Abstract
The evolutionarily conserved target of rapamycin complex 1 (TORC1) controls cell growth in response to nutrient availability and growth factors. TORC1 signaling is hyperactive in cancer, and regulators of TORC1 signaling may represent therapeutic targets for human diseases. To identify novel regulators of TORC1 signaling, we performed a genome-scale RNA interference screen on microarrays of Drosophila melanogaster cells expressing human RPS6, a TORC1 effector whose phosphorylated form we detected by immunofluorescence. Our screen revealed that the TORC1–S6K–RPS6 signaling axis is regulated by many subcellular components, including the Class I vesicle coat (COPI), the spliceosome, the proteasome, the nuclear pore, and the translation initiation machinery. Using additional RNAi reagents, we confirmed 70 novel genes as significant on-target regulators of RPS6 phosphorylation, and we characterized them with extensive secondary assays probing various arms of the TORC1 pathways, identifying functional relationships among those genes. We conclude that cell-based microarrays are a useful platform for genome-scale and secondary screening in Drosophila, revealing regulators that may represent drug targets for cancers and other diseases of deregulated TORC1 signaling.
The control of size in biological systems is an important and intricate process, comprising regulation of cell number and regulation of cell size. An individual cell grows as it incorporates raw nutrients into proteins and other macromolecules. For an organism to develop properly, cells of different tissues must grow at different rates during various stages of development. To this end, eukaryotes have evolved signaling networks that regulate cell growth in response to stimuli from growth factors as well as nutrient availability.
At the core of one such network is the target of rapamycin (TOR), a protein kinase that acts as part of multiple distinct and evolutionarily conserved protein complexes, as recently reviewed (Laplante and Sabatini 2009; Soulard et al. 2009). TOR complex 1 (TORC1) transduces signals to the ribosome and other translational machinery in response to amino acid levels and insulin or growth factors (Laplante and Sabatini 2009; Proud 2009). Several tumor suppressors regulate TORC1, and hyperactive TORC1 signaling contributes to the pathogenesis of many tumors, particularly those deficient in TSC1, TSC2, PTEN, STK11 (also known as LKB1), or VHL (Sabatini 2006; Huang and Manning 2009; Inoki and Guan 2009; Plas and Thomas 2009). Furthermore, TORC1 signaling is essential for the development of metazoans (Oldham et al. 2000; Murakami et al. 2004), as well as for organ-, tissue-, and cell-specific growth processes such as lymphocyte expansion during the adaptive immune response (Colombetti et al. 2006) and the growth of dendritic spines (Tavazoie et al. 2005). A more complete awareness of the cellular processes and genes that control TORC1 signaling, then, would benefit our understanding of these TORC1-dependent growth processes and diseases.
We previously developed a technique for efficient RNA interference screening by dsRNA reverse-transfection on living Drosophila cell microarrays (Wheeler et al. 2004); here we use that technique in the first such screen at genome scale, followed by secondary screening cell arrays interrogating nine phenotypes. We measured dTORC1 signaling by immunofluorescent staining of a phosphorylated effector of dTORC1, namely, the RPS6 ribosomal protein. The initial set of hits was highly enriched for known dTORC1–S6K pathway components, and we confirmed 70 genes as novel on-target phospho-RPS6 or phospho-S6K-regulating hits. We identified several unexpected and evolutionarily conserved subcellular complexes as regulators of the TORC1 signaling pathway.
Results
Development of a high-throughput-compatible Drosophila TORC1-reporter assay
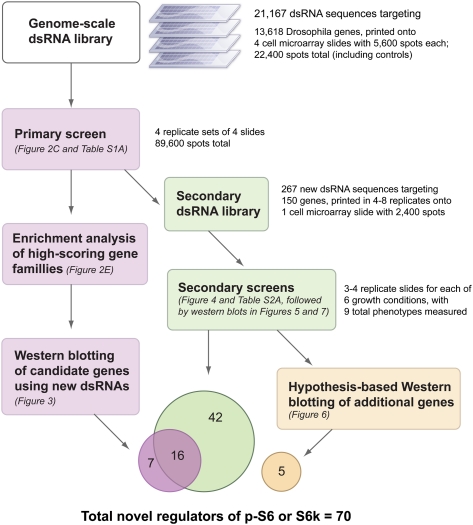
Drosophila melanogaster cells are a valuable model system for the study of the TORC1 pathway (Guertin et al. 2006a) and are very susceptible to RNA interference via high-throughput cell microarrays (Wheeler et al. 2004). We designed a systematic approach to identify novel TORC1 pathway regulators using a combination of genome-scale cell microarrays, bioinformatics analysis, smaller-scale secondary screening using cell microarrays, and low-throughput Western blotting (Fig. 1). Although the ideal readout for a TORC1-regulator screen would be phosphorylation of the TORC1 substrate S6K on its hydrophobic-motif site (T398 on dS6K, T389 in mammalian p70 S6K1, otherwise known as RPS6KB1), phospho-specific antibodies against S6K do not work for immunofluorescence. Therefore, we used a phospho-specific antibody against S6K1's substrate, S6 ribosomal protein (RPS6, or just S6). Due to the lack of commercially available antibodies against Drosophila S6 (dS6), we created a Drosophila S2R+ cell line that expresses the human S6 (hS6) under the control of the metallothionine promoter (see Methods), such that hS6 is induced upon copper sulfate treatment. As expected, TORC1-pathway perturbations yielded the expected effects on the S6 reporter: Rapamycin treatment and dRaptor knockdown ablate p-hS6 (and p-T398 dS6K), while dTSC2 knockdown increases p-hS6 (and p-T398 dS6K) by immunoblot (Fig. 2A).
Figure 1.
Experimental workflow for this study.
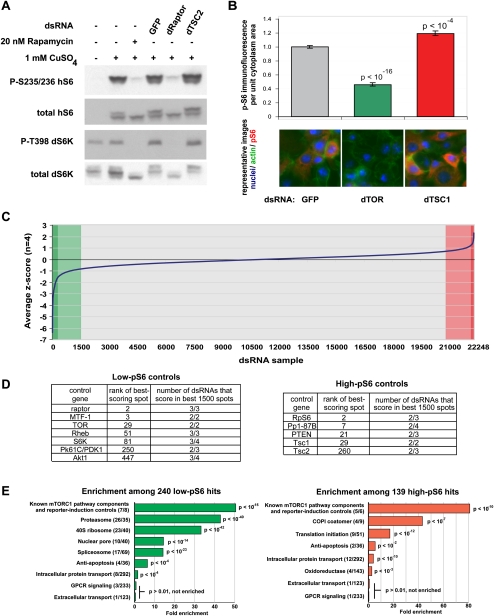
Figure 2.
Genome-scale RNAi screen using a TORC1–S6K pathway immunofluorescence reporter identifies families of genes that regulate phospho-S6 levels. (A) The Drosophila S2R+ cell line was transfected to express the human S6 ribosomal protein under a metallothionine promoter, allowing induction by CuSO4 treatment. In a stable clone of these cells, called S6_S2R+, Western blotting demonstrates that phosphorylation of human S6 is sensitive to rapamycin as well as to dTORC1-pathway RNA1 (dRaptor and dTSC2); the reporter responds similarly to the endogenous Drosophila S6K, for which immunofluorescence-compatible antibodies are not available. (B) This cell line was transfected on microarrays with the indicated dsRNAs, and pS6 immunofluorescence per unit cytoplasm area was quantified with CellProfiler software (data shown as mean ± standard error, n = 16 spots). The dsRNA labeled “GFP” has no identical 19+mers to any transcripts in the genome, has no effect on TORC1 signaling, and is used throughout this work as a transfection control. (C) We screened, in quadruplicate, a genome-scale RNAi library containing 22,248 microarray spots targeting 13,618 Drosophila genes, for regulators of pS6 immunofluorescence. Candidate high-pS6 (red) and low-pS6 (green) hits were determined by assigning a Z-score to each spot on the array and combining scores for replicates. Candidate hit genes showed at least one dsRNA having a very strong phenotype (deeply shaded) or multiple dsRNAs having a moderately strong phenotype (lightly shaded); see the Supplemental Methods for more details. We found 240 candidate low-pS6 hits and 139 candidate high-pS6 hits. (D) The candidate hits included all the expected nonredundant components of the TORC1–S6K pathway, as elaborated in the text. (E) Genes were grouped by GO annotation (and into several ad hoc groups; see Methods), and gene groups were screened for fold enrichment among low-pS6 and high-pS6 candidate hits. For a given group, parentheses denote (number of group members scoring)/(number of group members in library). P-values are corrected for multiple comparisons (Benjamini and Hochberg 1995).
This cell line is suitable for high-resolution image-based screening: Upon knockdown of TORC1 pathway genes, population-averaged increases and decreases in mean cytoplasmic p-hS6 immunofluorescence signal are detectable by microscopy (Fig. 2B; and controls described in the context of the primary screen, described next). Because antibodies against total hS6 were not robust enough to use for immunofluorescence, we predicted that the use of pS6 as a reporter might introduce three classes of genes: (1) those that, like the canonical TORC1 pathway members, regulate p-hS6 by regulating p-T398-dS6K; (2) those that regulate p-hS6 by regulating dS6K levels; and (3) those whose altered p-hS6 signal was due to altered levels of total hS6, without any effect on phospho-S6K or total S6K. Genes in the third class might represent artifacts of the hS6 reporter and would therefore not be of interest, but genes of the second class—the total-S6K regulators—are of some interest because they control growth by regulating levels of one of dTORC1's chief substrates, S6K. At various points in the screening process, we performed Western blotting to help classify candidate genes as regulators of phospho-dS6K, total dS6K, or S6 only.
Genome-scale RNAi library screen reveals groups of genes that regulate the phosphorylation and overall levels of S6
Cell microarrays use minuscule amounts of antibody as compared to screens in a traditional multi-well plate format, enabling large screens with many replicates. We assembled and screened a Drosophila genome RNAi library consisting of two commercially available libraries, including frequent positive and negative control samples, with a small set of handmade dsRNAs. We screened 22,248 samples, targeting 13,618 genes (roughly 97% of the currently defined Drosophila genome), in four replicates (Fig. 2C), as more fully described in the Supplemental Methods. The full genome screening data set is in Supplemental Table S1A.
Our primary screen identified 240 low-pS6 and 139 high-pS6 candidate genes, listed in Supplemental Table S1B. Among the candidate hits, we found many of the expected, nonredundant positive and negative regulators of the dTORC1 signaling pathway (Fig. 2D), with two exceptions. The dTORC1 component CG3004/GbL did not score, but this is not surprising because in vivo experiments have suggested GbL is essential only for TORC2 signaling and not TORC1 signaling (Guertin et al. 2006b). Likewise, the TORC1 inhibitor L (Lobe/PRAS40) has been shown to act only under conditions of low insulin signaling (Sancak et al. 2007), and it is therefore not surprising that the serum-containing conditions of the screen obscured an RNAi phenotype for Lobe. As expected, we found among the low-pS6 candidate hits the metallothionine transcription factor MTF-1, which induces the hS6 reporter. Among the high-pS6 candidates, we found Drosophila RpS6, which competes with the human S6 reporter, and Pp1-87B, an ortholog of protein phosphatase 1, which is known to dephosphorylate S6.
We grouped the genes in the library by their associated Gene Ontology terms and found that the sets of low-pS6 and high-pS6 candidate hits were markedly enriched for several groups of genes (Fig. 2E; Supplemental Table S1C). As mentioned, we wanted to determine whether these gene families' effects on pS6 were mere consequences of hS6 reporter levels, or whether the p-S6 phenotypes reflected effects on endogenous p-dS6K or total dS6K levels. We therefore investigated several implicated gene families' dS6K and hS6 phenotypes by Western blots. Per established DRSC guidelines, we used new dsRNAs designed to avoid significant sequence identity and thus off-target effects that might produce false-positives (Flockhart et al. 2006; Kulkarni et al. 2006; Ma et al. 2006).
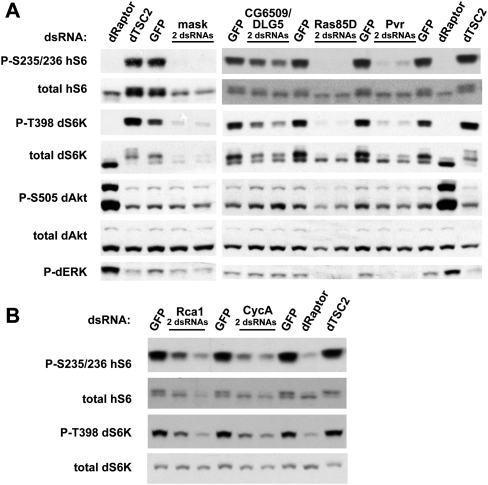
Four of nine genes encoding class I vesicle-coat proteins scored among the 139 high-pS6 candidate genes, representing a >40-fold enrichment versus the whole library (P < 1.3 × 10−8). In fact, a dsRNA targeting gammaCop was the best-scoring individual high-pS6 knockdown in the genome. To further characterize this screening result, we knocked down two COPI genes that scored highly, as well as one (alphaCop) that did not score. By Western blot, all three COPI component knockdowns showed not only increased pS6 but also increased p-T398-dS6K and decreased p-S505-dAKT (Fig. 3A), while total levels of these proteins were unaffected.
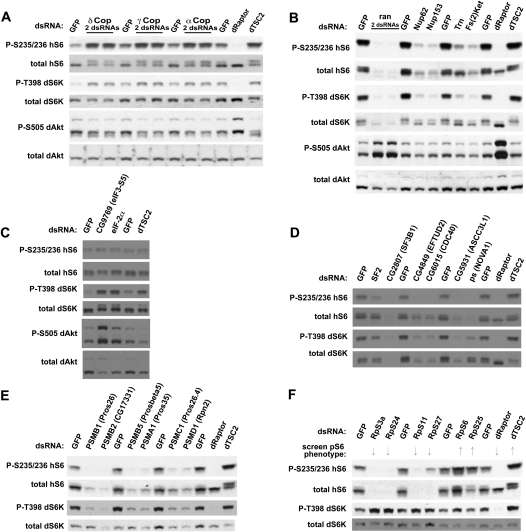
Figure 3.
Validation and functional characterization of enriched families of genes from genome-scale screen. (A–F) Members of enriched families of genes were validated by Western blotting. Drosophila S6_S2R+ cells were incubated with dsRNAs targeting the indicated genes from groups identified in Figure 2E, and lysates were analyzed by Western blotting. See also Supplemental Table S1 for results and analysis related to the genome screen.
Through similar analysis of enriched gene families, we found that knockdown of nuclear pore components decreased phospho-dS6K with corresponding decreases in total dS6K levels (Fig. 3B). Knockdown of the ran GTPase, which regulates nuclear transport, caused both the levels of S6K and the phosphorylation of T398 to decrease while AKT S505 phosphorylation increased (Fig. 3B). Knockdown of the translational-initiation machinery produced weak increases in phosphorylated hS6 by Western blot, but dramatic increases not only in dS6K phosphorylation but also dAKT phosphorylation, without changes in total dS6K levels (Fig. 3C). Proteasomal subunits and spliceosomal subunits produced decreases in both the phosphorylation and the levels of dS6K (Fig. 3D,E); some of these genes also affected levels of the hS6 reporter, but their effects on the endogenous dS6K validate these genes as bona fide p-S6 regulators. Lastly, knockdown of 40S ribosomal proteins typically decreased levels of hS6, presumably due to ribosomal instability (Fig. 3F). Knockdown of RpS6 itself is one exception: It scores among the highest p-hS6 knockdowns presumably because knockdown of dS6 allows incorporation of additional hS6 reporter into ribosomes (Fig. 3F). As most of these ribosomal genes affected only the hS6 reporter, we do not consider them hits; interestingly, though, RpS25 showed an increase in p-S6 despite a decrease in total S6 (Fig. 3F).
Validation, secondary screening, and Western blots to characterize functional relationships between S6 regulators
In order to efficiently characterize hits from the primary screen, we performed high-throughput secondary screens using customized, smaller-scale cell microarrays grown under multiple TORC1-relevant nutrient, growth, and double-knockdown conditions (Fig. 4), such that each gene was tested in eight to 16 replicates for each condition (see Methods for further details and for a description of how genes were selected for secondary screening). This also offered the opportunity to validate hits using alternate dsRNA sequences: Our original genome-scale library was designed prior to the establishment of guidelines for avoiding off-target effects (Kulkarni et al. 2006; Ma et al. 2006). In the secondary screens, we thus targeted the selected high-pS6 and low-pS6 hits from the primary screen by multiple novel dsRNAs free of significant sequence identity to all other Drosophila transcripts. In a few cases in which multiple subunits of a complex were included in the secondary screen (e.g., proteasome, nucleoporins, CSNs, Vha, splicing factors), we instead designed only one new dsRNA for each of multiple components of that complex.
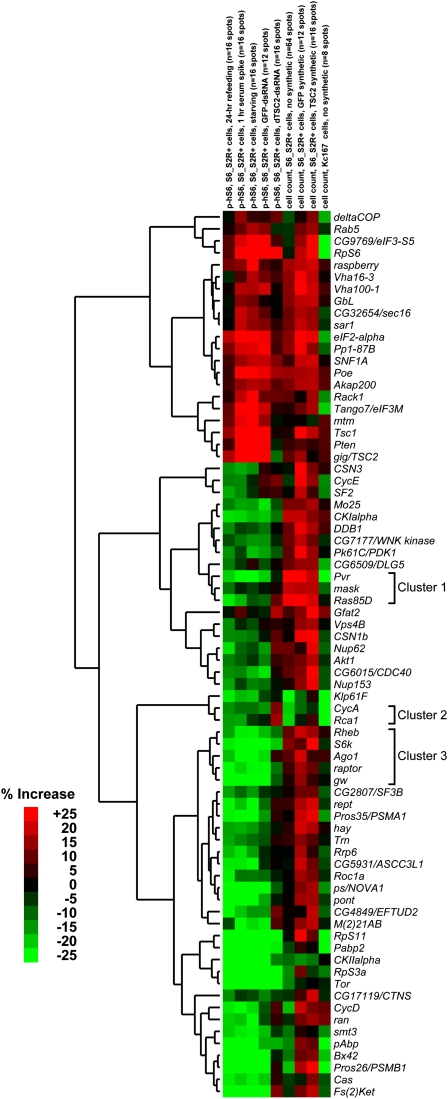
Figure 4.
Validation and functional characterization of on-target TORC1–S6K pathway regulators using secondary screening for multiple phenotypes. On-target hits from the primary screen were screened in several validation and secondary screens on microarrays, as described in the Methods. The results were subjected to hierarchical clustering, and several notable clusters are marked. Note that synthetic experiments (experiments where the cell population was treated with a dsRNA in addition to the dsRNA spots on the array) were performed by transfecting cells with the indicated dsRNA and transfection reagent prior to seeding on microarrays, such that culture conditions were different from nonsynthetic experiments. See Methods for details.
Overall, 46% of the low-pS6 genes and 30% of the high-pS6 genes that were tested in the secondary screens confirmed as hits in one or more phenotypes (Supplemental Fig. S1). Excluding canonical TORC1 components and excluding three genes that were revealed by Western blotting to affect only S6 levels, the secondary screens confirmed that the knockdown of 43 genes decreased pS6 and of 15 genes increased pS6 (58 pS6 regulators total); full rescreening data is in Supplemental Table S2A. Through the small-scale experiments described in earlier and later sections, we identified additional regulators of endogenous p-T398-dS6K or total dS6K; a final list of 70 confirmed noncanonical genes (51 low and 19 high) for which RNAi perturbs p-hS6, p-dS6K, or total dS6K is in Supplemental Table S2B.
To characterize and categorize these genes' effects on TORC1 signaling and cell proliferation under diverse nutrient conditions, we hierarchically clustered the data from the secondary screens (Fig. 4). Genes in established relationships tended to cluster together, and immunoblotting of correlated genes provided further functional insight into these correlations. For instance, we found a receptor-tyrosine-kinase signaling cluster (Fig. 4, cluster 1, correlation >0.85) comprising Pvr, mask, and Ras85D. RNAi to any of these genes, as well as the co-clustered gene CG6509/DLG5 (correlation > 0.8), decreased S6K levels, with Pvr and Ras85D also showing S6K laddering that correlates with reduced T398 phosphorylation (Fig. 5A). Similarly, we found that regulator of cyclin A (Rca1) and CycA form a tight cluster (Fig. 4, cluster 2, correlation > 0.85) marked by decreased phosphorylation of dS6K at T398 (Fig. 5B).
Figure 5.
(A,B) Validation and functional characterization of TORC1–S6K pathway regulators confirmed in the secondary screen (clusters 1 and 2). S6_S2R+ cells were incubated with dsRNAs targeting the indicated genes, and lysates were analyzed by Western blotting. See also Supplemental Table S2 for results and analysis related to the secondary screens and subsequent follow-up experiments.
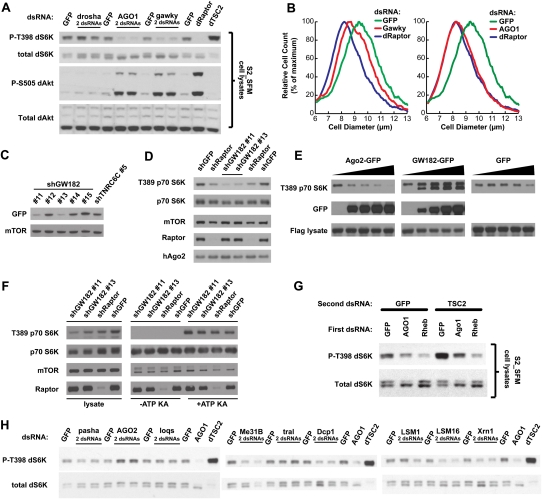
The TORC1 signaling axis of raptor, Rheb, and S6k formed a cluster (Fig. 4, cluster 3, correlation > 0.85) with two novel TORC1–S6K-pathway regulators, Argonaute-1 (AGO1) and gawky (gw). AGO1 and gw encode the essential protein components of an evolutionarily conserved miRNA-based translational-repression complex (Eulalio et al. 2008). In Drosophila S2 cells, AGO1 and gw knockdowns phenocopy raptor knockdowns with respect to S6K and AKT phosphorylation (Fig. 6A), as well as showing substantial decreases in cell size (Fig. 6B; Supplemental Fig. S2). We identified two shRNAs (Fig. 6C) that knocked down a recombinant GFP fusion of gw's human homolog GW182 (TNRC6A) (Bloch et al. 2006). When GW182 was knocked down in human HeLa cells for 3 d, we found significant decreases in p-T389-S6K and a shifting of p70S6K without any significant effect on p70S6K levels, consistent with the results for Drosophila gw (Fig. 6D). Furthermore, overexpression of the GW182-GFP fusion increased the p-T389 signal of Flag-purified S6K (as well as an unknown second band) in a dose-dependent manner (Fig. 6E). In order to determine whether GW182's effect on S6K phosphorylation occurred by regulation of mTORC1 kinase activity, we performed in vitro kinase assays on cells treated with shGFP, shRaptor, or shGW182; mTORC1 under all these treatments demonstrated identical capacity to phosphorylate S6K, suggesting that GW182 promotes T389 phosphorylation without regulating the kinase activity of mTORC1 (Fig. 6F).
Figure 6.
Validation and functional characterization of AGO1, gw, and related genes (cluster 3) in Drosophila and human cells. (A) Drosophila cells were transfected with the indicated dsRNAs, and lysates were analyzed by Western blotting. (B) Drosophila cells were transfected with dsRNAs targeting the indicated genes, and cell size distributions were obtained using a Coulter counter. Cell count is normalized to place the peaks of each distribution at equal heights; see also Supplemental Figure S2 for plots of absolute cell count as a function of cell diameter. (C) Human HeLa cells were transfected with a plasmid expressing a GFP-tagged GW182 and simultaneously infected with lentivirus expressing one of five GW182-targeting hairpins (#11–#15) or a hairpin targeting the related gene TNRC6C. Knockdown efficiency was determined by immunoblotting with an antibody recognizing GFP. (D) The shRNAs validated in C were used to knock down GW182 in HeLa cells. Lysates were prepared 3 d after infection and analyzed by Western blotting. The same shGFP and shRaptor samples on each side of the blot were loaded twice. (E) Human 293T cells were co-transfected with increasing amounts of AGO2-GFP, GW182-GFP, or pLJM1-GFP plasmids (0, 50, 100, 200, 400 ng) along with 2 ng of FLAG-S6K1-expressing plasmid. FLAG-S6K1 was purified, eluted off the Flag beads, and analyzed by Western blotting with an antibody against pT389. Lysates were also analyzed by immunoblotting. (F) mTORC1 was isolated through mTOR immunoprecipitations from HeLa cells 3 d after infection with shGW182 lentiviral constructs, and kinase assays were performed on S6K1 substrate. (G) Drosophila cells were incubated with the indicated dsRNAs in sequential order, with 2 d of incubation in the first dsRNA followed by 4 d of incubation in a mixture of the second and first dsRNA. (H) Drosophila cells were transfected with dsRNAs targeting the indicated regulators of RNA processing, and Western analysis revealed the relative effects on p-T398-dS6K of knockdown of these genes versus AGO1.
To identify a possible epistasis relationship between AGO1 and TSC2, we performed a combination-RNAi experiment. If AGO1 acted upstream of gig/TSC2, e.g., by producing miRNAs that regulate gig/TSC2, we would expect initial AGO1 knockdown to be overcome by subsequent gig/TSC2 knockdown, producing maximal T398 phosphorylation. In contrast, if AGO1 acted downstream from gig/TSC2, we would expect initial AGO1 knockdown to block any effect of gig/TSC2 knockdown, just as initial knockdown of Rheb does. However, the sequential RNAi experiment revealed an intermediate phenotype, as gig/TSC2 knockdown was able to increase p-T398 even after AGO1 knockdown, but not to the maximal extent seen following GFP knockdown (Fig. 6G). This result suggests that the effects of AGO1 and gig/TSC2 on p-dS6K are independent.
We tested a variety of other genes related to AGO1 and gw, such as other essential components of the miRNA pathway. Neither drosha (Fig. 6A), drosha's partner Pasha, nor loquacious (loqs), when knocked down, decreased pS6K to the extent that AGO1 knockdown did (Fig. 6H). Similarly, we tested knockdowns of various genes whose products act at GW-bodies (Eulalio et al. 2007). There was a slight decrease in p-T398-dS6K following knockdown of several regulators of 5′–3′ RNA decay (Me31B, Dcp1, LSM16/Edc3, pcm/XRN1) (Fig. 6H), although none had as strong an effect on p-T398-dS6K as did knockdown of AGO1. We also investigated proteins in the hAGO2 complex 1 (Hock et al. 2007). There are seven components with clear Drosophila orthologs, five of which scored as pS6 regulators in our genome-scale RNAi screen, a significant enrichment at P < 3 × 10−7. Other conserved genes that encode members of this complex include GW182/gawky, rept (RUVBL2/reptin), pic (DDB1), and eIF-4B, all of which scored, as well as TRBP/Loquacious and hnRNPF/glorund, neither of which scored.
Drosophila AGO2 serves distinct, nonoverlapping cellular functions from AGO1 (Forstemann et al. 2007). Interestingly, although our initial screen did not identify Drosophila AGO2 as a regulator of pS6, by Western blot its knockdown yielded a notable high-p-T398-dS6K phenotype (Fig. 6H). This effect was not seen in the equivalent experiment in human cells for the human homolog EIF2C2 (Argonaute2) (data not shown), possibly because of functional redundancy among the four human Argonautes, but overexpression of a recombinant GFP-fused AGO2 (Leung et al. 2006) in human cells decreased the phosphorylation of p70S6K on T389 in a dose-dependent manner (Fig. 6E), indicating conservation of AGO2's influence on S6K phosphorylation in mammalian cells.
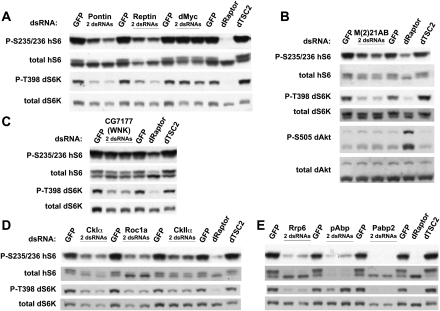
We confirmed S6 and/or S6K regulation by a number of additional genes using Western blotting. Nearby cluster 3 are two genes, pontin and reptin, that encode helicases homologous to bacterial RuvB and that have previously been proposed to regulate Drosophila growth through the transcriptional activity of Myc (Bellosta et al. 2005). Western blots confirmed that knockdowns of pont or rept decreased phosphorylation of hS6 and dS6K, while knockdown of dMyc (dm/diminutive) showed no change in these signals (Fig. 7A). This suggests that the RuvB-like helicases control cell growth through a mechanism distinct from MYC in these cells. We also confirmed methionine adenosyltransferase M(2)21AB (Fig. 7B), the WNK kinase CG7177 (Fig. 7C), casein kinases I and II (Fig. 7D), and Pabp2 and the exosome component Rrp6 (Fig. 7E). These results are also summarized in Supplemental Table S2B.
Figure 7.
(A–E) Validation and functional characterization of TORC1–S6K pathway regulators confirmed in the secondary screens. Drosophila S6_S2R+ cells were incubated with dsRNAs targeting the indicated genes, and lysates were analyzed by Western blotting. For results and analysis related to the secondary screens and subsequent follow-up experiments, see also Supplemental Table S2.
Meta-analysis of S6 and S6K regulators
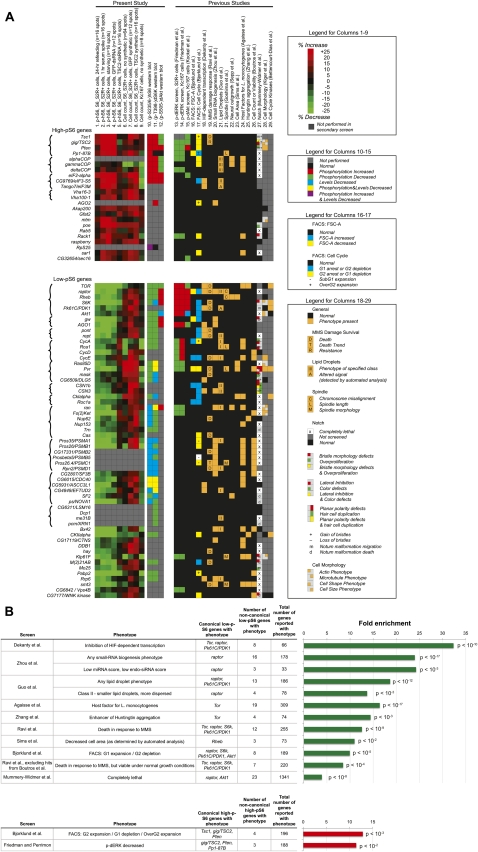
We performed a meta-analysis comparing hits between the present study and several other cell-growth-related Drosophila RNAi screens (Kiger et al. 2003; Bettencourt-Dias et al. 2004; Boutros et al. 2004; Bjorklund et al. 2006; Friedman and Perrimon 2006; Goshima et al. 2007; Sepp et al. 2008; Mummery-Widmer et al. 2009; Ravi et al. 2009; Sims et al. 2009; Dekanty et al. 2010; Kockel et al. 2010). We also examined prior Drosophila genome-scale RNAi screens whose measured phenotypes were not explicitly growth-related but whose hits included one or more canonical TORC1 pathway members (Agaisse et al. 2005; Guo et al. 2008; Zhou et al. 2008; Zhang et al. 2010). The results of this meta-analysis are illustrated in Figure 8, listed in Supplemental Table S2B,D, and discussed below.
Figure 8.
Meta-analysis of present study and prior related RNAi screens. (A) Columns 1–9 summarize the results of our secondary screens (as previously shown in Fig. 4), columns 10–12 indicate the results of our Western blot experiments, and columns 13–29 describe these genes' performance in other screens according to the legends shown. (B) Analysis of other studies' gene groups. Groups of genes are shown if they contained at least one canonical TORC1 pathway gene and are also significantly enriched among the 51 low-pS6 and 19 high-pS6 genes in our screen. The hypergeometric P-value of the fold enrichment is shown; it has been corrected for multiple comparisons (Benjamini and Hochberg 1995).
Discussion
In this study, we identified and confirmed 70 noncanonical regulators of the dTORC1 signaling pathway (Supplemental Table S2B). The network of genes and pathways that control cell growth and proliferation is gradually becoming better characterized, as systematic experiments by ourselves and others have recently probed various complementary aspects of these important biological processes. Here, we discuss how regulators of diverse biological processes fit with the regulators uncovered in our screen into a model for TORC1 pathway signaling.
TORC1 and MAPK pathways
Out of 70 noncanonical p-S6 regulators, 14 were genes found to regulate phospho-dERK in a screen by Friedman and Perrimon (2006). This is not surprising because that study found that canonical TORC1 agonists inhibit p-dERK while canonical TORC1 inhibitors agonize p-dERK (Supplemental Fig. 4C in Friedman and Perrimon 2006); furthermore, it is known that the Ras GTPase agonizes both the PI3K–AKT and Raf–MAPK pathways. A simple model might therefore place a dual p-S6, p-dERK regulator gene upstream of or parallel to Ras85D if the gene's effects on p-dERK and p-S6 are similar (e.g., Pvr), or along the PI3K branch of the cell growth pathway if its effects on p-dERK are opposite from p-S6 (e.g., alphaCop, AGO1).
TORC1 and HIF-dependent transcription
Several lines of evidence (for review, see Wouters and Koritzinsky 2008) suggest that under hypoxic conditions, decreased TORC1 signaling inhibits growth via HIF-1; accordingly, Tor, raptor, and Pk61C/PDK1 all scored in an RNAi screen for inhibitors of the HIF transcriptional response (Dekanty et al. 2010). Beyond these canonical TORC1 pathway members, the 66 hits from the HIF-dependent-transcription screen included eight of our low-pS6 hits (>30-fold enrichment) (Fig. 8); we tested seven of these—AGO1 (Fig. 6A); pont, rept (Fig. 7A); CG6509 (Fig. 5A); CG4849, CG5931 (Fig. 3D); and Rrp6 (Fig. 7E)—by Western blots, and all seven caused reductions in either dS6K phosphorylation or dS6K levels. It is possible that these noncanonical HIF-pathway regulators in the Dekanty et al. (2010) screen exert their effects on HIF-dependent transcription by modulating the TORC1–S6K axis.
TORC1 and lipid metabolism
In another prior RNAi screen (Guo et al. 2008), knockdown of raptor moderately decreased the size of lipid droplets formed after cellular uptake of oleate. Notably, four of our noncanonical low-p-S6 genes (rept, CSN1b, CSN3, me31B) shared this same “Class II” phenotype. In the same study, knockdown of COPI genes, which in the present study increased p-hS6 and p-dS6K (Fig. 3A), increased the size of droplets. Given the various actions of TORC1 in lipogenesis (for review, see Laplante and Sabatini 2009), we suspect that the results of the present p-S6 screen and the Guo et al. (2008) screen reflect a correlation between TORC1's pro-cell-growth signaling and the metabolic production of lipid droplets.
TORC1 and the cell cycle
Canonical TORC1 genes were found to regulate cell cycle progression in an FACS-based RNAi screen (Bjorklund et al. 2006), with knockdown of TORC1 agonists expanding the G1 population and knockdown of TORC2 inhibitors expanding the G2 population. Our noncanonical p-S6 genes were enriched for those respective phenotypes, with four of 19 high-pS6 genes showing G2 expansion (enrichment P < 10−3) and eight of 51 low-pS6 genes showing G1 expansion (P < 10−5). (The low-pS6 genes also showed an enrichment for G2-expansion genes, suggesting that some genes may independently agonize S6 phosphorylation and the G2/M transition.) Of note among the G1-expansion, low-pS6 genes is CkIalpha, which was implicated in a prior kinome-wide RNAi screen (Bettencourt-Dias et al. 2004) as a dTORC1 agonist because its knockdown, like that of Tor, S6k, and Pk61C/PDK1, produced G1 expansion and small cell size. However, our Western blotting clarifies that CkIalpha regulates S6K levels (Fig. 7D), not S6K phosphorylation, and CkIalpha is thus an agonist of the dTORC1 pathway but not dTORC1 itself.
TORC1 signaling and TORC2 signaling
Our list of phospho-S6 regulators does not share many hits with a recent RNAi screen for phospho-AKT regulators (Kockel et al. 2010). That study was designed to identify regulators of TORC2 signaling, and its most prominent hits were known components of the TORC1 pathway, underscoring the classic feedback loop in which TORC1 pathway signaling feeds back to inhibit p-AKT (for review, see Manning 2004). Among noncanonical TORC2 regulators, Kockel et al. (2010) identified zetaCop as a low-p-AKT hit. Our immunoblots for p-dS6K in COPI-RNAi cells (Fig. 3A) suggest that, in this instance, the Kockel screen for TORC2 regulators may have identified a noncanonical TORC1 regulator acting on AKT through feedback inhibition downstream from TORC1. Another gene for which this may have occurred is CG32654/sec16, whose knockdown increased p-S6 in our secondary screen and decreased p-AKT in the Kockel et al. screen.
TORC1 and small RNAs
Several prior genomic RNAi studies of phenotypes that are not explicitly growth-related nonetheless found canonical and noncanonical TORC1 pathway regulators among their hits. Particularly interesting were the results of a screen by Zhou et al. (2008) for genes involved in small RNA biogenesis. Among that screen's 33 hits with low endo-siRNA and low miRNA scores, four were present among our low-pS6 genes, including raptor, gw, CSN3, and CycE; this enrichment was significant at P < 10−3. Also in this group of 33 genes was Drosophila RpS6. The findings by Zhou et al. imply a possible role for the raptor-RpS6 axis in miRNA and endo-siRNA biogenesis. Conversely, our Western blots and cell size assays (Fig. 6) demonstrate a conserved role for the Argonaute–GW complex in regulation of the TORC1 pathway. This relationship is underscored by the results of other growth-related RNAi screens in which AGO1 and raptor (Friedman and Perrimon 2006; Dekanty et al. 2010) or gw and raptor (Bjorklund et al. 2006; primary screen of Friedman and Perrimon 2006) share a phenotype.
It will be interesting to tease apart the relationship between the AGO–GW and TORC1 pathways. Our combination-RNAi experiment suggests that AGO1 and gig/TSC2 act independently in regulating p-T398 (Fig. 6G). In our knockdowns of other miRNA and siRNA-processing genes (Fig. 6H), none produced as strong a p-dS6K phenotype as AGO1, suggesting that dS6K phosphorylation is not merely dependent on the production of miRNAs. These data, together with the enrichment for orthologs of hAGO2 complex I components (Hock et al. 2007) among our primary screen's p-S6 candidate hits (five out of seven genes, P < 3 × 10−7) lead us to suspect that the GW–AGO complex itself may interact with a component of the TORC1–S6K pathway. However, we were unable to immunoprecipitate mTORC1 with a recombinant GFP-GW182, and we found no evidence for colocalization between mTOR and GFP-GW182 in HeLa or HEK293T cells (data not shown). This does not rule out the possibility of interactions between GW182 and other components of the mTORC1 pathway.
TORC1 signaling and development/viability
A recent in vivo RNAi screen for Notch signaling regulators identified genes with diverse developmental phenotypes (Mummery-Widmer et al. 2009). However, it is difficult to correlate these phenotypes with the quantity of TORC1 pathway signaling; for instance, the category of “bristle morphology defects” encompasses genes with conflicting roles in p-S6 regulation, such as S6k and gig/TSC2. Nonetheless, we note that the group of “Completely lethal” genes found by Mummery-Widmer et al. included raptor and Akt1, and that group showed significant enrichment among noncanonical low-pS6 genes (Fig. 8B).
While TORC1 components in vivo are essential for survival (Gangloff et al. 2004; Guertin et al. 2006b), in vitro experiments suggest a complex relationship between TORC1 and cell viability. A screen for regulators of cell survival following DNA damage by methane methylsulfonate (MMS) demonstrated that knockdown of Tor, raptor, S6k, or Pk61C/PDK1 promoted cell death in response to MMS, while knockdown of gig/TSC2 promoted survival (Ravi et al. 2009). However, none of the canonical TORC1 pathway genes scored as hits in a prior Drosophila RNAi screen for regulators of cell count or viability under normal growth conditions (Boutros et al. 2004). In accord with these results, our noncanonical low-pS6 genes are significantly enriched for genes that scored in the Ravi et al. MMS screen but not the Boutros et al. general viability screen (seven out of 220, P < 10−4; Fig. 8B).
Notably, despite the absence of canonical TORC1 regulators from the hits found by Boutros et al. (2004), 15 of 51 noncanonical low-pS6 genes from the present study also scored as viability hits in that screen. These 15 genes have functions (e.g., splicing, proteasome, nuclear transport) that likely affect many cellular processes, but it is possible that their regulation of the TORC1 pathway combines with their other roles to produce a viability phenotype. Consistent with the results of the Ravi et al. MMS-response screen, there is substantial evidence in mammalian cells that mTORC1 regulates cell survival when other cellular processes are disrupted (MacKeigan et al. 2005; Pradelli et al. 2010), and that in such conditions, acute rapamycin treatment can reduce translation of anti-apoptotic proteins (Mills et al. 2008; Bhaskar et al. 2009).
Other screens
We found significant overlap among the low-p-S6 genes found in our screen and a screen for host factors that mediate Listeria monocytogenes infection (Agaisse et al. 2005), as well as a screen for regulators of Huntingtin aggregation (Zhang et al. 2010). However, the only canonical pS6 regulator to score in those screens was Tor itself, making it difficult to infer whether TORC1 has a specific role in these processes.
None of the hits from the present study were found in a prior study of Drosophila S2 cell growth regulators whose transcription was affected by rapamycin treatment (Guertin et al. 2006a). This is not surprising, however, as that study was designed to identify genes transcribed downstream from dTORC1, and the present study identifies genes that act upstream of S6. These two studies are complementary in design, and taken together, they provide a more complete perspective of growth control in Drosophila cells.
Analysis of “frequent hitters”
Qualitatively, some noncanonical p-S6 regulator genes appear to have been hits in prior RNAi screens more often than would be expected. We assessed this quantitatively by interrogating the DRSC database (Flockhart et al. 2006; Ramadan et al. 2007); computations are described in Methods, and results appear in Supplemental Table S2B. We found that our 70 noncanonical p-S6 regulators included eight genes that have scored in ≥30% of DRSC screens, and 25 genes that have scored in ≥20% of DRSC screens. This raised the possibility that such genes might be nonspecific “frequent hitters.” Frequent hitting could theoretically arise by multiple mechanisms such as off-target effects, artifacts related to cell viability, and pleiotropy, among others; these have not been completely characterized, although investigations have begun (Kulkarni et al. 2006). In our own study, we ruled out pervasive off-target effects or artifacts of the primary screen's immunofluorescence-based assay by using new, off-target-free dsRNAs to test seven of the top eight frequent-hitting p-S6 regulators, and 21 of the top 25, in Western blots; every one we tested was confirmed as a bona fide regulator of endogenous dS6K phosphorylation or levels (see Supplemental Table S2B for each gene's hit frequency; see blots in Figs. 3, 5–7). Although investigating the mechanism by which genes gain “frequent hitter” status is beyond the scope of our study, we suspect that TORC1–S6K-pathway components may more frequently score as hits in other screens in part because many prior screens have been related to growth processes. Indeed, given that the hits in prior DRSC screens have often included canonical p-S6 regulators, it is not surprising that our noncanonical p-S6 regulators regulate those same phenotypes. We also suspect that, if decreased cell viability does contribute to “frequent hitter” status, the decreased viability may in some cases be a consequence of altered TORC1–S6K signaling: It is known that perturbations that simultaneously affect the TORC1–S6K pathway and other cellular pathways can decrease cell viability (MacKeigan et al. 2005; Ravi et al. 2009; Pradelli et al. 2010). As an aside, this model connecting TORC1 and viability might account for the moderately decreased cell count among the low-pS6 and high-pS6 candidate hits found in our primary screen (Supplemental Fig. S4, left). We believe that the p-S6 regulators identified in this screen do not represent artifacts of viability phenotypes, for two reasons: (1) In the primary screen, there were many more genes that lacked any pS6 phenotype yet had lower cell counts than the candidate hits themselves (Supplemental Fig. S4, right); and (2) in the course of our experiments we used new, off-target-free dsRNAs and immunoblotting to test 11 of the 15 noncanonical pS6 regulators that were also found in the Boutros et al. (2004) viability screen (genes identified in Fig. 8; blots shown in Figs. 3, 5, 7), and every gene we tested was confirmed as a bona fide regulator of dS6K phosphorylation or levels.
Conclusion
Our success in finding novel TORC1 regulators was based on two important technical advances. One is the Drosophila cell microarray format (Wheeler et al. 2004), which eliminates the two greatest bottlenecks of the conventional, multi-well plate-based screening platform: Image acquisition time is reduced by 75%, and antibody quantity is reduced by 99%. The format of cell microarrays enabled a genome-scale screen in quadruplicate plus an unusually extensive secondary screen, collecting up to 16 replicates under each of several distinct growth conditions. The other major advance is the availability of image analysis software that improved the signal-to-noise ratio in the screen and allowed us to measure multiple phenotypes. The results of this work give us a broad and detailed view of those cellular processes and genes that regulate the TORC1 pathway, laying the foundation for a more thorough understanding of eukaryotic cellular growth in health and disease.
Methods
Drosophila S2R+ cell line expressing human S6 (S6_S2R+)
We obtained a puromycin-resistance fragment by restriction digest of pCoPuro (T. Iwaki, Notre Dame), then cloned this fragment into the pMTvector (Invitrogen), which contains a metallothionine promoter for inducible expression under CuSO4 treatment. Into this pMT-Puro vector we cloned the gene encoding the human S6 ribosomal protein, and we transfected the resulting construct into the Drosophila S2R+ cell line. After puromycin selection, we single-cell-cloned the surviving cells in a 1:1 mixture of conditioned medium to fresh medium. We identified a clone that expressed strongly enough for immunofluorescence studies and that performed RNAi efficiently on cell microarrays.
Genome-scale RNAi library
An RNAi library of 21,167 dsRNAs targeting 13,618 Drosophila genes was constructed by combining two commercially available libraries (dmRNAi, Open Biosystems: 7598 cDNAs that we transcribed in vitro into dsRNAs; and Silencer, Ambion, 13,067 dsRNAs) with 502 dsRNAs designed in our laboratory. The resulting library also contained 145 negative-control spots with dsRNA targeting GFP and 936 negative-control spots without any dsRNA. These samples together constitute the 22,248 samples listed in Supplemental Table S1A. Primer sequences for all dsRNAs are provided in Supplemental Table S1D. Finally, the library included 145 positive-control spots targeting DIAP1, which were used to verify transfection efficacy and assist in spot alignment but were omitted from Supplemental Table S1A for clarity's sake (DIAP knockdown induces apoptosis, which reduces cell count and causes extreme variability in pS6 signal). The whole library of 22,393 samples was stored in 384-well plates (67 total) and was printed as four microarrays (named A, B, C, and D), each containing 40 rows × 140 columns, for 5600 spots per microarray.
Cell microarrays
The printing, seeding, culture, fixation, staining, imaging, and image analysis of Drosophila cell microarrays is detailed in the Supplemental Methods and summarized only briefly here. dsRNAs were printed onto poly-lysine slides using a PixSys 5500 contact arrayer and SMP7 stealth microarray pins. One slide in each printed batch of 40 was sacrificed in order to identify dsRNA spot locations: it was imaged with an Axon slide-scanner and stained with SYBR Green I nucleic acid stain, and the spot locations were identified using two image analysis pipelines (Supplemental Pipelines 1 and 2) in CellProfiler software (Carpenter et al. 2006; Lamprecht et al. 2007; http://www.cellprofiler.org). These spot locations were fed to an Axiovert 200M microscope for high-magnification imaging of each spot using a 40× objective, such that each imaged field of view is just inside a dsRNA spot. We used spots with a strong cell-death phenotype (dsRNA against DIAP1) to align the arrays and estimate the transfection efficiency based on reduced cell count, >90% in Kc167 cells and >75% in S6_S2R+ cells. Images of spots were processed using CellProfiler software to identify nuclei based on DNA staining and cell borders based on actin staining. Phospho-S6 immunofluorescence was measured as the mean cytoplasmic fluorescence intensity of pS6, averaged across all cells in the imaged population. The metric is inherently normalized to cytoplasmic area and cell count and was subsequently normalized to correct for illumination variations across each cell array.
Computation of gene-family enrichments among primary screen candidate hits
We annotated each gene in the library with its associated Gene Ontology terms (as well as full name, homologs, and conservation information, as listed in screen results in Supplemental Tables) using the Entrez IEB Toolbox, which, in turn, drew its information from FlyBase 5.1. We then computed the fold enrichment and hypergeometric probability, with multiple-hypothesis correction (Benjamini and Hochberg 1995), for each GO term associated with those genes identified as candidate hits in the primary screen.
dsRNA design and primer selection for secondary screen and follow-up experiments
Supplemental Table S2C contains the primer sequences of all dsRNAs that we designed in the course of following up the genome screen. Primers were selected to target all transcripts of a gene as determined by alignment in ClustalX (http://www.clustal.org). We avoided regions of 19+mer sequence identity to other Drosophila transcripts (Kulkarni et al. 2006), using the appropriate DRSC tool (http://flyrnai.org/RNAi_find_frag_free.html; Flockhart et al. 2006). For most genes, we were able to design multiple dsRNAs targeting nonoverlapping transcript regions. dsRNAs were prepared using the OneStep RT-PCR kit (Promega) and the Megascript T7 in-vitro-transcription kit (Ambion). The library for the secondary screen contained 600 samples, of which 300 were GFP-dsRNA controls, and this library was printed four times per microarray slide, producing microarrays of 2400 spots (30 rows × 80 columns).
Hierarchical clustering of secondary screen data
We used Cluster 3.0 (Eisen Lab, Stanford University) to analyze possible functional relationships between those genes we identified in the secondary screens as on-target pS6 regulators. For hits targeted by two dsRNAs, we pooled results from both dsRNAs when computing the hit's average pS6 value and average cell count under each condition. For those hits targeted at only four spots per array, we assigned a weight of 0.5; for those hits and controls targeted at eight spots per array, we assigned weights of 1; for those controls targeted at 16 spots per array, we assigned weights of 2. These values were subjected to hierarchical clustering under the complete-linkage algorithm, using 1-Pearson correlation as the distance metric.
Cell culture and Western blotting for non-array-based experiments
Cells were grown in 12-well dishes from initial densities of 0.5 million to 1 million cells per well. S2R+ cells were grown in Schneider's medium supplemented by 10% IFS, while S2 cells were grown in Express Five serum-free medium (Invitrogen). For single-knockdown experiments, cells were transfected with 1 μg of dsRNA per well immediately following seeding and retransfected 24 h later with another 1 μg of dsRNA, except in the COPI-knockdown experiment when cells only received one dose of dsRNA. For the combination-knockdown experiment, cells were transfected with the first dsRNA at 1 μg per well immediately upon seeding, and after 48 h, medium was refreshed and cells were transfected with 1.5 μg of the second dsRNA and 0.5 μg of the first dsRNA. For experiments using the S6-reporter cell line (S6_S2R+), induction by addition of CuSO4 to 0.5 μg/μL occurred 24 h prior to lysis, except for the COPI-knockdown experiment when cells were induced upon seeding. Lysis and Western blotting were performed as previously described (Sancak et al. 2007). Primary antibodies were rabbit anti-phospho-S6 (Cell Signaling, #2211), mouse anti-total S6 (Cell Signaling, #2317), rabbit anti-phospho-T398-dS6K (Cell Signaling, #9209), rabbit anti-total-dS6K (gift of Mary Stewart, North Dakota State University), rabbit anti-phospho-S505-dAKT (Cell Signaling, #4054), rabbit anti-total-AKT (Cell Signaling, #9272), and rabbit anti-phospho-dERK (Promega, #V8031). Secondary antibodies were donkey anti-mouse HRP and donkey anti-rabbit HRP (Santa Cruz Biotechnology).
Human cell experiments
Lentiviral shRNA constructs for knockdown in human cells were obtained from the RNAi Consortium through Sigma-Aldrich (Moffat et al. 2006). TNRC6A #11 shRNA: TRCN0000127597, NM_014494.1-2478s1c1; TNRC6A #12 shRNA: TRCN0000129176, NM_014494.1-2622s1c1; TNRC6A #13 shRNA: TRCN0000147244, NM_014494.1-4312s1c1; TNRC6A #14 shRNA: TRCN0000147932, NM_014494.1-848s1c1; TNRC6A #15 shRNA: TRCN0000150256, NM_014494.1-685s1c1; EIF2C2 #6 shRNA: TRCN0000007864, NM_012154.2-1163s1c1; EIF2C2 #7 shRNA: TRCN0000007865, NM_012154.2-1219s1c1; EIF2C2 #8 shRNA: TRCN0000007866, NM_012154.2-150s1c1; EIF2C2 #9 shRNA: TRCN0000007867, NM_012154.2-719s1c1; EIF2C2 #10 shRNA: TRCN0000011203, NM_012154.2-2776s1c1; TNRC6C #5 shRNA: TRCN0000004494, NM_018996.x-5229s1c1.
shRNAs to human raptor and GFP were previously described (Sarbassov et al. 2005). shRNA-encoding plasmids were cotransfected with the Delta VPR envelope and CMV VSV-G packaging plasmids into actively growing HEK293T cells using FuGENE 6 transfection reagent (Ali and Sabatini 2005; Sarbassov et al. 2005). Virus-containing supernatants were collected 48 h after transfection and filtered, and HeLa cells were infected in the presence of 8 μg/mL polybrene. Twenty-four hours later, cells were selected with puromycin and analyzed beginning at the third day after infection and at subsequent time points. To prepare lysates, mammalian cells were rinsed once with ice-cold PBS and lysed in ice-cold lysis buffer (40 mM HEPES at pH 7.4, 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, and 0.3% CHAPS or 1% Triton X-100, and one tablet of EDTA-free protease inhibitors [Roche] per 25 mL). The soluble fractions of cell lysates were isolated by centrifugation at 13,000 rpm for 10 min by centrifugation in a microfuge. Proteins were denatured by the addition of sample buffer and boiling for 5 min, resolved by 8% SDS-PAGE, and analyzed by immunoblotting (Kim et al. 2002). For immunoprecipitations for kinase assays, antibody to human mTOR was added to lysates and incubated with rotation for 1.5 h at 4°C. Forty microliters of a 50% slurry of protein G-Sepharose was then added, and the incubation continued for an additional 1 h. Immunoprecipitates were washed three times with lysis buffer containing 150 mM NaCl. Kinase assays were performed as previously described (Sancak et al. 2007). Proteins were denatured by the addition of sample buffer and boiling for 5 min, resolved by 8% SDS-PAGE, and analyzed by immunoblotting (Kim et al. 2002). For Flag purifications, Flag M2 affinity resin was washed with lysis buffer three times. Twenty microliters of a 50% slurry of the resin was then added to pre-cleared cell lysates and incubated with rotation for 2 h at 4°C. The beads were washed three times with lysis buffer containing 150 mM NaCl. For elution of Flag-tagged proteins, beads were incubated in elution buffer (50 mM HEPES at pH 7.4, 500 mM NaCl, 0.5% CHAPS, 50 μg/μL l× Flag peptide) for 30 min at room temperature. The AGO2-GFP expression plasmid was a gift from P. Sharp (Leung et al. 2006), and the GW182-GFP expressing plasmid was a gift from D. Bloch (Bloch et al. 2006). pLJM1-GFP and the FLAG-S6K1 construct were described previously (Sancak et al. 2008). For cotransfection experiments, 200K HEK293T cells were plated in 6-well dishes. Twenty-four hours later, cells were transfected with DNA, as described below, and examined 2 or 3 d after transfection by Western blotting. To determine which shRNAs to human GW182 were most effective, 100 ng of GW182-GFP plasmid was transfected together with 200 ng of shRNA plasmid. To probe effects on phospho-T389 S6K1, increasing amounts (0, 50, 100, 200, 400 ng) of AGO2-GFP, GW182-GFP, and pLJM1-GFP plasmids were cotransfected with 2 ng of FLAG-S6K1. Transfection mixes were taken up to a total of 1 μg of DNA using empty pRK5. Lysates were examined by Western blotting, and phospho-T389 S6K1 status was determined by first immunoprecipitating S6K1 via the Flag epitope as described above, eluting off the Flag beads, and probing phospho-T389 by immunoblotting. For probing mammalian lysates, antibodies to human mTOR, Raptor, S6K1, and phospho-T389 S6K1 were obtained from Cell Signaling Technology; antibody to GFP from Abcam; Flag M2 antibody from Sigma-Aldrich. The 7C6 antibody to human AGO2 was a gift from T. Hobman (University of Calgary, Canada). Flag M2 affinity gel and ATP were obtained from Sigma-Aldrich, and protein G-Sepharose and immobilized glutathione from Pierce.
Meta-analysis of growth-related and TORC1-related Drosophila RNAi screens
We included in our meta-analysis previously published genome-scale Drosophila RNAi screens for phenotypes explicitly related to cell growth, as well as prior Drosophila RNAi screens that identified at least one canonical TORC1 regulator. The hits named in these screens are found in Supplemental Table S2D, along with the references to the tables that listed those genes. For screens that identified multiple classes of hits, e.g., the lipid droplets screen by Guo et al. (2008), we considered those multiple subclasses of hits as defined by the authors. For certain screens that did not clearly define subclasses of their hits, e.g., the p-dERK screen by Friedman and Perrimon (2006), we defined subclasses ad hoc according to criteria described in Supplemental Table S2D. Fold-enrichment and hypergeometric distributions were determined by comparing the 51 noncanonical low-pS6 genes, and 19 noncanonical high-pS6 genes, to the gene groups listed in Supplemental Table S2D. We used the Benjamini-Hochberg method to perform multiple hypothesis correction on the hypergeometric distributions (Benjamini and Hochberg 1995). For “hit frequency” analysis, we included all screens listed in the DRSC public database, including published and unpublished screens. If any DRSC amplicon targeting a given gene scored as a hit in a given screen, we assigned the gene a score of 1 in that screen; we then averaged the results for that gene over all the DRSC screens in which that gene was tested, producing the “hit frequency,” i.e., the proportion of DRSC screens identifying a given gene as a hit. Hit frequencies for each of the 70 noncanonical p-S6 regulators are listed in Supplemental Table S2B.
Acknowledgments
This work was funded in part by grants from the W.M. Keck foundation (D.M.S.), the National Institutes of Health (R01 GM072555 and CA103866 to D.M.S. and R01 GM089652 to A.E.C.), the DOD TSC Research Program (W81XWH-05-1-0318-DS to D.M.S.), the LAM Foundation (D.A.G.), the Society for Biomolecular Sciences (A.E.C.), the Whitehead Institute, and the Broad Institute. We thank Mary Stewart (Drosophila S6 Kinase antibody), Nadire Ramadan (S2R+ cells), Calvin Jan (S2 cells, adapted for serum-free conditions), Donald Bloch (GFP-GW182 construct), Philip Sharp (GFP-AGO2 construct), Tom Hobman (7C6 hAGO2 antibody), and T. Iwaki (pCoPuro) for their generous gifts of reagents. We thank David Bartel, Graham Ruby, Calvin Jan, Jason Moffat, Timothy Peterson, Yasemin Sancak, and Marvin Fritzler for helpful discussions and Peggy Anthony for helpful comments on the meta-analysis.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111492.110.
References
- Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309: 1248–1251 [DOI] [PubMed] [Google Scholar]
- Ali SM, Sabatini DM 2005. Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem 280: 19445–19448 [DOI] [PubMed] [Google Scholar]
- Bellosta P, Hulf T, Balla Diop S, Usseglio F, Pradel J, Aragnol D, Gallant P 2005. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc Natl Acad Sci 102: 11799–11804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. 2004. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432: 980–987 [DOI] [PubMed] [Google Scholar]
- Bhaskar PT, Nogueira V, Patra KC, Jeon SM, Park Y, Robey RB, Hay N 2009. mTORC1 hyperactivity inhibits serum deprivation-induced apoptosis via increased hexokinase II and GLUT1 expression, sustained Mcl-1 expression, and glycogen synthase kinase 3beta inhibition. Mol Cell Biol 29: 5136–5147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature 439: 1009–1013 [DOI] [PubMed] [Google Scholar]
- Bloch DB, Gulick T, Bloch KD, Yang WH 2006. Processing body autoantibodies reconsidered. RNA 12: 707–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Consortium HF, Paro R, Perrimon N 2004. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303: 832–835 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100 doi: 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombetti S, Basso V, Mueller DL, Mondino A 2006. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J Immunol 176: 2730–2738 [DOI] [PubMed] [Google Scholar]
- Dekanty A, Romero NM, Bertolin AP, Thomas MG, Leishman CC, Perez-Perri JI, Boccaccio GL, Wappner P 2010. Drosophila genome-wide RNAi screen identifies multiple regulators of HIF-dependent transcription in hypoxia. PLoS Genet 6: e1000994 doi: 10.1371/journal.pgen.1000994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E 2007. P bodies: At the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E 2008. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 15: 346–353 [DOI] [PubMed] [Google Scholar]
- Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, Richardson K, Xu A, Perrimon N, Mathey-Prevot B 2006. FlyRNAi: The Drosophila RNAi screening center database. Nucleic Acids Res 34: D489–D494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD 2007. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell 130: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A, Perrimon N 2006. High-throughput approaches to dissecting MAPK signaling pathways. Methods 40: 262–271 [DOI] [PubMed] [Google Scholar]
- Gangloff YG, Mueller M, Dann SG, Svoboda P, Sticker M, Spetz JF, Um SH, Brown EJ, Cereghini S, Thomas G, et al. 2004. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol Cell Biol 24: 9508–9516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316: 417–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Guntur KV, Bell GW, Thoreen CC, Sabatini DM 2006a. Functional genomics identifies TOR-regulated genes that control growth and division. Curr Biol 16: 958–970 [DOI] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM 2006b. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 11: 859–871 [DOI] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV 2008. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453: 657–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans 37: 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Guan KL 2009. Tuberous sclerosis complex, implication from a rare genetic disease to common cancer treatment. Hum Mol Genet 18: R94–R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger A, Baum B, Jones S, Jones M, Coulson A, Echeverri C, Perrimon N 2003. A functional genomic analysis of cell morphology using RNA interference. J Biol 2: 27 doi: 10.1186/1475-4924-2-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175 [DOI] [PubMed] [Google Scholar]
- Kockel L, Kerr KS, Melnick M, Bruckner K, Hebrok M, Perrimon N 2010. Dynamic switch of negative feedback regulation in Drosophila Akt-TOR signaling. PLoS Genet 6: e1000990 doi: 10.1371/journal.pgen.1000990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B 2006. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods 3: 833–838 [DOI] [PubMed] [Google Scholar]
- Lamprecht MR, Sabatini DM, Carpenter AE 2007. CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques 42: 71–75 [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM 2009. mTOR signaling at a glance. J Cell Sci 122: 3589–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci 103: 18125–18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA 2006. Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363 [DOI] [PubMed] [Google Scholar]
- MacKeigan JP, Murphy LO, Blenis J 2005. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol 7: 591–600 [DOI] [PubMed] [Google Scholar]
- Manning BD 2004. Balancing Akt with S6K: Implications for both metabolic diseases and tumorigenesis. J Cell Biol 167: 399–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills JR, Hippo Y, Robert F, Chen SM, Malina A, Lin CJ, Trojahn U, Wendel HG, Charest A, Bronson RT, et al. 2008. mTORC1 promotes survival through translational control of Mcl-1. Proc Natl Acad Sci 105: 10853–10858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124: 1283–1298 [DOI] [PubMed] [Google Scholar]
- Mummery-Widmer JL, Yamazaki M, Stoeger T, Novatchkova M, Bhalerao S, Chen D, Dietzl G, Dickson BJ, Knoblich JA 2009. Genome-wide analysis of Notch signalling in Drosophila by transgenic RNAi. Nature 458: 987–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Ichisaka T, Maeda M, Oshiro N, Hara K, Edenhofer F, Kiyama H, Yonezawa K, Yamanaka S 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol Cell Biol 24: 6710–6718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev 14: 2689–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plas DR, Thomas G 2009. Tubers and tumors: rapamycin therapy for benign and malignant tumors. Curr Opin Cell Biol 21: 230–236 [DOI] [PubMed] [Google Scholar]
- Pradelli LA, Beneteau M, Chauvin C, Jacquin MA, Marchetti S, Munoz-Pinedo C, Auberger P, Pende M, Ricci JE 2010. Glycolysis inhibition sensitizes tumor cells to death receptors-induced apoptosis by AMP kinase activation leading to Mcl-1 block in translation. Oncogene 29: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Proud CG 2009. mTORC1 signalling and mRNA translation. Biochem Soc Trans 37: 227–231 [DOI] [PubMed] [Google Scholar]
- Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B 2007. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc 2: 2245–2264 [DOI] [PubMed] [Google Scholar]
- Ravi D, Wiles AM, Bhavani S, Ruan J, Leder P, Bishop AJ 2009. A network of conserved damage survival pathways revealed by a genomic RNAi screen. PLoS Genet 5: e1000527 doi: 10.1371/journal.pgen.1000527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DM 2006. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer 6: 729–734 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell 25: 903–915 [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM 2005. Phosphorylation and regulation of Akt/PKB by the rictor–mTOR complex. Science 307: 1098–1101 [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Hong P, Lizarraga SB, Liu JS, Mejia LA, Walsh CA, Perrimon N 2008. Identification of neural outgrowth genes using genome-wide RNAi. PLoS Genet 4: e1000111 doi: 10.1371/journal.pgen.1000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Duchek P, Baum B 2009. PDGF/VEGF signaling controls cell size in Drosophila. Genome Biol 10: R20 doi: 10.1186/gb-2009-10-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulard A, Cohen A, Hall MN 2009. TOR signaling in invertebrates. Curr Opin Cell Biol 21: 825–836 [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alvarez VA, Ridenour DA, Kwiatkowski DJ, Sabatini BL 2005. Regulation of neuronal morphology and function by the tumor suppressors Tsc1 and Tsc2. Nat Neurosci 8: 1727–1734 [DOI] [PubMed] [Google Scholar]
- Wheeler DB, Bailey SN, Guertin DA, Carpenter AE, Higgins CO, Sabatini DM 2004. RNAi living-cell microarrays for loss-of-function screens in Drosophila melanogaster cells. Nat Methods 1: 127–132 [DOI] [PubMed] [Google Scholar]
- Wouters BG, Koritzinsky M 2008. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Natl Rev 8: 851–864 [DOI] [PubMed] [Google Scholar]
- Zhang S, Binari R, Zhou R, Perrimon N 2010. A genome-wide RNAi screen for modifiers of aggregates formation by mutant huntingtin in Drosophila. Genetics 184: 1165–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ 2008. Comparative analysis of Argonaute-dependent small RNA pathways in Drosophila. Mol Cell 32: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]