Abstract
Imprinted genes are critical for normal human growth and neurodevelopment. They are characterized by differentially methylated regions (DMRs) of DNA that confer parent of origin-specific transcription. We developed a new strategy to identify imprinted gene-associated DMRs. Using genome-wide methylation profiling of sodium bisulfite modified DNA from normal human tissues of biparental origin, candidate DMRs were identified by selecting CpGs with methylation levels consistent with putative allelic differential methylation. In parallel, the methylation profiles of tissues of uniparental origin, i.e., paternally-derived androgenetic complete hydatidiform moles (AnCHMs), and maternally-derived mature cystic ovarian teratoma (MCT), were examined and then used to identify CpGs with parent of origin-specific DNA methylation. With this approach, we found known DMRs associated with imprinted genomic regions as well as new DMRs for known imprinted genes, NAP1L5 and ZNF597, and novel candidate imprinted genes. The paternally methylated DMR for one candidate, AXL, a receptor tyrosine kinase, was also validated in experiments with mouse embryos that demonstrated Axl was expressed preferentially from the maternal allele in a DNA methylation-dependent manner.
The mammalian genome harbors imprinted genes that are critical for normal human growth and neurodevelopment (Ideraabdullah et al. 2008). Approximately 100 imprinted genes have been identified (Weaver et al. 2009), which represent ∼0.5% of the ∼20,000 protein-coding genes annotated in the human genome (IHGSC 2004). These genes, which are expressed exclusively or preferentially from a specific parental allele, are often clustered in a genomic region and respond to regulatory signals in cis from differentially methylated regions (DMRs). Such DMRs, comprised of CpG-rich regions of DNA, demonstrate ∼50% methylation because either the maternal or paternal allele is methylated. These regions of parental allele-specific methylation are generally maintained in all somatic tissues, whereas expression is occasionally cell type- or tissue-specific, possibly depending on the availability of specific proteins that are permissive for transcription.
Several approaches have been used previously in mice to identify imprinted genes and their associated DMRs. One approach that led to the identification of the first imprinted gene, Igf2r, linked phenotypes that were transmitted in a parent of origin-specific manner to a specific genomic region (Barlow et al. 1991). In addition, genome-wide screens for parental-specific methylation and computational analyses of specific DNA sequence features (Smith et al. 2003; Luedi et al. 2007; Hiura et al. 2010), as well as subtractive hybridization of cDNAs derived from parthenogenetic and androgenetic embryos (Kaneko-Ishino et al. 1995), have been informative. One of the limitations of expression-based approaches is the requirement for the correct tissue and the appropriate developmental stage to be tested to successfully identify new imprinted genes (Deltour et al. 1995). In contrast, approaches based on screening for an epigenetic feature, such as allele-specific DNA methylation, are applicable to all tissues at all developmental time points (Smith et al. 2003; Dockery et al. 2009; Hiura et al. 2010), and should provide a reliable estimate of the number of differentially methylated regions in humans or any model organism.
In humans, tissues of uniparental origin are rare. Only maternally derived chromosomes are found in mature cystic ovarian teratomas (MCT), which arise from a meiotic error during oocyte maturation. The result is the formation of a cyst containing tissues from each of the three embryonic germ cell layers. In contrast, an androgenetic complete hydatidiform mole (AnCHM) can develop from an abnormal conceptus carrying two paternally derived genomes. Such abnormal pregnancies are characterized by hydropic degeneration of all chorionic villi and the absence of an embryo, cord, and embryonic membranes. MCT and AnCHM display the extreme biological consequences of uniparental inheritance (Mutter 1997) and demonstrate that both parental genomes are required for the complete development of an embryo, with the paternal genome required for extraembryonic differentiation and maternal genome for proper embryonic development (Surani et al. 1993). Thus, it is not just two sets of chromosomes that are required for proper embryonic development; also necessary is the biparental origin of specific genomic regions that carry imprinted genes. Previously reported targeted methylation analyses, using bisulfite sequencing in AnCHM, revealed paternal-specific methylation for a subset of known imprinted DMRs (Judson et al. 2002; El-Maarri et al. 2003) demonstrating the validity of using this tissue for genome-wide screening for paternal-specific DNA methylation. In addition to MCT and AnCHM, tissues of uniparental origin can be obtained from rare individuals with uniparental disomy (UPD) who carry two copies of a particular chromosome or chromosomal region derived from only one parent, either mother or father. UPDs in imprinted genomic regions are associated with human diseases such as Angelman syndrome (paternal UPD of chromosome 15q11) (Buiting 2010) or Beckwith-Wiedemann syndrome (paternal UPD of chromosome 11p15) (Choufani et al. 2010). Notably, cell lines from individuals with UPD can also be used to generate parent of origin-specific DNA methylation profiles for specific genomic regions (Sharp et al. 2010). Therefore, tissues or cell lines from MCT, AnCHM, and UPD are valuable biological samples that can be used to screen for DMRs and imprinted genes in humans.
This is the first report of a genome-wide DNA methylation screen comparing human tissues with genomic regions of uniparental and biparental origin. Using this method, we were able to identify previously characterized DMRs associated with known imprinted genes, new DMRs in close proximity to known imprinted genes, and candidate DMRs that led to the discovery of a new imprinted gene, AXL, among several other candidates. Our approach to the identification of imprinted DMRs is unique, allowing the primary identification of imprinted domains in humans without depending on prior discoveries in other mammals.
Results
Identification of differentially methylated genomic regions using comparisons of biparental and uniparental human tissues
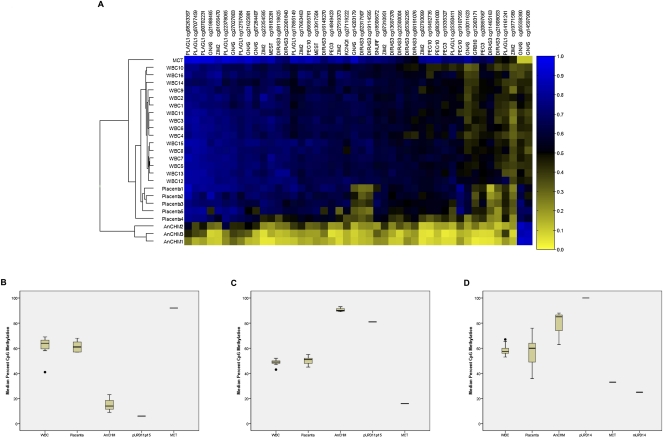
DMRs are an important hallmark of imprinted genes. To discover new imprinted genes, as well as new DMRs associated with known imprinted genes, we undertook a genome-wide search for DMRs with parent of origin-specific features. We compared DNA methylation profiles of gene promoters in tissues of uniparental origin to those of biparental origin. Tissues of known uniparental origin included AnCHM and MCT. The normal biparental tissues were placenta and white blood cell (WBC) samples. Our comparisons also included WBC from individuals with UPD carrying limited genomic regions of uniparental origin. Sodium bisulfite modified genomic DNA was hybridized to the Illumina Infinium Human Methylation27 microarray platform to compare single CpG site methylation among these samples. This array interrogates 27,578 CpG sites at single-nucleotide resolution, covering the promoter regions of 14,495 RefSeq genes, including most known imprinted DMRs (Grafodatskaya et al. 2009), and representing 0.1% of total CpG sites in the genome (Laurent et al. 2010). To determine whether our method could quantitatively detect parent of origin-specific differentially methylated CpG (DMCpG) sites, we assessed the methylation profiles of 718 CpG sites represented on the array that mapped within 48 known human imprinted domains (Kong et al. 2009). We determined the intraclass correlation coefficient (ICC) between the AnCHM and the MCT, representing paternal and maternal origins, respectively, and defined the methylation profiles for a subset of 45 DMCpG sites, corresponding to 10 known imprinted DMRs (Fig. 1A; Supplemental Table 1).These data showed that our microarray experiments captured all the known paternal and maternal DMRs, represented on the array, based on their DNA methylation levels. The analyses for known imprinted loci resulted in patterns consistent with the expected parent of origin-specific profiles (Fig. 1A).
Figure 1.
CpG methylation in differentially methylated regions (DMRs). (A) Heatmap showing the methylation levels of CpG sites in DMRs of known imprinted genes. The rows represent individual samples and the columns represent the 45 CpG sites (labeled with the CpG site unique ID and the gene associated with these sites). Euclidean clustering shows that samples from each tissue type are clustered together. The two paternally methylated CpG sites (in the paternally methylated GNAS/NESP55 DMR) are on the right of the heatmap and showed methylation patterns in uniparental tissues that are opposite to the other maternally methylated CpG sites. The samples are labeled white blood cell (WBC), placenta, androgenetic complete hydatidiform mole (AnCHM), and mature cystic ovarian teratoma (MCT). Note that each DMR is represented by one or several CpG sites and for detailed information about these CpG sites see Supplemental Table 1. (B) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent CpG methylation for five consecutive CpG sites within the maternally methylated KvDMR1 for each tissue. (C) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent CpG methylation for three consecutive CpG sites within the paternally methylated H19 DMR for each tissue. For B and C the bisulfite pyrosequencing data are derived from 15 blood samples, 10 placenta samples, three AnCHM samples, one paternal UPD11p15, and one MCT sample. (D) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent methylation for five consecutive CpG sites within the paternally methylated IG-DMR for each tissue. The bisulfite pyrosequencing data are derived from 15 blood samples, nine placenta samples, three AnCHM samples, one paternal UPD14 blood sample, one MCT sample, and one maternal UPD14 blood sample.
For validation of the microarray data, we developed pyrosequencing assays to assess methylation levels at the maternally methylated, KvDMR1 (imprinting center 2 [IC2]) on chromosome 11p15.5 (Fig. 1B) and the paternally methylated, H19 DMR (imprinting center 1 [IC1]), also on 11p15.5 (Fig. 1C). As predicted, the AnCHM and cells with paternal UPD for chromosome 11p15 showed high levels of methylation (>80%) for the H19 DMR and low levels of methylation for KvDMR1(<25%). In addition to KvDMR1 and H19 DMR, we also developed a pyrosequencing assay for IG-DMR, a paternally methylated DMR that controls imprinting of the DLK1 imprinting cluster (Kagami et al. 2008), which is not represented on the microarray. Median CpG methylation levels at five CpG sites overlapping the IG-DMR on human chromosome 14q32 exhibited patterns consistent with their parent of origin for both uniparental tissues and tissues from patients with maternal and paternal UPDs for chromosome 14, respectively (Fig. 1D). These genome-wide and supplementary targeted screens confirmed, for the first time, a genome-wide disruption of imprinting in uniparental tissues, extending the aberrant profile of DNA methylation in these tissues to all known imprinted loci that we assessed by microarray and by targeted pyrosequencing. We report here aberrant methylation at multiple DMRs not previously tested in AnCHM and MCT, including DIRAS3, PLAGL1, IG-DMR, and L3MBTL1. These data provide proof of principle for the utility of comparing genome-wide methylation profiles for genomic regions of uniparental versus biparental origin to identify new DMCpG sites associated with known imprinted genes as well as new candidate imprinted genes.
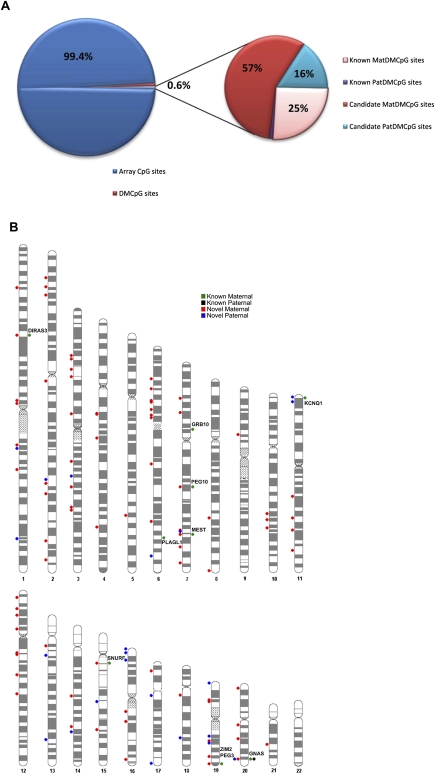
To identify novel DMCpG sites, we screened the 26,486 autosomal CpG sites represented on the microarray to find methylation patterns analogous to those identified at known imprinted DMRs in uniparental (AnCHM, MCT) and biparental tissues (placenta, WBC). Using criteria described in the Methods section, we identified a total of 166 DMCpG sites corresponding to ∼0.6% of the array CpG sites (Fig. 2A). Among those, 139 DMCpG sites were designated candidate maternally methylated DMRs (unmethylated in AnCHM and methylated in MCT). Of these, 43 DMCpG sites mapped to known DMRs (Supplemental Table 1). Further, 17 novel DMCpG sites mapped within 2 kb of known DMRs, likely expanding their currently annotated genomic boundaries (Supplemental Table 2). The remaining 79 DMCpG sites were considered new candidate maternal DMCpG sites overlapping the promoter regions of 77 transcripts and are listed in Supplemental Table 2. Similarly, 27 DMCpG sites were designated candidate paternally methylated DMRs (methylated in AnCHM, unmethylated in MCT), including six DMCpG sites mapping within 2 kb of known paternally methylated DMRs, H19 DMR, and GNAS (NESP55) DMR (Supplemental Table 3). Notably, the two DMCpG sites within 2 kb of H19 DMR are upstream of the currently annotated paternally methylated DMR, suggesting that the H19 DMR extends beyond the currently recognized boundaries (2–4.4 kb upstream of the H19 transcription start site) (Imprinted Gene Catalogue Records: www.otago.ac.nz/IGC). The newly identified DMCpG sites showed more maternal (57%) than paternal candidates (16%) similar to the known imprinted DMCpG sites (Fig. 2A). Notably, their distribution was not restricted to specific chromosomal regions; rather they were spread across most autosomes (Fig. 2B).
Figure 2.
Distribution of known and candidate DMCpG sites. (A) Pie chart showing the frequency of the identified (∼0.6%) DMCpG sites with the smaller pie chart illustrating the frequency of only the known and the candidate DMCpG sites. (B) Chromosome ideogram showing the distribution across all autosomes of known (chromosome ideogram, right) and candidate (chromosome ideogram, left) DMCpG sites. Green and black dots correspond to known maternally and paternally methylated CpG sites, respectively. Red and blue dots correspond to candidate maternally and paternally methylated CpG sites, respectively. The names of imprinted genes are displayed next to their associated known DMCpG sites.
New DMRs associated with known imprinted genes
The identification of new DMRs is important for the complete characterization of imprinted gene regulation. We identified among the new candidate DMCpG sites, several DMCpG sites that mapped within 2 kb of known imprinted DMRs (Supplemental Tables 2, 3). Interestingly, two candidate DMCpG sites overlapping the CpG island at the promoter of SNURF/SNRPN on chromosome 15 (Supplemental Table 2) were located in a maternally methylated DMR identified in a recent study (Sharp et al. 2010).
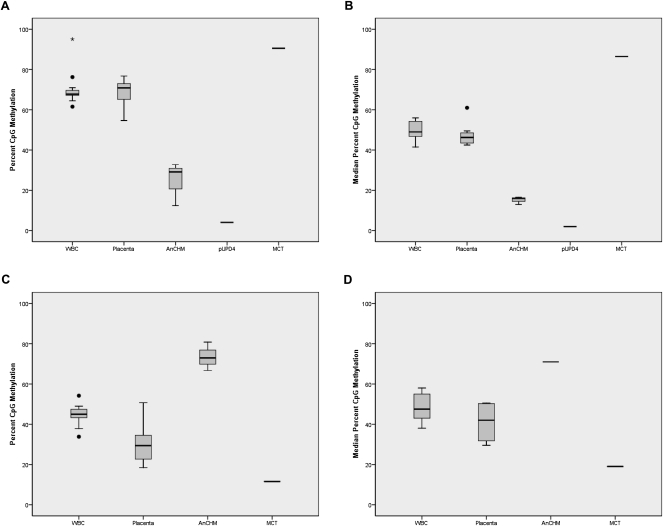
We selected three of these DMCpG sites that mapped to promoter regions of two known imprinted genes with previously unidentified DMRs, NAP1L5 and ZNF597, for further validation. Two DMCpG sites mapped to the CpG island located in the promoter region of NAP1L5, which encodes a nucleosome assembly protein. Data for these two DMCpG sites were consistent with a maternally methylated DMR, i.e., differential methylation in WBC and placenta (∼65%), increased methylation in the MCT (90%), reduced methylation in the AnCHMs (30%), and low methylation (4%) in the cell line with paternal UPD for chromosome 4 (Fig. 3A). Human NAP1L5 has been reported to have monoallelic paternal expression (Wood et al. 2007) but no DMR has previously been reported in humans. To map the extent of the DMR beyond the single CpG resolution of the microarray, we developed a pyrosequencing assay targeting six CpG sites in the promoter region of NAP1L5. The median methylation at the six CpG sites showed methylation patterns consistent with maternal-specific methylation in all tissues, including a paternal UPD for chromosome 4 (Fig. 3B) confirming our array data.
Figure 3.
CpG methylation in new candidate differentially methylated regions (DMRs) for the imprinted genes NAP1L5 and ZNF597. (A) Box-and-whisker plot showing the distribution of CpG methylation percent for each tissue at the NAP1L5 CpG site cg01026744 from the Illumina Infinium Human Methylation 27 BeadChip microarray. The methylation data are derived from 16 blood samples, five placenta samples, three AnCHM samples, one paternal UPD4 lymphoblastoid sample, and one MCT sample. (B) Box-and-whisker plot showing the median percent CpG methylation for six consecutive CpG sites at NAP1L5 DMR for each tissue. The bisulfite pyrosequencing data are derived from 15 blood samples, 10 placenta samples, three AnCHM samples, one paternal UPD4 lymphoblast sample, and one MCT sample. (C) Box-and-whisker plot showing the distribution of CpG methylation percent for each tissue at the ZNF597 CpG site cg14654875 from the Illumina Infinium Human Methylation 27 array. The methylation data are derived from 16 blood samples, five placenta samples, three AnCHM samples, and one MCT sample. (D) Box-and-whisker plot showing the median percent CpG methylation for seven consecutive CpG sites within the ZNF597 DMR for each tissue. The bisulfite pyrosequencing data are derived from 14 blood samples, four placenta samples, one AnCHM sample, and one MCT sample.
We also identified a new DMCpG site on chromosome 16, in the promoter region of the imprinted gene ZNF597, which encodes a zinc finger protein (Pant et al. 2006). This region was identified as a candidate paternally methylated DMR based on its high methylation level in the AnCHM, low methylation level in the MCT, and differential methylation in both biparental tissues (Fig. 3C). These data are consistent with the reported maternal expression of ZNF597 in humans (Pant et al. 2006). A targeted pyrosequencing assay for seven CpG sites mapping to the CpG island in the promoter region of ZNF597 validated the microarray data for this region (Fig. 3D). The data for these two new DMRs provided further evidence of the success of our approach for identifying new imprinted DMRs.
Identification of new candidate imprinted genes
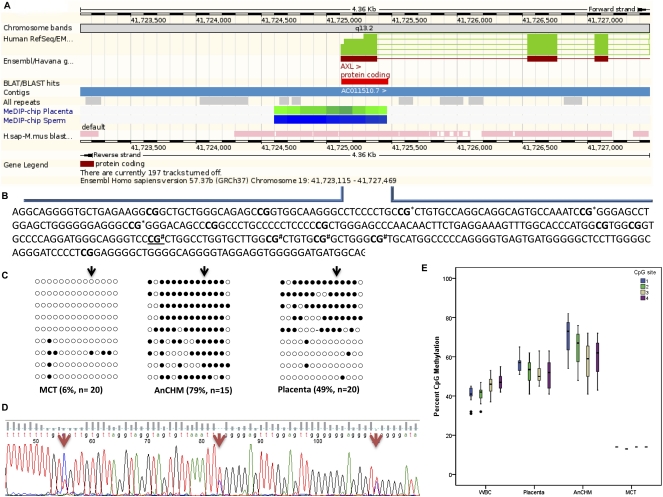
To demonstrate that our approach was capable of detecting new imprinted genes, we selected one candidate imprinted gene from the new predicted paternal DMCpG sites (Supplemental Table 3) for further validation. This candidate was chosen because of the high methylation difference at the DMCpG site observed between the AnCHMs and MCT (i.e., 70%) and because it was predicted to be a paternally methylated DMR, which is far less common in the genome than maternally methylated DMRs. This DMCpG site overlapped the promoter region of the AXL gene (Gene ID: 558) on chromosome 19q13.2 (Fig. 4A), which encodes a receptor tyrosine kinase belonging to the TAM subfamily of receptor tyrosine kinases.
Figure 4.
CpG methylation of the candidate differentially methylated region (DMR) for the new candidate imprinted gene AXL. (A) Genomic organization of the human AXL promoter region showing the relative location of the 355-bp region (displayed as a red rectangle) analyzed for methylation and overlapping the CG (cg14892768) identified by the array. Integration of placenta and sperm specific methylation values (Rakyan et al. 2008) into the Ensembl Genome Browser (www.ensembl.org). Green and blue represent intermediate and methylated regions, respectively. (B) Detailed sequence information of the region analyzed for methylation surrounding the AXL promoter region. (C) The 14 CGs analyzed are shown in bold in B and were assayed by standard bisulfite sequencing; the black arrow represents the same underlined CG in B and corresponds to the CG identified by the array. Filled black circles represent methylated CG and open circles represent unmethylated CG. (D) Direct bisulfite sequencing of a subset of CG sites marked by * in B in the same placental sample as C. (E) Box-and-whisker plot displaying the distribution of bisulfite pyrosequencing median percent CpG methylation for four consecutive CpG sites marked # in B within the new candidate AXL DMR for each tissue. The bisulfite pyrosequencing data are derived from 15 blood samples, 10 placenta samples, three AnCHM samples, and one MCT sample.
To validate the candidate paternally methylated DMR for AXL, we designed a pyrosequencing assay targeting four CpG sites that map to the non-CG island promoter region (a common feature of paternally methylated DMRs). At these four CpG sites, the median methylation levels were 45% and 55% in WBCs and placenta, respectively, consistent with differential methylation of the two parental alleles. The median methylation levels in AnCHMs (70%) and the MCT (18%) (Fig. 4E) were consistent with paternally methylated and maternally unmethylated alleles, respectively. These data suggested that the promoter region of AXL contains a paternally methylated DMR and that AXL is a candidate imprinted gene. To further validate the parent of origin-specific methylation patterns in this genomic region, we performed bisulfite sequencing of 14 CpG sites overlapping the transcription start site of AXL and found that DNA from the AnCHMs were almost completely methylated, whereas the DNA from the MCT was mostly unmethylated at all 14 CpG dinucleotides (Fig. 4B,C). Placental DNA showed a differentially methylated pattern that was demonstrated both by direct sequencing (Fig. 4D) and by bisulfite pyrosequencing of a subset of overlapping CpG sites (Fig. 4E). Interestingly, tissue-specific methylation patterns were previously reported for the same genomic region in human, i.e., intermediate methylation in the placenta and high levels of methylation in sperm (Fig. 4A; Rakyan et al. 2008).
Since imprinted genes are often clustered within chromosomal domains, we examined the genomic sequence surrounding AXL and found that it is proximal to the cytochrome P450 gene cluster. Interestingly, this P450 gene cluster is contained in a genomic region recently suggested to constitute an “epihaplotype” that confers sequence dependent allele-specific DNA methylation (Kerkel et al. 2008).
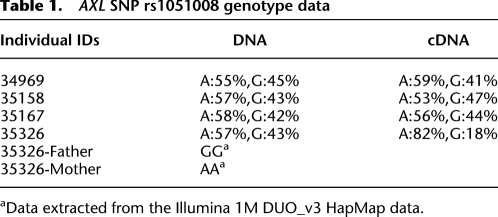
For imprinted genes, differential methylation of parental alleles is often associated with preferential parent of origin-specific expression in some tissues. We examined human WBCs and placenta for the presence of transcribed polymorphisms in the AXL coding region and were unable to identify such polymorphisms in our tissue samples. We then reviewed a large sample of HapMap genotype data generated on the Illumina Human 1M-Duo v3.0 (kindly provided by www.TCAG.ca). We analyzed available genotype data from 90 individuals of Northern and Western European ancestry (CEU) and 90 individuals from the Yoruba of Ibadan, Nigeria (YRI). We searched for SNPs mapping to the AXL genomic region (46435769-46451873, Genome build 36); and identified one that mapped to the 3′ UTR (untranslated region) of AXL (rs1051008). Analysis of the genotyping data showed one CEU individual and 33 YRI individuals from 20 different pedigrees with heterozygous genotypes for this SNP. These data showed that the YRI are more polymorphic for this genomic region than CEU, which explained our initial lack of success in identifying polymorphic SNPs in this region in our Caucasian cohort. We performed genotyping analysis using pyrosequencing and confirmed the heterozygous genotype for the selected individuals at this SNP. We selected four informative pedigrees for allelic expression studies. Pyrosequencing of double-stranded cDNA was used to interrogate the expression level of each allele for this SNP (rs1051008) in the coding region of AXL. This allelic discrimination assay identified one of four heterozygous individuals with skewed allelic expression of AXL. Parental genotypes at this SNP confirmed preferential maternal expression in this sample (Table 1). These data indicate that AXL demonstrates preferential maternal expression in some individuals, suggesting polymorphic imprinting in WBC similar to what has been previously reported for IGF2 in human blood (Giannoukakis et al. 1996).
Table 1.
AXL SNP rs1051008 genotype data
aData extracted from the Illumina 1M DUO_v3 HapMap data.
In summary, we demonstrated AXL to be a new maternally-expressed, imprinted gene that harbors a parent of origin-specific DMR in its promoter region, detected first by our microarray analysis and then validated by bisulfite pyrosequencing.
Axl allelic expression and DNA methylation in mice
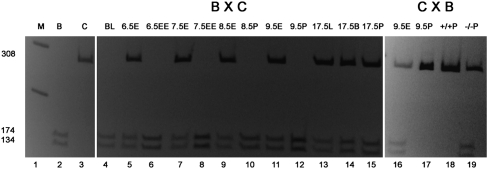
Given the challenges in accessing multiple human tissue types across different developmental stages to assess allelic expression patterns of AXL, we decided to perform these experiments in mice. Axl allelic transcription can be comprehensively interrogated in the murine model system because it provides access to a large number of transcribed polymorphisms in all tissues at multiple developmental stages. Axl has not previously been shown to be imprinted in mice; however, it is located on mouse chromosome 7, which harbors approximately half of the mouse imprinted genes (Williamson 2010). Using C57BL/6 and Mus castaneus F1 hybrid crosses (Mann et al. 2003), we assayed allelic expression of Axl in blastocysts and post-implantation stages from embryonic day (E) 6.5 to E17.5 dpc. Blastocysts expressed Axl exclusively from the maternal allele (Fig. 5). At E6.5 dpc, Axl was transcribed in extra-embryonic tissues either exclusively or predominantly from the maternal allele while embryonic tissues showed largely unbiased allelic expression. The imprinting patterns in extra-embryonic tissues became more variable, with a proportion of tissues retaining preferential maternal expression for a few additional days. The majority of embryonic tissues analyzed between E6.5 and E17.5 dpc showed equal maternal and paternal expression. To determine whether the preferential maternal expression of Axl is DNA methylation-dependent, we analyzed E9.5 placental tissues from mice carrying an engineered mutation in the DNA methyltransferase 1 gene (Dnmt1) (Li et al. 1993). Our data demonstrated that while most wild-type placentas showed preferential maternal expression of Axl, all mutant Dnmt1 placentas had equivalent parental expression in the absence of DNA methylation. That is, DNA methylation was critical for Axl imprinted expression in the mouse placenta.
Figure 5.
Allelic expression analysis for the new candidate imprinted gene Axl in mouse. Shown here are cDNA amplified using Axl primers digested with Hpy188III enzyme that cuts a polymorphic sequence specifically on the C57BL/6 allele. Direction of the cross (Maternal X Paternal) is indicated as BXC or CXB (B = C57BL/6; C = Cast). The lanes are as follows: 1: M (marker); 2: B, for digestion (control); 3: C, for no digestion (control); 4: BL (blastocyst); 5: 6.5E (6.5 dpc embryonic tissue); 6: 6.5EE (6.5 dpc extra-embryonic tissue); 7: 7.5E (7.5 dpc embryonic tissue); 8: 7.5EE (7.5 dpc extra-embryonic tissue); 9: 8.5E (8.5 dpc embryo); 10: 8.5P (8.5 dpc placenta); 11 and 16: 9.5E (9.5 dpc embryo); 12 and 17: 9.5P (9.5 dpc placenta); 13: 17.5L (17.5 dpc liver); 14: 17.5B (17.5 dpc brain); 15: 17.5P (17.5 dpc placenta); 18: +/+P (9.5 dpc Dnmt1 +/+ placenta); and 19: −/−P = 9.5 dpc Dnmt1 −/− placenta. Exclusive maternal expression of Axl was detected starting in blastocyst (n = 2), in the extra-embryonic tissues of all examined E6.5 concepti (n = 6), and subsequently in variable proportions of extra-embryonic tissues of 7.5 dpc (2/4), 8.5 dpc (1/4), and 9.5 dpc (6/9) mice. Embryonic tissues from 6.5 dpc (4/4), 7.5 dpc (4/4), 8.5 dpc (4/4), 9.5 dpc (8/9), and 17.5 dpc (2/2) concepti, as well as from 17.5 dpc placentas (2/2), showed equal maternal and paternal expression. While 4/6 placentas from 9.5 dpc Dnmt1 (n allele) +/+ mice showed biased maternal expression, all Dnmt1 −/− placentas exhibited equal expression from the parental alleles (n = 3).
Because of the dependence of Axl monoallelic expression on DNA methylation, we examined the methylation of a 428 bp region of the mouse Axl gene (Chr7: 26,573,331–26,573,758 [NCBI37/mm9]). The region includes sequence that is syntenic to the region of human AXL containing the newly identified DMCpGs. We performed bisulfite sequencing (a total of 16 CpG sites) in mouse sperm, E9.5 placenta, and E9.5 embryo but found the region to be largely unmethylated in all samples (data not shown). Thus, these data did not indicate the presence of a conserved DMR. We also designed a combined bisulfite restriction analysis (COBRA) in E6.5 extraembryonic tissues that showed the most consistent Axl imprinting, but this assay also failed to detect DNA methylation. Finally, we also assayed methylation in mouse sperm and oocytes of a 330 bp region downstream from Axl (Chr 7:26,541,083–26,541,412 [NCBI37/mm9]), which harbors a CTCF binding site, and found no evidence of DMR.
Discussion
We report a successful new strategy for the identification of novel imprinted DMCpG sites. This strategy robustly targeted known imprinted genes, expanding the boundaries of some of their known DMRs, and led to the discovery of a new DMR and a new imprinted gene that were validated in both human and mouse. This approach, which uses a comparison of genome-wide DNA methylation profiles from uniparental versus biparental tissues, was able to identify more known imprinted DMRs than did other studies (Strichman-Almashanu et al. 2002; Luedi et al. 2007; Maynard et al. 2008; Pollard et al. 2008). Prediction of the total number of human imprinted loci varies according to the method. Approximately 154 high-confidence candidate imprinted human genes were identified using a computational approach looking for specific features of DNA sequence (Luedi et al. 2007), another 26 genes with parent of origin-dependent allelic expression were identified by sequencing the transcriptome of mouse neonatal brains (Wang et al. 2008). Furthermore, allelic expression studies by Pollard et al. (2008) identified 61 candidate imprinted genes (Pollard et al. 2008), and only 26 candidate imprinted DMRs were identified in a screen for maternal methylation in parthenogenetic mouse embryos (Smith et al. 2003). In our study we identified 166 DMCpG sites, including 45 DMCpG sites that map to known imprinted DMRs and 26 candidate DMCpG sites that map within 2 Kb of known imprinted loci. These data generated a total of 110 candidate DMCpG sites corresponding to the promoter regions of 108 genes. The number of candidate imprinted genes identified in different studies is variable and very few candidates have been identified in more than one study. This is due in part to the diverse experimental approaches used, to the tissue specificity of imprinting and the developmental timing of imprinting. Our approach, based on the identification of putative allelic and parent of origin-specific DNA methylation marks, provided a robust method for the successful identification of candidate DMCpG sites in close proximity to most known as well as novel imprinted genes.
Studies employing the uniparental tissues AnCHM and MCT have demonstrated the importance of parent-of-origin specific contributions in normal development, but actual data regarding the extent of involvement of altered methylation at known imprinted genes was limited to a small number of sites identified by targeted bisulfite PCR (Judson et al. 2002; El-Maarri et al. 2003). The DNA methylation profiles generated by our microarray and pyrosequencing experiments are concordant with the targeted DNA methylation data for known imprinted genes reported in these studies (Judson et al. 2002; El-Maarri et al. 2003). More importantly, our data have substantially expanded the number of documented dysregulated parental-specific methylation marks in AnCHM and MCT supporting the designation of these tissues as uniparental in origin.
Of the 166 DMCpG sites identified, we identified a considerable number of new candidate maternal DMCpGs (57%) versus paternal DMCpGs (16%). The low percentage of new candidate paternally DMCpGs is not surprising because there appears to be a significant bias toward maternally methylated DMRs among the well characterized primary DMRs associated with imprinted genes in human and mouse. Perhaps, very few paternally methylated DMRs are present in the genome. Another possible explanation for the maternal bias could be the array platform used in this study, which is enriched mostly in promoter CpGs, whereas primary paternal DMRs tend to map to intergenic regions (Weaver et al. 2009).
A number of studies have suggested that imprinted genes have characteristic structural features (Hurst et al. 1996; Greally 2002). With respect to primary DMRs, paternally methylated DMRs tend to contain fewer CpGs than maternally methylated DMRs (Kobayashi et al. 2006). In our candidates we found an enrichment of CpG-rich paternal DMRs and CpG-poor maternal DMRs. This could be explained by the fact that our array targets mostly promoter regions of transcribed genes, whereas many of the known maternal DMRs overlap promoter regions of noncoding RNAs that map to intragenic regions (for example, KCNQ1OT1) (Sleutels et al. 2002; Mancini-DiNardo et al. 2003). For the paternal DMRs, another possible explanation is that the subset of candidate paternal DMRs that we identified are secondary DMRs (imprinted post-fertilization), which are known to overlap CpG-rich promoter regions, as is the case for MEG3 and NESP55 DMRs (Peters et al. 1999; Kagami et al. 2010).
One of the most important aspects of the analyses of this data set was that it led to the identification of all known DMRs associated with imprinted genes that were represented on the microarray; as well as the identification of a number of novel DMCpG sites extended the currently known boundaries of imprinted DMRs, including the H19 DMR, the promoter of SNURF/SNRPN, and the very complex GNAS imprinting cluster. Of interest, two new candidate DMCpG sites overlapping the CpG island at the promoter of SNURF/SNRPN that were identified in our study are located in a region that has been recently reported as a novel DMR in a comparison of methylation profiles of maternal versus paternal UPD 15 (Sharp et al. 2010).
The analyses of our data also permitted the identification and validation of two new DMRs that are associated with known imprinted genes. Overlapping the promoter region of the imprinted transcript NAP1L5 on chromosome 4q22.1, we found differential methylation at several CpG sites. A mouse ortholog of NAP1L5 has been described (Smith et al. 2003) and the maternally methylated mouse DMR has been reported (Smith et al. 2003). These results are in concordance with the known paternal expression of this gene in human and mouse (Wood et al. 2007). Furthermore, we have identified and validated the differential methylation at the paternally methylated DMR overlapping the promoter region of ZNF597 on chromosome 16. The observed paternal methylation is in concordance with the known maternal expression of this gene in human (Pant et al. 2006). Our map of new imprinted DMCpGs and new boundaries for known imprinted DMRs will be of particular interest to scientists who are dissecting the structure and function of these DMRs with respect to the transcriptional regulation of imprinted genes.
A third novel imprinted DMR identified in this investigation is the DMR overlapping the promoter region of AXL on chromosome 19q13.2. We validated imprinted features of the human AXL DMR and gene using several experimental approaches. We performed methylation analyses using multiple methodologies all based on sodium bisulfite modified DNA and were able to assess DNA methylation at 14 CpG sites overlapping the transcription start site of AXL. Allelic expression studies in humans identified preferential maternal monoallelic expression in one of four informative individuals, suggesting polymorphic imprinting of the gene in human peripheral blood. These data are similar to those found for the IGF2R gene, which is imprinted in mouse but biallelically expressed in humans (Killian et al. 2001) despite having an intronic DMR (Smrzka et al. 1995). We assayed allelic expression in mouse and showed that the mouse Axl gene exhibits exclusive maternal expression in the early embryo and in extraembryonic tissues during early development. However, we were not successful in finding differential DNA methylation in mouse at the sequence orthologous to the human DMR (data not shown). Nevertheless, the maintenance DNA methyltransferase DNMT1, which is known to ensure the faithful copying of germline imprinted methylation marks through subsequent cell divisions (Constancia et al. 2004), does indeed regulate the monoallelic expression of Axl in mice. Dnmt1 mutant mice lost the monoallelic expression of Axl in extraembryonic tissues suggesting that Axl imprinting in mouse is also DNA methylation dependent. Several reports have suggested that genes that show preferential extraembryonic imprinted expression can be located further away from the imprinting control region (for recent review, see Hudson et al. 2010) suggesting that the Axl imprinting control region in mice might be some distance away. The developmental timing and tissue-specific profile of Axl gene expression is analogous to other reported imprinted genes (Yu et al. 1998; Hayward et al. 2001); that is monoallelic expression occurs only at certain stages of development in mouse and later imprinting is confined to extraembryonic tissue. The persistence of the differential methylation mark beyond the narrow developmental time frame and tissue-specific pattern of monoallelic expression permitted the identification of this new imprinted gene in humans first and then in mice. The human AXL gene is located on chromosome 19q13.2, which is centromeric to the known cluster of imprinted genes PEG3/ZIM2 (chromosome19q13.43). Interestingly, among all human chromosomes, chromosome 19 is the only chromosome for which no cases of UPD have been reported to date (Liehr 2010). This raises the possibility that uniparental inheritance of two copies of chromosome 19 may be lethal and that biparental chromosome 19 carries epigenetic marks indispensable for normal development. Of potential interest in this regard is the recent identification of mutations in NALP7, a gene on chromosome 19q, in recurrent familial hydatidiform moles (Murdoch et al. 2006).
The identification of new imprinted genes is important in furthering our understanding of normal human growth and neurodevelopment. With respect to AXL, an essential epithelial-to-mesenchymal transition-induced regulator (Gjerdrum et al. 2010), this gene has also been shown to play a critical role in human cancer pathogenesis (Linger et al. 2008). These data are consistent with the general hypothesis that imprinted genes involved in regulating early growth and development are also potential human cancer susceptibility loci (Feinberg et al. 2006). Understanding that AXL expression is regulated in some tissues by DNA methylation in a parent of origin-specific manner could facilitate the development of treatment modalities relevant to cancers that involve dysregulation of AXL.
In addition, the lack of DMRs in mice for Axl could suggest that the latter has developed alternative imprinting mechanisms that differ between species as has been previously proposed for Igf2r (Monk et al. 2006) or that the mouse DMR is yet to be identified and does not necessarily overlap with the orthologous human region. Furthermore, we have shown that imprinting of Axl in mice is mostly specific to extraembryonic tissues in early gestation whereas in humans we have clearly demonstrated differential DNA methylation and a skewed allelic expression suggesting polymorphic imprinting of AXL in peripheral blood. These data suggest that AXL imprinted status is tissue- and species-specific; further studies are needed to elucidate the range of mechanisms (for example, histone modifications), that might participate in imprint regulation at AXL in humans and mice.
We have shown that our unique approach of identifying CpG sites that demonstrate differential methylation in tissues of biparental origin and parent of origin-specific methylation in uniparental tissues can be used to identify new DMRs associated with novel imprinted genes. In particular, this approach permitted the identification of new candidate DMRs as well as the characterization of a new candidate imprinted gene, AXL, in humans. This is one of the few instances of an imprinted gene being identified first in humans and then in mice. Importantly, it sets the stage for the identification and characterization of genes that might be imprinted in humans but not in mice or other model organisms. One potential caveat is that the array platform that we used identified differential CpG methylation at individual loci, which requires extensive downstream validation. Nonetheless, we have proven that although we are assaying differential methylation at single CpG resolution, we were successful in detecting significant differential methylation at all known DMRs and identifying new DMRs associated with known and new imprinted genes. Therefore, our approach using profiling of DNA methylation in uniparental and biparental tissues revealed greater promise in identifying DMRs that could reside in close proximity to as yet unidentified imprinted genes. We expect that applying this particular approach in concert with recent sequencing technologies will lead to the full characterization of the imprintome in humans.
Methods
Tissue samples and DNA preparation
Uniparental tissues
Three androgenetic complete hydatidiform moles (AnCHMs) were analyzed in the study. DNA samples from two AnCHMs (AnCHM1 [also known as AnCHM23] [El-Maarri et al. 2003] and AnCHM2) were obtained from McGill University Health Center, Montreal, Canada, and 1 (AnCHM3) from Mt Sinai Hospital, Toronto, Canada. All three were identified as complete androgenetic hydatidiform moles by microsatellite analyses (data not shown). Mature cystic ovarian teratoma (MCT also known as P315) tissue was obtained from the Biopathology Center, CHTN Pediatric Division, Nationwide Children's Hospital, Columbus, Ohio.
Samples were collected from individuals with chromosomes of uniparental origin as follows: WBC from one case of maternal uniparental disomy 14 (UPD14) and one case of paternal UPD14 as well as two lymphoblastoid cell lines, one paternal UPD11p15 and one paternal UPD4.
Biparental tissues
Five placenta samples (greater than 25 wk gestation) were collected through the placental Biobank at Mount Sinai Hospital, Toronto, Canada, as previously described (Guo et al. 2008). Sixteen WBC samples (age and sex matched) were collected from healthy individuals at the Hospital for Sick Children, Toronto, Canada, following informed consent. Placental and peripheral blood DNA were extracted as previously described (Guo et al. 2008).
Methylation analysis
Bisulfite modification
Genomic DNA (1μg) was sodium bisulfite modified using EpiTect Bisulfite kit (Qiagen). Bisulfite modified DNA was hybridized to the Illumina Infinium Human Methylation27 array and used as the template for targeted pyrosequencing assays.
Microarray analyses
The Infinium Human Methylation27 BeadChip supports single CpG-site resolution, using two site-specific probes (both 50 base pairs long) for each targeted CpG site. One site-specific probe targets the methylated CpG site and the other site-specific probe targets the unmethylated CpG site. The targeted CpG sites are located at the ends of the 50 base pair oligonucleotides allowing quantification of methylation by single-base extension of the probes using a labeled ddNTP. This array targets CpG sites located within the proximal promoter regions of transcription start sites of 14,475 consensus coding sequences (CCDS) in the NCBI Database (Genome Build 36). In addition, 254 array CpG sites cover 110 miRNA promoters. On average, each CCDS gene represented on the array is covered by two array CpG sites. Over 200 cancer-related genes and imprinted genes are represented on the array by a variable number of CpG sites (3–20/gene).
All data were analyzed using the BeadStudio Methylation Module 3.2.0 from Illumina. The CpG methylated percentage for the interrogated locus was determined by dividing the intensity of the C (methylated) signal by the total intensity of the C (methylated) and T (unmethylated) signal. Average normalization was performed to allow comparisons between samples run on different microarray chips. The AnCHM group was chosen as the reference group because it consists of a uniparental tissue group with more than one sample.
Microarray statistical analysis
For differential methylation analysis we performed Mann-Whitney tests with correction for multiple testing using q-value False Discovery Rate, for a false positive rate of <1%. Prior to further analyses, probes with high background hybridization signals (detection p-value>0.05) and sex chromosome CpG sites were filtered out. Approximately 26,486 CpG sites remained in each sample. We calculated the median and the interquartile ranges for each CpG site for the AnCHMs, WBCs, and placentas within each group followed by the calculation of CpG methylation differences between the median of each biparental group and the AnCHM at each CpG site. We then selected the AnCHM sample that had the highest level of disagreement (i.e., highest methylation difference as measured by the most negative intraclass correlation coefficient [ICC]) with the MCT for the probes mapping to known imprinted DMRs (see below). We then chose this AnCHM sample (AnCHM1) to calculate the methylation difference between the two opposite uniparental tissues (AnCHM and MCT) at each CpG site.
The list of Illumina Infinium Human Methylation27 BeadChip microarray CpG sites located in DMRs known to help regulate genomic imprinting was compiled using journal articles listed in the OTAGO imprinted gene catalogue (www.otago.ac.nz/IGC). CpG sites were removed from this list if their Mann-Whitney statistical test comparing methylation in the AnCHMs to control WBCs was not significant (i.e., p-value > 0.05). CpG sites were also removed from this list if they had unexpected methylation patterns. In paternally methylated DMRs we expect AnCHMs to have 100% CpG methylation, MCTs to have 0% CpG methylation, WBCs and placentas to have 50% CpG methylation. However, the CpG methylation values observed for one paternally methylated DMR, NESP55, were 87% in AnCHMs, 11% in MCTs, 42% in WBCs, and 55% in placentas. Then for each CpG site, we measured the differences between the selected AnCHM and the MCT. Using the range of the observed methylation difference values at known imprinted DMRs, we established the minimum and maximum values for microarray CpG sites in the compiled list of CpG sites in known DMRs (with respect to whether the DMR is maternally/paternally methylated). This analysis provided us with the cutoffs (maximum and minimum methylation difference) for maternally and paternally methylated DMRs and were used as criteria for selection of candidate differentially methylated CpG sites. We tested these criteria by their ability to pick all CpG sites on our list of probes mapped to known imprinted DMRs (Supplemental Table 1). We then measured the methylation levels, for the candidate regions, in blood and placenta, in order to confirm the expected 50% methylation levels in these tissues.
To identify candidate paternally methylated CpG sites we adapted the criteria for candidate maternally methylated CpG sites because the criteria for candidate paternally methylated CpG sites were too strict (allowed less variation). This may be due to the scarcity of probes in known paternally methylated DMRs.
DNA methylation analysis by sodium bisulfite sequencing
A total of three samples (MCT, AnCHM1, placenta2) were investigated for AXL promoter region DNA methylation by sequencing of sodium bisulfite-modified DNA. Fourteen CpG sites were evaluated for methylation within a 355-bp amplicon spanning the AXL transcription start site (chr19: 41725115–41725469; UCSC Genome Browser at http://genome.ucsc.edu/). Primers are listed in Supplemental Table 4. DNA was treated with sodium bisulfite as described above. PCR was performed for 45 cycles using HotStar Taq DNA polymerase (Qiagen, Hilden, Germany). PCR products were cloned into the TA vector pCR4-TOPO (Invitrogen) and plasmid DNA was extracted from the resulting clones with the use of a GeneElute Plasmid Miniprep kit (Sigma). Fifteen to 20 independent plasmid clones were selected and their nucleotide sequences were determined. We also compared the sequencing after subcloning of bisulfite-modified DNA to the direct sequencing approach and obtained similar results for both methods for the same placental sample at selected CpG sites.
Pyrosequencing analysis
Quantitative sodium bisulfite pyrosequencing was performed for KvDMR1, H19 DMR, and for IG-DMR (not covered on Illumina array) and candidate DMRs for genes NAP1L5, ZNF597, and AXL. All targeted assays were designed using the PyroMark Assay Design Software 1.0 (Qiagen). All primer sets are listed in Supplemental Table 4. Sodium bisulfite modified genomic DNA was amplified using Hot-Start Taq master mix (Qiagen) as previously described (Guo et al. 2008). Regions of interest were amplified by PCR and pyrosequencing was carried out using the PyroMark Q24 pyrosequencer (Qiagen) according to the manufacturer's protocol (PyroGold reagents). Output data were analyzed using PyroMark Q24 1.0.10 Software (Qiagen), which calculates the methylation percentage (mC/(mC+C)) for each CpG site, allowing quantitative comparisons.
Allelic expression analysis in human
We performed allelic expression analysis for AXL using available genotyping data from HapMap samples. RNA from four informative heterozygous individuals identified on the Illumina 1M DUO_v3 was kindly provided by the Center for Applied Genomics, Toronto, Canada. cDNA was synthesized with SuperScript III reverse transcriptase (Invitrogen) following the manufacturer's protocol. Primers used for AXL-SNP genotyping are listed in Supplemental Table 4. Pyrosequencing was done on genomic DNA and cDNA as described above. The relative level of the two parental alleles was quantified by the PyroMark Q24 1.0.10 Software (Qiagen) using the allele quantification method.
Allelic expression analysis in mice
RNA from blastocysts was isolated using Dynabeads (Dynal Biotech) and post-implantation embryos using High Pure RNA Tissue Kit (Roche Molecular Biochemicals) according to manufacturer's recommendation. cDNA was generated using Superscript III Reverse Transcriptase and random primers. Axl primers (final concentration 0.375 μM), AxlL (5′TCCACTTGACTGGCATCTTG3′), and AxlR (5′CCCAGAACCTGTGGTCATCT3′) were used in PCR (94°C for 2 min followed by 35 cycles at 94°C for 10 sec, 60°C for 15 sec, 72°C for 1 min, and 72°C for 2 min) to amplify a 308 bp fragment containing a polymorphism between B6 (A) and C7 (G) [position 26,548,329 UCSC Genome Browser Mouse July 2007]. Restriction digestion with Hpy188III resulted in 134 and 174 bp fragments in B6 and a 308 bp fragment in C7. Parental specific expression patterns for Axl were calculated as the percentage expression of the B6 or C7 alleles relative to the total expression of both alleles based on DNA band intensity using a digital gel documentation system. Biased maternal expression was defined as ≥75% of total expression.
Data release
The WBC, placenta, AnCHM, and MCT raw and analyzed DNA methylation data generated from this project can be downloaded from NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE22091.
Acknowledgments
We thank Chunhua Zhao, Yi-An Chen, and Sarah Goodman for their technical assistance. We thank the donors, and the Research Centre for Women's and Infants' Health BioBank of Mount Sinai Hospital for the human specimens used in this study. J.S.S. is funded by a Canadian Institute for Health Research (CIHR) training scholarship. D.T.B. is an Ontario Mental Health Foundation (OMHF) scholar. J.C.F. is supported by a scholarship from FCT (Science and Technology Foundation), Lisbon, Portugal (SFRH / BD / 28642 / 2006) funded by POPH (Operational Program for Human Potential) coparticipated by FSE (European Social Fund) and by national funds from MCTES (Ministry of Science, Technology, and Higher Education). Funding for D.G. was provided by the Autism Training Research Program (McGill University, Montreal). S.W.S. holds the GlaxoSmithKline-CIHR Pathfinder Chair in Genetics and Genomics at the University of Toronto and the Hospital for Sick Children. R.S. is supported by the Fonds de la recherche en Santé (FRSQ) and CIHR (MOP86546). M.S.B. is supported by the National Institute of Health (NIH) grant GM51279. R.W. is supported by CIHR (MOP86758; MOP89933).
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE22091.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.111922.110.
References
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87 [DOI] [PubMed] [Google Scholar]
- Buiting K 2010. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet 154C: 365–376 [DOI] [PubMed] [Google Scholar]
- Choufani S, Shuman C, Weksberg R 2010. Beckwith-Wiedemann syndrome. Am J Med Genet C Semin Med Genet 154C: 343–354 [DOI] [PubMed] [Google Scholar]
- Constancia M, Kelsey G, Reik W 2004. Resourceful imprinting. Nature 432: 53–57 [DOI] [PubMed] [Google Scholar]
- Deltour L, Montagutelli X, Guenet JL, Jami J, Paldi A 1995. Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Developmental Biology 168: 686–688 [DOI] [PubMed] [Google Scholar]
- Dockery L, Gerfen J, Harview C, Rahn-Lee C, Horton R, Park Y, Davis TL 2009. Differential methylation persists at the mouse Rasgrf1 DMR in tissues displaying monoallelic and biallelic expression. Epigenetics 4: 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Seoud M, Coullin P, Herbiniaux U, Oldenburg J, Rouleau G, Slim R 2003. Maternal alleles acquiring paternal methylation patterns in biparental complete hydatidiform moles. Hum Mol Genet 12: 1405–1413 [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S 2006. The epigenetic progenitor origin of human cancer. Nat Rev Genet 7: 21–33 [DOI] [PubMed] [Google Scholar]
- Giannoukakis N, Deal C, Paquette J, Kukuvitis A, Polychronakos C 1996. Polymorphic functional imprinting of the human IGF2 gene among individuals, in blood cells, is associated with H19 expression. Biochem Biophys Res Commun 220: 1014–1019 [DOI] [PubMed] [Google Scholar]
- Gjerdrum C, Tiron C, Hoiby T, Stefansson I, Haugen H, Sandal T, Collett K, Li S, McCormack E, Gjertsen BT, et al. 2010. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proc Natl Acad Sci 107: 1124–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, Scherer SW, Weksberg R 2009. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics 95: 73–83 [DOI] [PubMed] [Google Scholar]
- Greally JM 2002. Short interspersed transposable elements (SINEs) are excluded from imprinted regions in the human genome. Proc Natl Acad Sci 99: 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, Uxa R, Keating S, Kingdom J, Weksberg R 2008. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol 320: 79–91 [DOI] [PubMed] [Google Scholar]
- Hayward BE, Barlier A, Korbonits M, Grossman AB, Jacquet P, Enjalbert A, Bonthron DT 2001. Imprinting of the G(s)alpha gene GNAS1 in the pathogenesis of acromegaly. J Clin Invest 107: R31–R36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, Miyanari Y, Horiike T, Li Y, Yaegashi N, Sasaki H, et al. 2010. A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mosuse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res 38: 4929–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson QJ, Kulinski TM, Huetter SP, Barlow DP 2010. Genomic imprinting mechanisms in embryonic and extraembryonic mouse tissues. Heredity 105: 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, McVean G, Moore T 1996. Imprinted genes have few and small introns. Nat Genet 12: 234–237 [DOI] [PubMed] [Google Scholar]
- Ideraabdullah FY, Vigneau S, Bartolomei MS 2008. Genomic imprinting mechanisms in mammals. Mutat Res 647: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IHGSC 2004. Finishing the euchromatic sequence of the human genome. Nature 431: 931–945 [DOI] [PubMed] [Google Scholar]
- Judson H, Hayward BE, Sheridan E, Bonthron DT 2002. A global disorder of imprinting in the human female germ line. Nature 416: 539–542 [DOI] [PubMed] [Google Scholar]
- Kagami M, Sekita Y, Nishimura G, Irie M, Kato F, Okada M, Yamamori S, Kishimoto H, Nakayama M, Tanaka Y, et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat Genet 40: 237–242 [DOI] [PubMed] [Google Scholar]
- Kagami M, O'Sullivan MJ, Green AJ, Watabe Y, Arisaka O, Masawa N, Matsuoka K, Fukami M, Matsubara K, Kato F, et al. 2010. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 6: e1000992 doi: 10.1371/journal.pgen.1000992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Ishino T, Kuroiwa Y, Miyoshi N, Kohda T, Suzuki R, Yokoyama M, Viville S, Barton SC, Ishino F, Surani MA 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat Genet 11: 52–59 [DOI] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40: 904–908 [DOI] [PubMed] [Google Scholar]
- Killian JK, Nolan CM, Wylie AA, Li T, Vu TH, Hoffman AR, Jirtle RL 2001. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet 10: 1721–1728 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Suda C, Abe T, Kohara Y, Ikemura T, Sasaki H 2006. Bisulfite sequencing and dinucleotide content analysis of 15 imprinted mouse differentially methylated regions (DMRs): paternally methylated DMRs contain less CpGs than maternally methylated DMRs. Cytogenet Genome Res 113: 130–137 [DOI] [PubMed] [Google Scholar]
- Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Frigge ML, et al. 2009. Parental origin of sequence variants associated with complex diseases. Nature 462: 868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L, Wong E, Li G, Huynh T, Tsirigos A, Ong CT, Low HM, Kin Sung KW, Rigoutsos I, Loring J, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res 20: 320–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R 1993. Role for DNA methylation in genomic imprinting [see comments] Nature 366: 362–365 [DOI] [PubMed] [Google Scholar]
- Liehr T 2010. Cytogenetic contribution to uniparental disomy (UPD). Mol Cytogenet 3: 8 doi: 10.1186/1755-8166-3-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linger RM, Keating AK, Earp HS, Graham DK 2008. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res 100: 35–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ 2007. Computational and experimental identification of novel human imprinted genes. Genome Res 17: 1723–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-DiNardo D, Steele SJ, Ingram RS, Tilghman SM 2003. A differentially methylated region within the gene Kcnq1 functions as an imprinted promoter and silencer. Hum Mol Genet 12: 283–294 [DOI] [PubMed] [Google Scholar]
- Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS 2003. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol Reprod 69: 902–914 [DOI] [PubMed] [Google Scholar]
- Maynard ND, Chen J, Stuart RK, Fan JB, Ren B 2008. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat Methods 5: 307–309 [DOI] [PubMed] [Google Scholar]
- Monk D, Arnaud P, Apostolidou S, Hills FA, Kelsey G, Stanier P, Feil R, Moore GE 2006. Limited evolutionary conservation of imprinting in the human placenta. Proc Natl Acad Sci 103: 6623–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch S, Djuric U, Mazhar B, Seoud M, Khan R, Kuick R, Bagga R, Kircheisen R, Ao A, Ratti B, et al. 2006. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 38: 300–302 [DOI] [PubMed] [Google Scholar]
- Mutter GL 1997. Role of imprinting in abnormal human development. Mutat Res 396: 141–147 [DOI] [PubMed] [Google Scholar]
- Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA 2006. Analysis of allelic differential expression in human white blood cells. Genome Res 16: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Wroe SF, Wells CA, Miller HJ, Bodle D, Beechey CV, Williamson CM, Kelsey G 1999. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc Natl Acad Sci 96: 3830–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Serre D, Wang X, Tao H, Grundberg E, Hudson TJ, Clark AG, Frazer K 2008. A genome-wide approach to identifying novel-imprinted genes. Hum Genet 122: 625–634 [DOI] [PubMed] [Google Scholar]
- Rakyan V, Down T, Thorne N, Flicek P, Kulesha E, Graf S, Tomazou E, Backdahl L, Johnson N, Herberth M, et al. 2008. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res 18: 1518–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp AJ, Migliavacca E, Dupre Y, Stathaki E, Sailani MR, Baumer A, Schinzel A, Mackay DJ, Robinson DO, Cobellis G, et al. 2010. Methylation profiling in individuals with uniparental disomy identifies novel differentially methylated regions on chromosome 15. Genome Res 20: 1271–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Smith RJ, Dean W, Konfortova G, Kelsey G 2003. Identification of novel imprinted genes in a genome-wide screen for maternal methylation. Genome Res 13: 558–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrzka OW, Fae I, Stoger R, Kurzbauer R, Fischer GF, Henn T, Weith A, Barlow DP 1995. Conservation of a maternal-specific methylation signal at the human IGF2R locus. Hum Mol Genet 4: 1945–1952 [DOI] [PubMed] [Google Scholar]
- Strichman-Almashanu LZ, Lee RS, Onyango PO, Perlman E, Flam F, Frieman MB, Feinberg AP 2002. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res 12: 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA, Sasaki H, Ferguson-Smith AC, Allen ND, Barton SC, Jones PA, Reik W 1993. The inheritance of germline-specific epigenetic modifications during development. Philos Trans R Soc Lond B Biol Sci 339: 165–172 [DOI] [PubMed] [Google Scholar]
- Wang X, Sun Q, McGrath SD, Mardis ER, Soloway PD, Clark AG 2008. Transcriptome-wide identification of novel imprinted genes in neonatal mouse brain. PLoS ONE. 3: e3839. doi: 10.1371/journal.pone.0003839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver JR, Susiarjo M, Bartolomei MS 2009. Imprinting and epigenetic changes in the early embryo. Mamm Genome 20: 532–543 [DOI] [PubMed] [Google Scholar]
- Williamson CM, Blake A, Thomas S, Beechey CV, Hancock J, Cattanach BM, Peters J 2010. MRC Harwell, Oxfordshire. World Wide Web Site—Mouse Imprinting Data and References. http://www.har.mrc.ac.uk/research/genomic_imprinting/
- Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ 2007. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet 3: e20 doi: 10.1371/journal.pgen.0030020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Yu D, Lee E, Eckhaus M, Lee R, Corria Z, Accili D, Westphal H, Weinstein LS 1998. Variable and tissue-specific hormone resistance in heterotrimeric Gs protein alpha-subunit (Gsalpha) knockout mice is due to tissue-specific imprinting of the gsalpha gene. Proc Natl Acad Sci 95: 8715–8720 [DOI] [PMC free article] [PubMed] [Google Scholar]