Abstract
Regulation of gene transcription in diverse cell types is determined largely by varied sets of cis-elements where transcription factors bind. Here we demonstrate that data from a single high-throughput DNase I hypersensitivity assay can delineate hundreds of thousands of base-pair resolution in vivo footprints in human cells that precisely mark individual transcription factor–DNA interactions. These annotations provide a unique resource for the investigation of cis-regulatory elements. We find that footprints for specific transcription factors correlate with ChIP-seq enrichment and can accurately identify functional versus nonfunctional transcription factor motifs. We also find that footprints reveal a unique evolutionary conservation pattern that differentiates functional footprinted bases from surrounding DNA. Finally, detailed analysis of CTCF footprints suggests multiple modes of binding and a novel DNA binding motif upstream of the primary binding site.
Demarcation of transcription factor binding is key to the understanding of gene expression and whole regulatory networks within a cell. This is a particularly daunting task since it is estimated that there are approximately 1500 transcription factors in the human genome (Vaquerizas et al. 2009). A number of methods have been developed to identify the location of transcription factor binding, such as chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq), position weight matrices (PWMs), electrophoretic mobility shift assays (EMSAs), and footprinting using DNase I or dimethylsulfate.
While these methods are extremely powerful and complementary, each method has limitations. For example, ChIP-seq requires a large number of cells and a high-quality antibody (or epitope tagged version) and is unable to resolve DNA–protein interactions at base-pair resolution. PWMs model DNA binding site sequence preferences, commonly referred to as “motifs,” for different transcription factors. Since most transcription factor motifs are four to eight bases in length, these annotations often include large numbers of predicted sites with low specificity. In addition, PWMs are only available for a fraction of factors. EMSAs test whether any fragment of DNA can bind to nuclear extracts or purified single proteins. However, this in vitro assay may not be accurate if multiple factors or DNA segments are required for binding. Traditional footprinting assays accurately identify the precise binding sites of any factor. However, this low-throughput method is highly technical and can only analyze a single small region (<1 kb) at a time. Together, this indicates that additional methods are clearly needed to better understand global gene regulation.
Mapping DNase I hypersensitive (HS) sites across the genome using a deep sequencing approach (DNase-seq) identifies a broad variety of active cis-regulatory elements (Wu 1980; Gross and Garrard 1988; Boyle et al. 2008a). DNase-seq identifies sites of DNase I digestion at single base resolution, even though these data are typically smoothed to identify larger DNase I HS sites (∼200 bp). Previously it was shown that similarly derived DNase I digestion data could identify individual binding sites in Saccharomyces cerevisiae based on the protection of short stretches of nucleotides with the larger HS sites (Hesselberth et al. 2009). Likewise, in humans we observed that the raw distribution of sequence tag locations within each HS site is not uniform reflecting in vivo protection of DNA by individually bound proteins, similar to traditional DNase I footprinting assays.
Here, we describe DNase I footprints identified from DNase-seq data generated from seven similar (lymphoblastoid cell lines) (McDaniell et al. 2010) and five diverse (K562, HeLaS3, HUVEC, NHEK, and embryonic stem cell) human cell types (available at http://www.genome.duke.edu/labs/furey/datasets/). We show that DNase I footprints are reproducible, robust, and accurate at identifying and annotating hundreds of thousands of putative protein binding sites genome-wide. Footprinting data alone cannot annotate every site for every known and unknown factor, but they are an important complement to ChIP-seq and conservation data that provides valuable protein/DNA interaction information. Together, these enable an even more comprehensive accounting and characterization of active cis-elements with base-pair resolution.
Results
Transcription factor binding sites are depleted for DNase I cleavage sites
DNase-seq data was generated and uniformly processed from multiple independent replicates as part of the human ENCODE project (Supplemental Table 1A; The ENCODE Project Consortium 2004). To assess the ability of DNase-seq to identify footprints, we first investigated general digestion patterns around published motifs for transcription factors with known PWMs (Matys et al. 2006; Kim et al. 2007; Bryne et al. 2008; Newburger and Bulyk 2009). We determined all positions in the genome matching these motifs, referred to from here on as motif predicted binding sites. We then calculated the number of DNase I cuts at each base pair within and surrounding all motif predicted binding sites for a particular factor across the genome. At this cumulative level, we clearly detected a footprint characterized by a lack of DNase I digestion within these sites for many individual transcription factors (for CTCF, see Fig. 1A; for multiple other factors, see Supplemental Fig. 1A). These footprints were detected at the cumulative level even though presumably many of the motif predicted binding sites were not bound by transcription factors. This aggregate detection of footprints was only seen for factors with high information content motifs whose predictions are significantly enriched for functional sites in the genome (Supplemental Fig. 1A). Approximately 30% of motif predicted CTCF binding sites corresponded with a footprint signal. Factors with low information content motifs (shorter and/or less complex) generate many false positives that mask the cumulative footprinting signal. For example, less than an estimated 0.1% of over 40 million short (four bases), information-poor GATA1 motif predicted binding sites showed evidence of footprinting (data not shown).
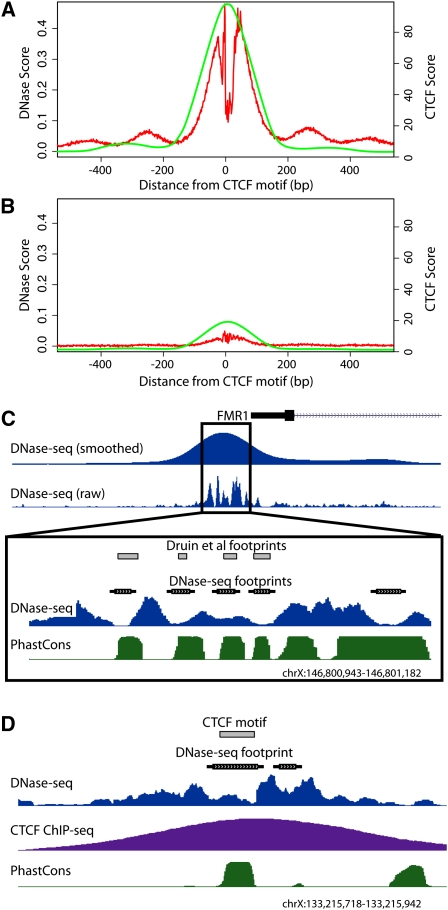
Figure 1.
DNase-seq identifies protein–DNA footprints. All potential CTCF binding sites were identified genome-wide using motif matching and compiled such that their 5′ end was set at position zero. Cumulative DNase-seq and CTCF ChIP-seq signals within 500 bp of each site in both directions were determined. (A) CTCF motifs that have a DNase I footprint (red) also display high CTCF ChIP-seq signal (green). (B) CTCF motifs that have no footprint have greatly reduced CTCF ChIP-seq signal. (C) Footprinting using DNase-seq accurately identified footprints within the FMR1 promoter region previously mapped using traditional in vitro DMS footprinting. Dips in raw DNase-seq signal and annotated footprints correspond perfectly with previously identified footprints (gray boxes) (Drouin et al. 1997). The phastCons annotation shows increased levels of evolutionary conservation within called footprints. (D) A representative individual region displaying a DNase I footprint matching a known CTCF binding motif (gray box) with a strong corresponding CTCF ChIP-seq signal. See also Supplemental Figure 1A.
To determine whether functional binding sites could be determined based solely on DNase-seq data, we used k-means clustering to divide up motif predicted CTCF binding sites that overlap or do not overlap a footprint (see Methods). We compared these sets to CTCF ChIP-seq data collected from the same cell growth. Motif predicted CTCF binding sites with a footprint signal were highly enriched for ChIP-seq signal (Fig. 1A), whereas those without footprint evidence displayed almost no ChIP-seq signal (Fig. 1B).
ChIP-seq signal was significantly stronger in regions that overlapped footprints compared with ChIP-seq peaks without footprints (P < 2.2 × 10−16; Supplemental Fig. 1B). Similarly, motif predicted sites that overlapped footprints had higher PWM scores than those without footprints (P < 2.2 × 10−16; Supplemental Fig. 1C). It is important to note that the strength of the PWM score only partially predicts in vivo binding by ChIP-seq (Supplemental Fig 1D) compared with using footprint data as a guide (Fig. 1A,B). General correspondence of footprints, ChIP-seq signal, and PWM strength indicates that all three data describe biologically relevant and important characteristics of transcription factor binding, which is likely related to increased protein binding affinity and/or increased occupancy throughout the cell population.
Identification of individual footprints
While cumulative plots provide summary validation for DNase I footprinting, we also developed a five-state hidden Markov model (HMM) (see Methods) in order to identify individual footprints throughout the genome (Supplemental Fig. 1E). This HMM identified small regions within DNase I HS sites where there was reduced DNase I digestion (footprints) compared with adjacent bases (see Methods). Footprints were identified in individual cell types as well as in pooled lymphoblastoid data. In general, we found that signals in all lymphoblastoid cell lines were extremely similar (Supplemental Fig. 2). We were conservative in our delineation of footprints to reduce the number of false positives. The number of footprints per cell type ranged from 100,000–325,000. Variation in the number of footprints identified appears to be primarily due to differences in the number and average size of DNase I HS sites annotated in each cell line (Supplemental Table 1).
We identified the putative factors bound to each footprint using STAMP (Mahony and Benos 2007) in conjunction with motifs that are publicly available in the JASPAR (Bryne et al. 2008), TRANSFAC (Matys et al. 2006), and UniPROBE (Newburger and Bulyk 2009) PWM databases. In total, there were 476 PWMs representing 398 distinct factors, some of which represent the binding of multi-protein complexes. We required motif matches with P-values < 10−6 and allowed footprints to be labeled with multiple factors if each separate motif matched with scores below this threshold. Using this strict criterion, only 21%–26% of predicted footprints were annotated by a currently known motif (Supplemental Table 1). Our strict motif criteria and, more generally, the incomplete knowledge of sequence preferences for all DNA binding proteins likely contributed to this low rate of annotation. Using more lenient motif match criteria would increase the number of annotated sites but would also increase the rate of incorrect annotations. We note that these annotations represent candidate binding factors. Those factors with large, information-rich motifs, like CTCF and REST, are more likely to be correctly labeled. In many cases, multiple distinct factors have very similar binding site motifs (Mahony et al. 2007). Thus, annotations of factors with smaller, information-poor motifs or whose motifs are similar to those of other factors may be less accurate.
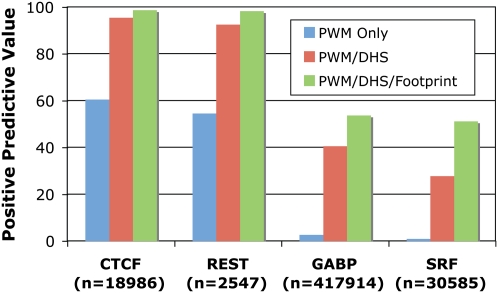
To demonstrate the accuracy of our model, we show that predicted footprints from DNase-seq data pooled from seven distinct lymphoblastoid cell lines perfectly match previously identified NRF1, SP1, AP-2, and MYC footprints near the FMR1 (fragile X mental retardation 1) promoter (Fig. 1C; Drouin et al. 1997). We also found that CTCF footprints corresponded extremely well to individual CTCF binding sites detected both by ChIP-seq as well as by motif prediction (Fig. 1D). To more globally determine the accuracy of our model, we used ChIP-seq data for CTCF, REST, GABP, and SRF and determined the positive predictive value (PPV) of motifs that were (1) present across the entire genome, (2) found within a DNase I HS site, or (3) found within a footprint (Fig. 2; Supplemental Table 2). The motifs with a corresponding ChIP-seq peak were considered functional (true positives), while the motifs with no ChIP evidence were considered not functional (true negatives). Predicted CTCF and REST footprints had a PPV of >98%, while predicted GABP and SRF footprints had a PPV of >50%. The reduced PPV for GABP and SRF footprints may be due to DNase I and ChIP data originating from nonmatched cell types (Valouev et al. 2008) or may be due to these factors having binding motifs with lower information content. However, we note that for GABP and SRF, the footprint PPV significantly outperforms the PPV using a purely sequence-based motif approach by 20- to 50-fold (Fig. 2). Using stricter PWM criteria to identify positives and negatives does not significantly affect the PPV for CTCF and REST footprints, but it does increase the PPV for GABP and SRF footprints to ∼80% (Supplemental Fig. 3). Footprints also are much more accurate at identifying ChIP-seq peaks compared with simply using motifs that are present in a DNase I HS site (Fig. 2; Supplemental Fig. 3). Sensitivity and specificity measurements show similar results (Supplemental Table 2). These observations indicate that DNase-seq footprinting accurately identifies active transcription factor binding sites.
Figure 2.
Accuracy of footprinting model. Positive predictive value (PPV) was calculated for predictions of four factors: CTCF, REST, GABP, and SRF. True-positives were determined by ChIP-seq peaks with a matching motif, while true-negatives were determined by motifs without corresponding ChIP-seq peaks. PPV is shown for predictions using only PWMs (all PWMs are considered an actual binding site), PWMs that map within DHS sites (all PWMs within a DNase I hypersensitive site are considered actual binding sites, while those PWMs outside of DNase I hypersensitive sites are considered negatives), and PWMs that map within footprints (all PWMs within a footprint are considered actual binding sites while those PWMs outside of footprints are considered negatives). The total number of PWMs mapped to the genome for each factor is listed in parentheses.
The reproducibility of our DNase I footprinting method is evident by the footprint annotations across two lymphoblastoid lines being much more correlated than between cell lines of different lineages (Supplemental Fig. 4A). In fact, these two lines showed higher correlation based on footprint annotations than based on gene expression levels (Supplemental Fig. 4B). Additionally, DNase I footprint annotations and CTCF ChIP-seq data were also generated for K562, HelaS3, NHEK, HUVEC, and embryonic stem cells. Even though there were less DNase I sequences generated than for the combined lymphoblastoid data (Supplemental Table 1), we found that the accuracy of predicted CTCF footprints remained high in all cell lines, as evidenced by strong PPV rates (94%-99%) for CTCF ChIP-seq signals (Supplemental Table 3).
As mentioned previously, the number of footprints identified in each cell type is dependent on the DNase I HS site annotation. Higher numbers of DNase I sequences improves the accuracy of the footprint annotation within these HS sites. For example, the PPV for CTCF is higher and more footprints are annotated with factors by STAMP in the combined lymphoblastoid data compared with individual lymphoblastoid cell lines (Supplemental Tables 3, 1, respectively).
Preferences in footprint locations relative to genes and each other
Many transcription factors have been more closely associated with binding proximal promoter regions, while others have shown a preference for distal regions. Using the distribution of footprints from 89 factors that are annotated in at least 100 distinct locations in the combined lymphoblastoid data, we determined the expected number and enrichment/depletion of footprinted sites for a single factor in promoter or nonpromoter categories (Supplemental Table 4). Not surprisingly, several well-known factors, including SP1, AP-2, USF, and GABPA, were enriched in promoter regions. These have been previously associated with basal promoter activity in a large variety of genes with diverse functions. In contrast, other factors are depleted in promoters and enriched in nonpromoter regions, including FOS, STAT1, IRF1, IRF2, HSF, and CTCF. Many of these latter factors are involved in more specialized cellular functions, in this case in lymphoblastoid cells, and are often activated in response to an external stimuli.
Since many factors bind within complexes or work co-operatively, we asked if any two factors were preferentially bound within the same DNase I HS site more often than chance. When analyzing each pair of factors, individual footprints labeled with both factors were discarded to correct for factors with very similar motifs. We found 35 combinations of factors that significantly colocalize in lymphoblastoid cells (Supplemental Table 5). Colocalization graphs depicting these relationships show that the factors divide neatly into two clusters. The first, larger cluster consists of 12 factors, including SP1, AP-2, MYCN, and GABP (Supplemental Fig. 5A). Approximately 87% of co-occurrences for these factors map within promoter regions in lymphoblastoid cells. In addition, on average in each of the other five diverse cell types, over 97% of the DNase I HS sites containing these co-occurrences were also annotated. In contrast, the second, smaller cluster consists of six factors known to regulate genes in response to external stimuli (Supplemental Fig. 5B). Interestingly, 87% of the second cluster instances are found in nonpromoter regulatory regions. Only 33% of the DNase I HS sites containing these colocalized footprints were similarly identified on average in nonlymphoblastoid cell types. Therefore, very distinct combinations of factors appear to colocalize in ubiquitously open promoters in contrast to cell type–specific nonpromoter regions.
We repeated these analyses on each of the five other cells lines. Interestingly, we found a core set of nine combinations involving seven factors (SP1, AP-2, MYCN, PAX4, USF, ARNT, RREB1) that significantly colocalized in essentially all cell types (Supplemental Table 6; Supplemental Fig. 5A, bold lines). Another 22 pairs of factors were commonly found in two or more cell lines, and several co-occurrences were limited to a single cell-type (Supplemental Table 6). As mentioned previously, greater sequencing depth appears to influence the ability to identify footprints and the accuracy of footprint annotations. Besides the combined lymphoblastoid data, only the H1 embryonic stem cells were sequenced to a depth of at least 100 million sequences. Thus, the combined lymphoblastoid and H1 lines had far greater numbers of significantly co-occurring factors. In the H1 data, we also see colocalized factors for which footprints for both are primarily found (>89%) in nonpromoter distal regions (Supplemental Table 6; Supplemental Fig. 5C). Interestingly, these primarily consist of NFKB or its subunits (REL family, specifically c-REL) in combination with another factor. Presumably, deeper sequencing in other cell lines would likewise reveal more cell type–specific combinations in this and other cell types.
Cell type–specific footprinting patterns
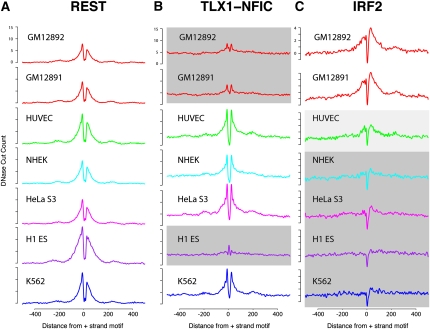
By mapping footprints across various cell lines, we used cumulative plots similar to Figure 1 to detect factors similarly utilized in all cell types as well as those that differed in a cell type–specific manner. Factors used in all cell types displayed consistent footprinting signals in all cell types, while cell type–specific footprints showed a diminished or distinct lack of a footprint signal in one or more cell types. For example, REST showed a footprint pattern in all cell types (Fig. 3A), while TLX1-NFIC and IRF2 displayed cell type–specific patterns (Fig. 3B,C). These cell type–specific footprinting patterns are highly correlated with gene expression differences (data not shown) and are supported by previous studies. For example, REST is known to repress neuronal genes in all non-neuronal cell types, and IRF2 is an interferon regulatory transcription factor known to be involved in the development of immune-related cells, including B cells (Tamura et al. 2008). The homeoprotein TLX1 is known to interact with the CCAAT binding transcription factor NFIC (N Zhang et al. 1999). We clearly see footprints for the TLX1/NFIC complex in K562, HeLaS3, HUVEC and NHEK cells but not lymphoblastoid or embryonic stem cells. We do not detect a difference in the mean expression of TLX1 across these cell lines (μ1 = 5.47 log2 expression for cells with footprints, μ2 = 5.41 for cells without footprints), but we do see a nearly threefold increase in the expression of NFIC in those cell types with footprints (μ1 = 9.23, μ2 = 7.73). This suggests that the differential binding of the TLX1/NFIC complex in these cell types identified by the footprinting data is likely mediated by NFIC expression.
Figure 3.
Identification of cell type–specific footprints. Cumulative DNase-seq footprinting signals were determined across seven different cell lines for REST (A), TLX1-NFIC (B), and IRF2 (C). For each factor, the same set of motifs was used for all seven cell types. DNase-seq read counts were calculated in the regions surrounding these motifs, similar to Figure 1. Regions shaded in gray represent cell types that display reduced footprinting signal. HUVEC IRF2 shows moderate footprinting signal (light gray). Note that for REST, all cell lines display consistent signals.
Relationship to gene expression
We compared DNase-seq footprints to gene expression patterns using RNA isolated from the same growths of the six diverse cell lines. Transcription factors with low expression signals in a cell line had fewer predicted footprints, while highly expressed factors were overrepresented in the number of predicted footprints (Supplemental Fig. 4C). This trend was especially striking in the top quantile of highly expressed factors where significantly higher numbers of footprints were predicted than for factors in the second quantile (P < 2.381 × 10−15) and even more so compared with factors in the bottom quantile (P < 8.381 × 10−16) (Supplemental Fig. 4D). Interestingly, CTCF was one of the most highly expressed genes and displayed the most footprints in all cell types (Supplemental Fig. 4C). However, we find that high levels of transcription factor expression do not necessarily imply a corresponding high number of annotated footprints.
Evolutionary conservation of footprints
Previous studies have shown that many functional binding sites are more conserved than background at the sequence level (Liu et al. 2004; Hesselberth et al. 2009). Many computational predictors of novel regulatory elements often attempt to incorporate this information (Boffelli et al. 2003). To date, this has proved difficult, likely due to the short, degenerate nature of binding motifs for many factors and the inability to precisely locate their positions.
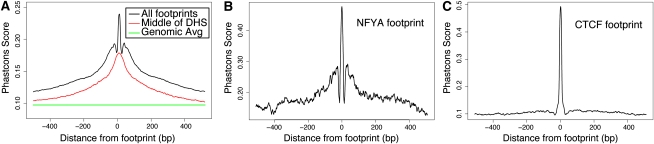
To assess patterns of evolutionary conservation within our predicted footprints, we analyzed the conservation profiles using phastCons (Siepel et al. 2005) for sites corresponding to each specific factor. We found a significant increase in conservation directly within the footprint, which is above the average level of conservation across an entire DNase I HS site (Fig. 4A). For most factors, we detected a marked drop in conservation ∼10 bp immediately flanking the footprint. Beyond this drop, conservation increased again before gradually decreasing to background levels, creating a “shoulder” in this signal. This unusual conservation pattern was not observed in footprints identified by DNase-seq in S. cerevisiae (Hesselberth et al. 2009) and was detected for most individual factors (Fig. 4B; Supplemental Fig. 6C; data not shown). This shoulder is notably absent from CTCF, which only displays a single peak at the footprint (Fig. 4C). This is not an artifact of the large number of annotated CTCF sites as the shoulder is still absent when considering only CTCF footprints at promoters or a small fraction of random CTCF sites (Supplemental Fig. 6D). The sharp conservation pattern in footprints can also be seen for individual footprinted regions (Fig. 1C–D). The drop in conservation between the footprint and shoulder can also be seen by analyzing conservation patterns in the DNA that corresponded to each state in our footprint HMM model (Supplemental Fig. 1E). DNA labeled by the “footprint” state (“FP”) is highly conserved, whereas DNA adjacent to footprints but still within regulatory regions (“UP” and “DOWN”) shows much lower conservation (Supplemental Fig. 6A). In general, higher evolutionary conservation in DNase I HS regions (Supplemental Fig. 6B) is likely due to the presence of multiple functional sites in the larger cis-regulatory modules or promoter regions. The drop in conservation immediately adjacent to most footprints suggests that steric hindrance prevents the binding of nearby factors and may have resulted in relaxed selection pressure for those bases.
Figure 4.
Conservation of sequence in and around DNase I footprints. (A) In general, footprints contain a strong sequence conservation signal with a nearby “shoulder” of conservation around all footprinted regions (black). Between the conservation peak and shoulder is a region with a marked decrease in conservation. This conservation pattern is not detected when the signal is centered on DNase I hypersensitive sites (red). The average conservation signal across the genome is shown in green. (B) The conservation pattern for a single factor, NFYA, displays the characteristic drop in conservation surrounding the footprint. (C) This conservation pattern is not detected around CTCF footprints, which shows relatively little conservation outside the highly conserved footprint. See also Supplemental Figure 6.
CTCF footprints display unique binding characteristics
CTCF is a unique transcription factor that has been shown to display diverse regulatory roles including insulator, enhancer, and repressor activity (Phillips and Corces 2009). CTCF contains 11 zinc fingers, and several studies have discovered a highly conserved GC-rich 20-bp motif associated with many, but not all, detected CTCF binding sites (Kim et al. 2007). Mutational studies have demonstrated that in at least some instances, additional zinc fingers contact sequence upstream of this GC-rich core that may be necessary for binding (Filippova et al. 1996) or appear to stabilize binding (Quitschke et al. 2000) of CTCF. A significant fraction of CTCF binding sites do not contain this primary motif or any other motif, indicating that CTCF binds indirectly to some regions of the genome.
It has been demonstrated that footprint digestion patterns can reflect the actual structural interaction of a factor with the DNA (Hesselberth et al. 2009). Orienting and aligning CTCF footprints based on the direction of the 20-bp motif, we found that CTCF has a very unique footprinting profile (Fig. 5A). CTCF footprints were extremely depleted for DNase I digestion in the 20-bp region that corresponds to the previously characterized GC-rich binding motif. Unlike other factors where the frequency of DNase I cuts rises sharply at the boundaries of the motif, the signal upstream region of the CTCF motif increases more gradually, indicating that CTCF protection extends beyond the 20-bp motif. A similar phenomenon was detected to a lesser extent in the 3′ downstream direction. The total length of these combined protected regions agrees with a single footprinted CTCF site that displayed 50–60 bases of general protection (Phillips and Corces 2009).
Figure 5.
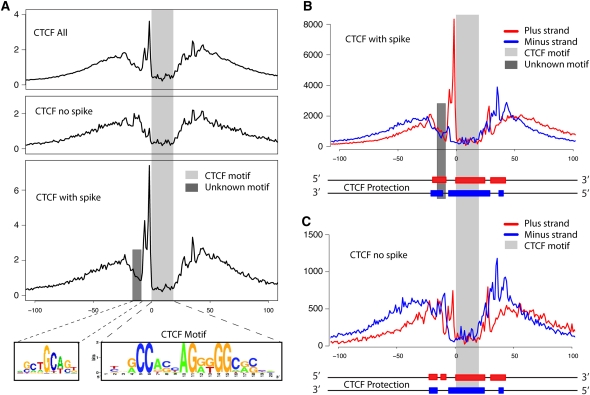
High-resolution analysis of CTCF binding sites. (A) Cumulative footprinting signal at all CTCF motif predicted sites that includes sites with and without a large increase in DNase I digestion upstream of the CTCF motif. The light gray bar indicates the location of the known CTCF motif. The dark gray bar represents the location of a novel binding motif. The novel binding motif was only detected in CTCF footprints that contain the small upstream region with a spike in DNase I hypersensitivity (HS). Note that the entire protected region is approximately 50–60 bases. (B) Strand-specific DNase-seq signal for the subset of CTCF motif identified sites that contain the upstream DNase I HS spike. The DNase I HS spike is only detected on the positive strand. (C) Similarly for the CTCF motif identified sites without the upstream DNase I HS spike. The diagram below each plot in B and C illustrates the estimated strand-specific protected regions surrounding the CTCF motif predicted sites.
Interestingly, we detected a 10-bp spike in DNase I hypersensitivity immediately upstream of the primary motif that is only present on the positive strand (Fig. 5B). This spike was previously reported by traditional DNase I footprinting on a single CTCF binding site in the promoter of the Amyloid precursor protein gene but was not shown to be strand-specific (Quitschke et al. 2000). We found that this spike was not present in all footprints. Of the 4122 CTCF footprints where we additionally required the footprint to overlap 95% of the 20-bp motif, ∼80% showed evidence of a spike in hypersensitivity, while the remaining 20% did not (Fig. 5A). Approximately 60%–80% of CTCF footprints that contained the spike in lymphoblastoid cells also contained the spike in the other five diverse cell types. DNA strand-specific footprint analysis displays very distinct patterns of digestion within and across these two sets of footprints (Fig. 5B,C) and suggests alternative ways that CTCF associates with DNA.
We analyzed different portions of the larger CTCF footprint for secondary motifs and identified SCTGCAST, a motif similar to that recently described for the zinc finger protein ZBTB3 (Badis et al. 2009), ∼10 bp (or one DNA helical turn) upstream of the 5′ end of the CTCF motif (Fig. 5A). This motif appears in 10%–20% of sites with the spike in hypersensitivity but is not present in the set of CTCF footprints lacking the upstream spike. Excluding CTCF motifs that have an adjacent ZBTB3 motif does not eliminate the upstream footprint, indicating that ZBTB3 is not the only putative factor that binds upstream of CTCF, and it is not likely contributing to the spike digestion pattern upstream of the main CTCF motif.
Discussion
We have shown that genome-wide in vivo DNase I footprinting can precisely identify a large number of specific cis-regulatory protein binding events in human in a single experiment. DNase-seq works well with as little as 1 million cells, which will be useful in studying rare primary cell types that are limited in number. We have generated publicly available footprint annotations for seven similar and six diverse cell lines for which DNase-seq data was generated as part of the ENCODE project. Even though comprehensiveness of footprint annotations is likely dependent on the number of sequences from DNase-seq and the level of background noise in the experiment, we show evidence that even a relatively small number of sequences provide a good initial annotation. This indicates that high-throughput DNase-footprint identification is possible for genomes much larger than yeast. As sequencing continues to get less expensive, we expect to be able to obtain even more accurate footprint maps.
Since the binding affinity of a factor to DNA can affect the relative amount of protection from DNase I digestion, this may affect our ability to annotate all transcription factor binding sites. This is supported by our observation that ChIP-seq sites that do not overlap DNase I footprints have weaker ChIP signals. Relative intensity of footprints may therefore provide another source of data to measure binding affinity and occupancy for various factors across the genome. We have also shown that DNase-seq footprints can distinguish more precisely how factors like CTCF may interact with DNA in different ways and may bind in conjunction with other novel factors. These types of information are distinctly different than that typically extracted from ChIP-seq data.
DNase I footprint annotation relies on known PWMs to predict what individual factors are bound within each footprint. Since DNA binding preferences are available for only a fraction of known factors in the public JASPAR (Bryne et al. 2008), TRANSFAC (Matys et al. 2006), and UniPROBE (Newburger and Bulyk 2009) PWM databases, we believe this contributes to our ability to only annotate 25% of footprints. We anticipate that continued PWM discovery using both in vivo and in vitro assays (ChIP, SELEX, dsDNA arrays, etc.) will increase our knowledge of binding preferences for factors, enabling a more accurate and complete annotation of DNase I footprints. We believe that identifying de novo computational motifs in unlabeled footprints will also be an important part of this endeavor.
While DNase-seq footprinting offers a powerful method to identify transcription factor binding sites, it has a number of limitations. For example, it will not likely be able to precisely identify different transcription factors that bind to the same motif. It will also not identify proteins that indirectly bind DNA via interactions with other DNA binding proteins. This indirect binding likely explains why many binding sites identified by ChIP experiments often lack a recognizable motif. Therefore, to more fully identify and understand how complexes bind DNA, future studies will need to integrate multiple complementary genome-wide data sets, including DNase I footprints, ChIP-seq for many factors, PWMs, and other data sets such as those that have recently identified combinatorial binding interactions between large numbers of transcription factors (Ravasi et al. 2010).
DNase-seq footprinting represents a significant advance toward the better understanding of the location, identity, and affinity for hundreds of thousands of cis-regulatory elements genome-wide. For most footprints identified here, it is unknown what trans factor binds to each site. While these data provide a scaffold to more fully annotating the genome, other complementary methods will be required for complete annotation. However, regardless of their identity, we now have evidence of specifically where factors are interacting with the DNA, which is an important step in understanding the components that regulate global gene expression.
Methods
Cell line growth
The source of cells, catalog numbers, and extensive cell growth protocols for all cell types described here can be found on the UCSC Genome Browser ENCODE website (http://genome.ucsc.edu/ENCODE/cellTypes.html).
RNA expression
Total RNA was isolated from these cells using trizol extraction followed by cleanup on RNEasy column (QIAGEN) that included a DNase I step. The RNA was checked for quality using a nanodrop and an Agilent Bioanalyzer. RNA (1 μg) was then processed according to the standard Affymetrix Whole Transcript Sense Target labeling protocol that included a riboreduction step. The fragmented biotin-labeled cDNA was hybridized over 16 h to Affymetrix Exon 1.0 ST arrays and scanned on an Affymetrix Scanner 3000 7G using AGCC software. The resulting cell files were analyzed for quality using Affymetrix Expression Console software. All expression data were submitted to the Gene Expression Omnibus (GSE15805).
DNase I assay
DNase-seq was performed as previously described (Song and Crawford 2010). Briefly, cells were lysed with NP40 and digested with optimal amounts of DNase I enzyme. DNase I ends were made blunt and ligated to biotinylated linkers containing an MmeI restriction site. After digesting with MmeI, DNase I digested ends were enriched on streptavidin magnetic beads (Invitrogen) and ligated to a second set of linkers. DNA was lightly amplified and sequenced using the Illumina GAII. All DNase-seq data have been made publicly available on the UCSC Genome Browser (Regulation Group, Open Chromatin track, March 2006 [hg18] assembly).
CTCF ChIP-seq assay
We fixed 108 cells for 10 min at room temperature by adding formaldehyde (1% final concentration). Formaldehyde was deactivated with 2.5 M glycine (125 mM final concentration). The cross-linked cells were lysed and sonicated three times for 10 min with a Bioruptor (Diagenode), which generated an average size of 500-bp DNA fragments. Chromatin immunoprecipitation was performed with the sonicated cell lysate to purify CTCF-DNA complexes, using an anti-CTCF antibody (Millipore 07-729). Crosslinks in the immunoprecipitated DNA protein complexes were reversed by incubation overnight at 65°C. These samples were then treated with RNase A (Ambion) and proteinase K (Invitrogen), followed by a phenol-chloroform extraction and ethanol precipitation. Thirty nanograms of immunoprecipitated DNA was used to construct the library for Illumina sequencing.
Raw sequence processing
Raw sequence data from technical replicates for each cell type were combined and aligned to the March 2006 (hg18) assembly with MAQ (Li et al. 2008). We retained all sequences that aligned to, at most, four genomic locations and that contained, at most, two mismatches. Those sequences that aligned to multiple genomic locations were randomly assigned one of these locations by MAQ.
Aligned sequences were filtered using three different methods. First, all sequences that fall into regions where the human genome assembly underrepresents the true amount of a particular sequence, namely, satellites in pericentromeric and subtelomeric regions, were removed. These locations can be found on the UCSC browser (table wgEncodeDukeRegionsExcluded). Next, when more than five sequences aligned to a single location, this count was reduced to five as based on fitting the sequence data to a Poisson distribution; it is highly unlikely that more than five of the same sequence would be present and likely represents experimental artifact. Finally, we removed all sequences from regions where 70% of sequences in a 31-bp region fall in a 5-bp window. We believe these represent experimental artifacts likely due to incorrect PCR amplification.
DNase I HS regions identified by DNase-seq
Regions of significant enrichment of DNase I tags were identified using the F-seq peak caller (Boyle et al. 2008b). F-seq identifies regions of high density of sequence reads as compared to a random background distribution of reads. We adjusted the background of F-seq based on input sequence from each cell line and an alignability background (UCSC Browser, table wgEncodeDukeUniqueness20bp). The distribution of base pair F-seq scores was fit to a gamma distribution, and the score corresponding to a P-value of 0.05 was used to discretely define DNase I HS sites.
CTCF, SRF, REST, and GABP binding sites identified by ChIP-seq
CTCF binding sites defined by significant enrichment of sequences from ChIP-seq were similarly identified as for DNase-seq using F-seq. A P-value cut-off was also used to define discrete binding regions, and the maximum F-seq score was assigned to each peak. Factor binding sites for SRF, REST, and GABP defined in their original publication within Jurkat human T lymphoblasts were used for these factors (Valouev et al. 2008).
Mapping motif binding sites
PWMs from the JASPAR (Bryne et al. 2008), free TRANSFAC (Matys et al. 2006), and UniPROBE (Newburger and Bulyk 2009) databases, as well as the Ren laboratory CTCF motif (Essien et al. 2009), were used to initially annotate potential factor binding sites. There is redundancy between these sets, but because of slight motif differences, all PWMs were used.
Each mapping produced a bit-score that was used to filter the motif matches. We set the cutoff for matches as the lowest of either 70% of maximum possible bit score or 90% of functional depth, where the functional depth is defined as the difference between the maximum possible and minimum possible score for a particular PWM. Finally, if the PWM had a 0 value for any base (AGCT) in a particular position, this was respected, meaning no mappings for this motif may contain this 0-scored base in this position.
Clustering CTCF motifs based on footprinting data
For each base ±200 bases surrounding the 20-bp CTCF motif, we determined the number of DNase I cut sites. Using this 420-value vector, CTCF motifs were split into two clusters using k-means clustering implemented in R (kmeans package, k = 2).
HMM to identify footprints
Prior to input to the HMM, aligned raw sequence data were preprocessed by normalizing and smoothing the input sequences as follows. First, the number of sequence tags at each base was normalized. This normalization consisted of dividing counts at each base by the mean of all non-zero sequence counts in a 1-kb region surrounding that base. Next, these values were fit to a second-order polynomial based on the eight surrounding bases using the Savitzk-Golay filter. In this way, these values corresponded to the slope of a curve representing the relative change in the density of DNase I cuts.
The HMM consisted of five states where emissions from each state corresponded to features of the values described above (Supplemental Fig. 1E). Each footprint was expected to be in a region of low DNase I digestion with a reduced density of DNase I cuts surrounded by increased densities of cuts to either side. This pattern was captured by a path through the model states starting with the background DNase I HS state (“HS”), transitioning to the state with increasing DNase I digestion (“UP”), followed by the state with decreasing DNase I digestion (“DOWN”) and then the footprint state (“FP”) and again through the UP and DOWN states. The transition and emission probabilities were initially trained for the combined lymphoblastoid cell lines with experimental data from the previously studied promoter of the FMR1 gene. The footprinting of the FMR1 region was small and specific to lymphoblastoid cells; therefore, we chose to use a set of predicted sites from the combined lymphoblastoid data to train each additional line. Footprints from the 1000 highest scoring DNase I HS sites from chromosome 6 were chosen with the assumption that these strong sites will primarily consist of ubiquitous regulatory features based on our analyses of DNase I HS sites across cell lines (data not shown). Emission probabilities for all other cell lines were trained using these sites using maximum likelihood training. Emission and transition probabilities for all models are listed in Supplemental Table 7. HMM software used to implement these models can be found at http://www.kanungo.com/software/software.html (Kanungo 1999).
Each identified DNase I HS region were annotated with footprints using posterior decoding based on the trained HMM model. For display purposes, 3 bp was added to either side of the called footprint. STAMP was then used to label each footprint based on known PWMs. Each motif label required a STAMP calculated P-value match of <1 × 10−6 to be labeled with a particular factor. All factors meeting this threshold were included in the factor label and sorted by significance. Note that the motif was required to be fully contained within the footprinted region in order to be annotated by STAMP.
Conservation values and plots
The first base of all footprints was set to the 0 position. Footprint conservation scores were then down-sampled or up-sampled to create 20 bp of discrete values for each footprint (essentially shrinking or spreading each footprint conservation scores to the same genomic size). The upstream values were then calculated starting at the beginning of the footprint, and the downstream values were calculated starting at the end of the footprint. Vertebrate plots were created using the UCSC table phastCons44way, mammal plots were created using the UCSC table phastCons44wayPlacental, and primate plots were created using the UCSC table phastCons44wayPrimates.
Acknowledgments
We thank Lisa Bukovnik, Tonya Severson, and Fangfei Ye at the Duke sequencing core facility. We thank Sridar Chittur and Scott Tenenbaum at the University of Albany-SUNY for help with RNA expression studies. This research is supported by a NHGRI grant U54HG004563 to T.S.F., V.R.I., E.B., and G.E.C.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession nos. GSE19622 and GSE25442.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.112656.110.
References
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324: 1720–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, Pachter L, Rubin EM 2003. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299: 1391–1394 [DOI] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE 2008a. High-resolution mapping and characterization of open chromatin across the genome. Cell 132: 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS 2008b. F-Seq: A feature density estimator for high-throughput sequence tags. Bioinformatics 24: 2537–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne JC, Valen E, Tang ME, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A 2008. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res 36: D102–D106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin R, Angers M, Dallaire N, Rose TM, Khandjian EW, Rousseau F 1997. Structural and functional characterization of the human FMR1 promoter reveals similarities with the hnRNP-A2 promoter region. Hum Mol Genet 6: 2051–2060 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306: 636–640 [DOI] [PubMed] [Google Scholar]
- Essien K, Vigneau S, Apreleva S, Singh LN, Bartolomei MS, Hannenhalli S 2009. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol 10: R131 doi: 10.1186/gb-2009-10-11-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol 16: 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross DS, Garrard WT 1988. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem 57: 159–197 [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanungo T 1999. UMDHMM: Hidden Markov model toolkit. In Extended finite state models of language (ed. Kornai A). Cambridge University Press, Cambridge: http://www.kanungo.com/software/software.html [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B 2007. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu XS, Wei L, Altman RB, Batzoglou S 2004. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res 14: 451–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Benos PV 2007. STAMP: A web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35: W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Auron PE, Benos PV 2007. DNA familial binding profiles made easy: Comparison of various motif alignment and clustering strategies. PLoS Comput Biol 3: e61 doi: 10.1371/journal.pcbi.0030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. 2006. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res 34: D108–D110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, et al. 2010. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newburger DE, Bulyk ML 2009. UniPROBE: An online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res 37: D77–D82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG 2009. CTCF: Master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke WW, Taheny MJ, Fochtmann LJ, Vostrov AA 2000. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res 28: 3370–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140: 744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrich AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE 2010. DNase-seq: A high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor Protoc 2010: doi: 10.1101/pdb.prot5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T 2008. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol 26: 535–584 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM 2009. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet 10: 252–263 [DOI] [PubMed] [Google Scholar]
- Wu C 1980. The 5′ ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature 286: 854–860 [DOI] [PubMed] [Google Scholar]
- Zhang N, Shen W, Hawley RG, Lu M 1999. HOX11 interacts with CTF1 and mediates hematopoietic precursor cell immortalization. Oncogene 18: 2273–2279 [DOI] [PubMed] [Google Scholar]