al. 1997). Thus, sequences conserved on the X in seven eutherian species, but absent at the autosomal locus, could potentially facilitate an increase in expression on the active X and/or silencing on the inactive X. Clcn4-2 decreased expression on chromosome 7 could result from adaptation involving mutations in its promoter region, as suggested by low conservation of promoters P1 and P2 and/or in regulatory elements/motifs such as enhancer elements. Our findings of differential enrichment in the active histone mark H3K4me3 at the 5′end of the autosomal gene compared with the X-linked gene in a cell line suggest different chromatin configurations and possibly a different promoter usage. However, additional studies will be needed to fully define these chromatin differences in vivo.
The dramatic reduction in intron size that we observed at the autosomal locus is due to large deletions centered within Clcn4-2 introns. There is no evidence of significant differences between average intron length of autosomal genes versus X-linked genes (Ross et al. 2005), suggesting that the reduction in intron size is unique to Clcn4-2 and may have played a role in lowering its expression after translocation. Sequences conserved between the autosomal and X-linked Clcn4 loci (most exons and parts of promoters) are probably critical for proper expression and function of the gene, which is most highly expressed in brain as confirmed by the presence of binding sites for brain-specific transcription factors. Nonetheless, differences at the 5′UTR and in promoters may explain some degree of tissue specificity: In M. musculus, expression is detected in brain and to a lesser extent in heart, while in M. spretus, expression is mainly confined to brain (Adler et al. 1997). Little is known about the functions of the CLCN4 protein, a chloride channel that mediates voltage-dependent electrogenic Cl−/H+ exchange (Jentsch 2008). In human, CLCN4 is expressed in multiple tissues, especially in excitable tissues, such as heart, brain, and skeletal muscle (van Slegtenhorst et al. 1994).
We have found a high density of repeats at the X-linked Clcn4 loci, which may facilitate their silencing by X inactivation. LINE1 elements have been proposed to play a role in X inactivation in eutherian mammals (Lyon 1998), but not in marsupials (Mikkelsen et al. 2007). A recent study further implicates LINE1 elements in formation of heterochromatin of the inactive mouse X chromosome (Chow et al. 2010). LINE1 and LTR repeats are depleted in regions containing genes that escape X inactivation in human and in mouse (Bailey et al. 2000; Tsuchiya et al. 2004). In fact, DNA sequence features that include specific repeats and selected 3- and 5-base sequences have been used to classify human genes in terms of X inactivation or escape (Wang et al. 2006). Interestingly, genes moved to a different chromosomal location after an ancient evolutionary duplication often have shorter introns, fewer LINE elements, higher GC content, and large CpG islands, all characteristics of the translocated M. musculus Clcn4-2 gene (Rayko et al. 2006).
Intronic regions conserved in the X-linked Clcn4 loci of five eutherian species, but deleted in the autosomal locus in M. musculus, retained specific DNA sequence motifs. A subset of AT-rich motifs was enriched on the whole X in mouse and rat, and to a lesser extent in human, dog, and cow, suggesting a role in X regulation. Enrichment in specific dinucleotide repeats [AT]n, [AC]n, [AG], and in [GATA]n has been previously reported on the human X (McNeil et al. 2006). The eutherian X has a lower overall GC content than the rest of the genome (Rhead et al. 2010). In contrast, the opossum X, which we found depleted in AT motifs, has a high GC content, possibly due to a high rate of recombination (Mikkelsen et al. 2007). Following the hypothesis that GC-rich isochores are associated with higher recombination (Montoya-Burgos et al. 2003), it could be argued that lower recombination on the X in eutherian mammals resulted in depletion in these isochores, hence, the observed enrichment in AT motifs. In turn, deletion of AT-rich regions due to high recombination could have shaped the autosomal Clcn4-2 locus in M. musculus. However, this locus is located very close to the centromere of chromosome 7, where recombination is low (Cox et al. 2009). In addition, we observed a lower enrichment in AT motifs in mouse and human genes that escape X inactivation, which would not be a priori expected to have a higher rate of recombination than genes subject to X inactivation.
Alternatively, the overall enrichment in AT motifs on the eutherian X chromosome may be interpreted in relation to the molecular mechanisms of dosage compensation, especially X inactivation. Our analyses clearly indicate that the rodent X especially is unique in comparison to any autosome. It is fitting that, compared with the mouse X, the human X is both less completely inactivated and also less enriched in AT motifs (Carrel and Willard 2005; Yang et al. 2010). There is prior indication that AT motifs may be important for X inactivation. Indeed, Wang et al. (2006) reported that the majority of 3-mers and 5-mers enriched around human escape genes are GC-rich, while those enriched around genes subject to X inactivation are AT-rich, despite a similar overall GC content for each type of gene. This is consistent with our observations of a greater difference between the whole human p and q arms than between escape genes and genes subject to inactivation, suggesting that the intergenic distribution of motifs is important.
Specific sequence motifs may attract Xist RNA (X-inactive-specific transcript), a noncoding RNA essential for the onset of X inactivation via the recruitment of protein complexes that implement repressive epigenetic modifications (Payer and Lee 2008). Consistent with this idea, we have found a lesser enrichment in AT motifs at escape genes, perhaps explaining a lack of Xist coating (Murakami et al. 2009). AT motifs may also be important for maintenance of silencing by recruiting AT-binding proteins involved in chromatin scaffolding. Both SATB1 (SATB homeobox 1) and HNRNPU (heterogeneous nuclear ribonucleoprotein U) associate with the inactive X, suggesting that AT motifs could facilitate changes in chromatin conformation (Helbig and Fackelmayer 2003; Agrelo et al. 2009). In contrast to eutherians, the opossum X has a high GC content and yet is subject to X inactivation; this apparent discrepancy may reflect the known differences in molecular mechanisms of X inactivation in marsupials, despite some common features (Duret et al. 2006; Koina et al. 2009; Mahadevaiah et al. 2009). One important difference is the absence of Xist in marsupials (Duret et al. 2006).
Whether AT motifs may also be involved in X up-regulation remains to be determined. AT-binding proteins influence chromatin structure by binding to the base of chromatin loops, and thus could facilitate increased gene expression on the active X compared with autosomes. For example, SATB1 has been implicated in regulation of actively transcribed chromatin, and HNRNPU has been proposed as a factor that helps in the formation of functional domains within the nucleus (Cai et al. 2006; Malyavantham et al. 2008). HNRNPU has also been implicated in RNA elongation and stability (Kukalev et al. 2005; Yugami et al. 2007; Obrdlik et al. 2008).
Methods
BAC library screening, BAC sequencing, and FISH
The M. spretus BAC library was constructed as previously described (http://bacpac.chori.org/library.php?id=170) (Osoegawa and de Jong 2004). Probes generated by PCR were labeled prior to hybridization to the high-density colony arrays. The BACs were characterized using PCR and BAC-end sequencing. Two BACs, Ch35-246O15 (BAC31) and Ch35- 316H16 (BAC29), which overlapped by 48,468 bp and covered the Clcn4-2 locus, were completely sequenced after BAC-end analysis to map them by alignment to the rat sequence (Supplemental Fig. S1). DNA sequencing and assembly of the BAC clones were carried out using methods established in our Genome Center (Gregory et al. 2006; Muzny et al. 2006). FISH using a probe for Mid1 labeled with Texas red and BAC29 labeled with fluorescein was done on M. spretus chromosome preparations using standard procedures.
Sequence analyses
Promoters P1, P2, and P3 and associated transcription factors were identified using Genomatix tools (www.genomatix.de). Each promoter region represents ∼600 bp. The software Sequencer ver. 4.1.4 was used to define the breakpoints by aligning sequences from different species (http://www.genecodes.com). Clcn4 sequences from seven mammalian species downloaded from the UCSC Genome Browser (Rhead et al. 2010) and from Ensembl (Flicek et al. 2010) were aligned to the M. spretus sequence using CLUSTALW software (http://www.clustal.org) (Larkin et al. 2007). Regions of high homology were found using VISTA (http://pipeline.lbl.gov/cgi-bin/gateway2) (Frazer et al. 2004). CpG islands were searched using the CpG island searcher (http://cpgislands.usc.edu) (Takai and Jones 2003).
Repeat and oligomer analyses
Repeat density was determined using RepeatMasker (http://www.repeatmasker.org, Institute for Systems Biology). ExactPlus (http://research.nhgri.nih.gov/exactplus) was used to screen for intron sequences uniquely present in X-linked Clcn4 genes and absent in the autosomal gene (Antonellis et al. 2006). Oligomers (6-mers) uniquely enriched in these regions were then tested for their distribution in the entire mouse, human, rat, dog, and cow genome, either unmasked or masked by RepeatMasker (Rhead et al. 2010). To evaluate escape from X inactivation, 13 mouse escape genes were considered versus 259 mouse genes subject to X inactivation (Yang et al. 2010). Comparisons were also done between the short and long arms of the human X chromosome and between 79 human escape genes and 247 human genes subject to X inactivation (Carrel and Willard 2005). Motif logos were generated using WebLogo (Crooks et al. 2004).
Chromatin analyses
Chromatin immunoprecipitations were done using antibodies for histone H3 trimethylated at lysine 4 (Millipore) and RNA polymerase II phosphorylated at serine 5 (Abcam), following established methods (Nelson et al. 2006). Quantitative PCR analyses were done using primers that distinguish M. musculus and M. spretus loci in Patski cells (Yang et al. 2010; Supplemental Table S6). ChIP fractions were normalized to the input fractions prior to calculating the ratios between enrichments on the M. musculus and M. spretus loci. Published chromatin data for the mouse and human Clcn4-2/CLCN4 genes were downloaded to the UCSC browser (Celniker et al. 2009; Gupta et al. 2010).
Acknowledgments
This work was supported by National Institutes of Health Grants GM046883 and GM079537 (to C.M.D.), 3U54HG002043 (to R.K.), and NS060983 (to A.A.). BAC library construction was funded by NIH grants HG01165-07SI and HG025323-01 (P.J.d.J.) as part of the NIH-funded BAC Resource Network (http://www.genome.gov/page.cfm?pageID=10001844).
Footnotes
[Supplemental material is available for this article. The sequencing data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. HM053970 and HM053971.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.108563.110.
References
- Adler DA, Rugarli EI, Lingenfelter PA, Tsuchiya K, Poslinski D, Liggitt HD, Chapman VM, Elliott RW, Ballabio A, Disteche CM 1997. Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc Natl Acad Sci 94: 9244–9248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrelo R, Souabni A, Novatchkova M, Haslinger C, Leeb M, Komnenovic V, Kishimoto H, Gresh L, Kohwi-Shigematsu T, Kenner L, et al. 2009. SATB1 defines the developmental context for gene silencing by Xist in lymphoma and embryonic cells. Dev Cell 16: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alekseyenko AA, Peng S, Larschan E, Gorchakov AA, Lee OK, Kharchenko P, McGrath SD, Wang CI, Mardis ER, Park PJ, et al. 2008. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell 134: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Bennett WR, Menheniott TR, Prasad AB, Lee-Lin SQ, Green ED, Paisley D, Kelsh RN, Pavan WJ, Ward A 2006. Deletion of long-range sequences at Sox10 compromises developmental expression in a mouse model of Waardenburg-Shah (WS4) syndrome. Hum Mol Genet 15: 259–271 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE 2000. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: The Lyon repeat hypothesis. Proc Natl Acad Sci 97: 6634–6639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434: 400–404 [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. 2009. Unlocking the secrets of the genome. Nature 459: 927–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, et al. 2010. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141: 956–969 [DOI] [PubMed] [Google Scholar]
- Cox A, Ackert-Bicknell CL, Dumont BL, Ding Y, Bell JT, Brockmann GA, Wergedal JE, Bult C, Paigen B, Flint J, et al. 2009. A new standard genetic map for the laboratory mouse. Genetics 182: 1335–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE 2004. WebLogo: A sequence logo generator. Genome Res 14: 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Ellis N, Goodfellow PN 1989. The mammalian pseudoautosomal region. Trends Genet 5: 406–410 [DOI] [PubMed] [Google Scholar]
- Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. 2010. Ensembl's 10th year. Nucleic Acids Res 38: D557–D562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I 2004. VISTA: Computational tools for comparative genomics. Nucleic Acids Res 32: W273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA, Wakefield MJ, Toder R 1998. The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet 7: 1991–1996 [DOI] [PubMed] [Google Scholar]
- Gregory SG, Barlow KF, McLay KE, Kaul R, Swarbreck D, Dunham A, Scott CE, Howe KL, Woodfine K, Spencer CC, et al. 2006. The DNA sequence and biological annotation of human chromosome 1. Nature 441: 315–321 [DOI] [PubMed] [Google Scholar]
- Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B 2006. Global analysis of X-chromosome dosage compensation. J Biol 5: 3 doi: 10.1186/biol30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Wikramasinghe P, Bhattacharyya A, Perez FA, Pal S, Davuluri RV 2010. Annotation of gene promoters by integrative data-mining of ChIP-seq Pol-II enrichment data. BMC Bioinformatics (Suppl 1) 11: S65 doi: 10.1186/1471-2105-11-S1-S65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig R, Fackelmayer FO 2003. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma 112: 173–182 [DOI] [PubMed] [Google Scholar]
- Jentsch TJ 2008. CLC chloride channels and transporters: From genes to protein structure, pathology and physiology. Crit Rev Biochem Mol Biol 43: 3–36 [DOI] [PubMed] [Google Scholar]
- Koina E, Chaumeil J, Greaves IK, Tremethick DJ, Graves JA 2009. Specific patterns of histone marks accompany X chromosome inactivation in a marsupial. Chromosome Res 17: 115–126 [DOI] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P 2005. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol 12: 238–244 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lyon M 1961. Gene action in the X-chromosome of the mouse (Mus musculus L). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Lyon MF 1998. X-chromosome inactivation: A repeat hypothesis. Cytogenet Cell Genet 80: 133–137 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM 2009. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19: 1478–1484 [DOI] [PubMed] [Google Scholar]
- Malyavantham KS, Bhattacharya S, Barbeitos M, Mukherjee L, Xu J, Fackelmayer FO, Berezney R 2008. Identifying functional neighborhoods within the cell nucleus: Proximity analysis of early S-phase replicating chromatin domains to sites of transcription, RNA polymerase II, HP1gamma, matrin 3 and SAF-A. J Cell Biochem 105: 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ 2006. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 444: 614–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil JA, Smith KP, Hall LL, Lawrence JB 2006. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res 16: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177 [DOI] [PubMed] [Google Scholar]
- Montoya-Burgos JI, Boursot P, Galtier N 2003. Recombination explains isochores in mammalian genomes. Trends Genet 19: 128–130 [DOI] [PubMed] [Google Scholar]
- Murakami K, Ohhira T, Oshiro E, Qi D, Oshimura M, Kugoh H 2009. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet Genome Res 125: 19–25 [DOI] [PubMed] [Google Scholar]
- Muzny DM, Scherer SE, Kaul R, Wang J, Yu J, Sudbrak R, Buhay CJ, Chen R, Cree A, Ding Y, et al. 2006. The DNA sequence, annotation and analysis of human chromosome 3. Nature 440: 1194–1198 [DOI] [PubMed] [Google Scholar]
- Nelson JD, Denisenko O, Bomsztyk K 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc 1: 179–185 [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53 [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P 2008. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol 28: 6342–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S 1967. Sex chromosomes and sex linked genes. Springer; Verlag, Berlin, Germany [Google Scholar]
- Osoegawa K, de Jong PJ 2004. BAC library construction. Methods Mol Biol 255: 1–46 [DOI] [PubMed] [Google Scholar]
- Palmer S, Perry J, Ashworth A 1995. A contravention of Ohno's law in mice. Nat Genet 10: 472–476 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT 2008. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Perry J, Palmer S, Gabriel A, Ashworth A 2001. A short pseudoautosomal region in laboratory mice. Genome Res 11: 1826–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayko E, Jabbari K, Bernardi G 2006. The evolution of introns in human duplicated genes. Gene 365: 41–47 [DOI] [PubMed] [Google Scholar]
- Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. 2010. The UCSC Genome Browser database: Update 2010. Nucleic Acids Res 38: D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. 2005. The DNA sequence of the human X chromosome. Nature 434: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugarli EI, Adler DA, Borsani G, Tsuchiya K, Franco B, Hauge X, Disteche C, Chapman V, Ballabio A 1995. Different chromosomal localization of the Clcn4 gene in Mus spretus and C57BL/6J mice. Nat Genet 10: 466–471 [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA 2007. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat Rev Genet 8: 424–436 [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA 2003. The CpG island searcher: A new WWW resource. In Silico Biol 3: 235–240 [PubMed] [Google Scholar]
- Tsuchiya KD, Greally JM, Yi Y, Noel KP, Truong JP, Disteche CM 2004. Comparative sequence and x-inactivation analyses of a domain of escape in human Xp11.2 and the conserved segment in mouse. Genome Res 14: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst MA, Bassi MT, Borsani G, Wapenaar MC, Ferrero GB, de Conciliis L, Rugarli EI, Grillo A, Franco B, Zoghbi HY, et al. 1994. A gene from the Xp22.3 region shares homology with voltage-gated chloride channels. Hum Mol Genet 3: 547–552 [DOI] [PubMed] [Google Scholar]
- Wang Z, Willard HF, Mukherjee S, Furey TS 2006. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol 2: e113 doi: 10.1371/journal.pcbi.0020113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yugami M, Kabe Y, Yamaguchi Y, Wada T, Handa H 2007. hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett 581: 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]