Abstract
Cortical interneurons originate from subpallial precursors and migrate into the cortex during development. Using genetic lineage tracing in transgenic mice we examine the contribution of two germinal zones, the septum and the lateral ganglionic eminence/caudal ganglionic eminence (LGE/CGE) to interneurons of the cortex. We find that the septal neuroepithelium does not generate interneurons for the neocortex. There is, however, clear migration of cells from the LGE/CGE to the cortex. Comparison of the dynamics of cortical colonization by the two major cohorts of interneurons originating in the medial ganglionic eminence (MGE) and the LGE/CGE has shown differences in the timing of migration and initial route of entry into the cortex. LGE/CGE-derived interneurons enter the cortex later than the MGE-derived ones. They invade the cortex through the subventricular/intermediate zone route and only later disperse within the cortical plate and the marginal zone. During the first postnatal week MGE interneurons move extensively to acquire their laminar position within the cortical plate whereas LGE/CGE-derived cells remain largely within the upper layers of the cortex. The two populations intermingle in the adult cortex but have distinct neurochemical properties and different overall distributions. LGE/CGE-derived interneurons account for one third of the total GABAergic interneuron population in the adult cortex.
Introduction
GABAergic interneurons in the adult cortex display a large diversity in axonal morphology, neurochemical content and physiological properties. The identity of these cells is specified during development when different neuroepithelial precursor pools generate distinct interneuron subtypes (Batista-Brito and Fishell, 2009). A large number of cortical GABAergic interneurons are born in the medial ganglionic eminence (MGE). Transplantation studies and mouse mutant analysis (Wonders and Anderson, 2006), as well as more recent genetic fate-mapping work (Fogarty et al., 2007; Miyoshi et al., 2007; Q. Xu et al., 2008; Sousa et al., 2009), have clearly defined the spectrum of interneurons generated from MGE precursors.
Another identified source of cortical interneurons is the caudal ganglionic eminence (CGE) (Nery et al., 2002; Butt et al., 2005; Yozu et al., 2005). The lateral ganglionic eminence (LGE), and its caudal extension, the dorsal CGE (dCGE), are molecularly distinct from the MGE (Flames et al., 2007). The identity of interneurons generated from the LGE/dCGE has been more difficult to define by transplantation studies due to extensive cell migration and mixing in the subpallium. The lack of molecular markers that uniquely identify the LGE/dCGE and distinguish it from the MGE had precluded any comprehensive genetic lineage tracing. More recently, however, an inducible genetic labeling strategy allowed labeling of CGE-derived cells and identification of interneuron subtypes generated from this region (Miyoshi et al., 2010).
The MGE and the dCGE are the two major sources of GABAergic interneurons for the cortex. Other potential sources include the preoptic area (POA) and the septum. A small number of cortical interneurons that represent a homogenous population of cells are indeed generated from the POA and migrate to the cortex (Gelman et al., 2009). Evidence supporting the idea that the septum may be a major source of cortical interneurons has come from mice lacking the transcription factor VAX1, which show complete loss of septal structures and severe reduction of interneurons in the cortex (Taglialatela et al., 2004).
Once they reach the cortex, interneurons from the subpallium continue to migrate tangentially to disperse within the cortex (Ang et al., 2003; Tanaka et al., 2003, 2006, 2009) before switching to a radial migration mode to enter the cortical plate and acquire their final position (Nadarajah et al., 2002; Tanaka et al., 2003; Yokota et al., 2007). Some of the signals that guide their movements within the cortex have been identified (Stumm et al., 2003; Flames et al., 2004; Tiveron et al., 2006; Li et al., 2008; López-Bendito et al., 2008; Stanco et al., 2009). It is unknown whether interneurons with distinct origins follow different routes and cues to reach the cortex.
Using genetic lineage tracing we examined the contribution of septal precursors to cortical neurons. To label LGE/dCGE-derived cells we used a subtractive fate-mapping approach. This allowed us to visualize migrating LGE/dCGE-derived cortical interneurons separately from the MGE-derived ones and follow their routes through the cortex as well as their fate in the adult brain.
Materials and Methods
Transgenic mice.
The Nkx2.1-CreTg and Lhx6-CreTg transgenic mice have been described previously (Kessaris et al., 2006; Fogarty et al., 2007). Mice expressing Cre under control of the Zic4 gene and the Dlx1-Venusfl mice were generated using bacterial artificial chromosome (BAC) transgenic technology. The codon-improved Cre recombinase (iCre) (Shimshek et al., 2002) containing a nuclear localization signal was fused to the translation initiation codon of Zic4 using a PCR-based approach. This was followed by a simian virus 40 (SV40) polyadenylation signal and a bacterial kanamycin selection cassette that was flanked by FRT sites. For Dlx1-Venusfl a loxP site was inserted in exon 1, 345 bp upstream of the ATG. The Venus open reading frame (ORF) (Nagai et al., 2002; Shimshek et al., 2002) was fused to the ATG of Dlx1, followed by a downstream loxP site, an SV40 polyA and the removable bacterial selection cassette. Recombination resulted in removal of 310 bp downstream of the Dlx1 ATG in exon 1. BAC modification was performed in a bacterial system as described previously (Lee et al., 2001). The bacterial antibiotic selection cassettes were removed before microinjection of the constructs into fertilized eggs. All mice used in this study were maintained on a mixed C57BL/6/CBA background. Mouse colonies were maintained at the Wolfson Institute for Biomedical Research, University College London, in accordance with United Kingdom legislation.
Three reporter mice for Cre recombinase have been used in this study: Rosa26R–green fluorescent protein (GFP) (Mao et al., 2001) (obtained from The Jackson Laboratory), Rosa26R-LacZ (Soriano, 1999), and Rosa26R–yellow fluorescent protein (YFP) (Srinivas et al., 2001). Upon Cre-mediated recombination, the three mice express GFP, LacZ and YFP, respectively, under control of the Rosa26 promoter.
Tissue preparation.
The day of the vaginal plug was considered embryonic day 0.5 (E0.5). For postnatal studies, the birth date was considered day 0. Unless otherwise stated, whole embryo heads (for embryos younger than E13.5) or isolated brains (for embryos older than E13.5) for immunohistochemistry or in situ hybridization were fixed overnight by immersion in 4% (w/v) paraformaldehyde (PFA) in PBS. Postnatal animals were anesthetized and perfused with 4% (w/v) PFA through the left ventricle of the heart. Adult brains [older than postnatal day 30 (P30)] were removed from surrounding tissue, sliced into 2 mm slices using a rodent brain matrix (Agar Scientific), and immersed in 4% PFA overnight. Fixed samples were cryoprotected overnight by immersion in 20% (w/v) sucrose/PBS that had been pretreated with diethyl pyrocarbonate (DEPC) (Sigma), embedded in optimal cutting temperature (OCT) compound (Tissue Tek; Raymond Lamb Ltd Medical Supplies) and frozen on dry ice. All samples were stored at −80°C until needed.
In situ hybridization.
Solutions used for in situ hybridization were pretreated with 0.1% DEPC. Sections 18 μm in thickness were collected onto Superfrost Plus microscope slides (VWR International) and allowed to air dry. Hybridization buffer containing digoxigenin (DIG)-labeled antisense RNA probe (prepared according to manufacturer's instructions and diluted 1:1000 in hybridization buffer), was applied onto the slides and sections were covered with a coverslip. The hybridization buffer was made up with 50% v/v deionized formamide (Sigma), 1× “Salts” [2 m NaCl, 50 mm EDTA, 100 mm Tris-HCl, pH7.5, 50 mm NaH2PO4 · 2H2O, 50 mm Na2HPO4, 0.1 mg/ml tRNA from baker's yeast (phenol chloroform extracted, Roche Diagnostics)], 1× Denhardt's solution (Sigma), and 10% w/v dextran sulfate (predissolved in DEPC-treated water and maintained at 4°C as 50% stock). Hybridization was performed at 65°C overnight in a chamber humidified with 50% v/v deionized formamide containing 1× SSC buffer. Sections were washed three times at 65°C for 30 min each in prewarmed wash solution (50% v/v formamide, 1× SSC, 0.1% Tween 20), followed by two washes in 1× MABT (100 mm maleic acid, 150 mm NaCl, pH 7.5, 0.1% Tween 20) at room temperature also for 30 min each. Blocking was performed using 2% w/v Blocking Reagent (Roche Diagnostics), 10% v/v heat-inactivated sheep serum (Sigma) in 1× MABT for 1 h at room temperature and anti-DIG antibody conjugated with alkaline phosphatase (AP) (Roche Diagnostics) diluted 1:1500 in blocking solution was applied overnight at 4°C. Excess anti-DIG antibody was washed 3 times for 15 min each at room temperature in 1× MABT and sections were equilibrated in 100 mm NaCl, 100 mm Tris-HCl, pH 9.5, 0.1% Tween 20 for 10 min. Development was performed at 37°C for 4–8 h with nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate in freshly prepared staining solution containing 100 mm NaCl, 50 mm MgCl2, 100 mm Tris-HCl, pH 9.5, and 0.1% Tween 20. To increase sensitivity, 5% (w/v) polyvinyl alcohol was included during staining. Sections were washed in tap water and gradually dehydrated in ethanol before being mounted in DePex mounting medium (VWR International).
To detect Nkx2.1 and Zic4 transcripts simultaneously, the sections were hybridized with Nkx2.1-DIG and Zic4-FLUO-labeled RNA probes. Following washes and blocking of the sections as described above, anti-DIG-AP antibody was applied and developed using Fast Red (Roche) as the substrate. The sections were washed in PBS, endogenous peroxidase activity was blocked by incubation with 3% H2O2 in PBS for 15–30 min and incubated with anti-FLUO-peroxidase (POD)-conjugated antibody (1:1500, Roche), for 1 h at room temperature. Sections were washed 3 times 10 min each in 0.1 m Tris-HCl, pH7.5, 0.15 m NaCl, 0.05% Tween 20, and the POD signal was developed by incubating the sections with Tyramide-FITC amplification buffer (1:50, TSA-Plus, PerkinElmer) for 10 min, at room temperature. Sections were washed 3 times 5 min each in PBS-Triton X-100 and mounted with Dako Fluorescence Mounting Medium.
For in situ hybridization followed by immunohistochemistry (for simultaneous detection of Gad67 transcripts and Venus) on adult brain sections, the Gad67-DIG RNA probe was detected with an anti-DIG-POD-conjugated antibody followed by incubation with Tyramide-Cy3 (1:100, TSA-Plus, PerkinElmer) for 10 min at room temperature. The reaction was stopped with 3% H2O2 before the sections were used for immunodetection of Venus (see Immunohistochemistry, below).
The following plasmids were used to generate RNA probes: IMAGE clone 5694793 for Zic1 (linearized with AscI and transcribed with T3 RNA polymerase), IMAGE clone 2779975 for Zic2 (linearized with EcoRI and transcribed with T3 RNA polymerase), IMAGE clone 6400880 for Zic4 (linearized with EcoRI and transcribed with T3 RNA polymerase), IMAGE clone 374236 for Gad1 (Gad67) (linearized with XhoI and transcribed with T3) and a fragment from the Nkx2.1 cDNA (linearized with EcoRI and transcribed with T7 RNA polymerase) (a kind gift from S. Kimura, National Cancer Institute, National Institutes of Health, Bethesda, MD).
Immunohistochemistry.
Immunohistochemical detection was performed as described previously (Fogarty et al., 2007). Briefly, immunolabeling of calbindin (CB), calretinin (CR), parvalbumin (PV), somatostatin (SST), neuropeptide Y (NPY), reelin (RLN), vasoactive intestinal peptide (VIP), and Venus on adult mice was performed on 30 μm floating cryosections (postnatal day 30 or older). Sections from embryonic brains (18 μm thickness) were collected directly onto Superfrost plus slides. All sections were blocked at room temperature in PBS containing 10% heat-inactivated sheep serum (Sigma) and 0.1% Triton X-100 (Sigma) for a minimum of 1 h. For immunodetection of Venus or GFP using the rat anti-GFP antibody, sections were not allowed to dry out at any stage before application of the primary antibody. Primary antibodies were applied overnight at 4°C. Secondary antibodies were applied for >60 min at room temperature. All antibodies were diluted in block solution. Following antibody treatment, sections were treated with Hoescht 33258 (1:104, Sigma) to detect cell nuclei and washed with PBS. Immunostained floating sections were transferred onto Superfrost Plus slides and air-dried. For detection of Venus on slides containing sections previously treated for in situ hybridization for Gad67, the sections were incubated with a chick anti-GFP antibody. The immunosignal was detected with a biotin-conjugated secondary antibody, amplified using the ABC kit (Vector Labs) and developed using Tyramide-FITC as described above. ABC amplification followed by Tyramide-Cy3 development was also used for detection of LHX6. All sections were mounted in Dako Fluorescent Mounting Medium. Antibodies requiring only short fixation of the tissue were the anti-β-gal, anti-VIP and anti-PDGFRa (30 min fixation at room temperature). Antigen retrieval using citrate buffer (Sigma) was performed for detection of Ki67.
Primary antibodies used were the following: rabbit anti-GFP (1:6000, Abcam); rat anti-GFP IgG2a (1:1000, Nacalai Tesque); chicken anti-GFP (1:500, Aves Labs); rabbit anti-β galactosidase (1:2000, MP Biomedicals); rabbit anti-NKX2.1 (1:100, Santa Cruz Biotechnology); rabbit anti-calbindin (1:2000, Swant); rabbit anti-calretinin (1:2000, Swant); mouse anti-parvalbumin (1:1000, Millipore Bioscience Research Reagents); rabbit anti-neuropeptide Y (1:2000, ImmunoStar), rabbit anti-somatostatin (1:200, Peninsula Laboratories), mouse anti-reelin (kindly provided by A. Goffinet, Université Catholique de Louvain, Brussels, Belgium) (1:2000), rabbit anti-vasoactive intestinal peptide (1:500, Insight Biotechnology/ImmunoStar), rabbit anti-neuronal nitric oxide synthase (1:2000, Insight Biotechnology), rabbit anti-collagen IV (1:200, AbD, Serotec), rat anti-PDGFRa (1:500, BD Biosciences), rabbit anti-OLIG2 (1:4000) (kind gift from C. Stiles, Duna-Farber Cancer Institute, Harvard Medical School, Boston, MA, and D. Rowitch, Howard Hughes Medical Institute, University of California, San Francisco, CA), mouse anti-GFAP (1:400, Sigma), mouse anti-NEUN (1:500, Millipore Bioscience Research Reagents), mouse anti-S100β (1:400, Sigma), rat anti-BrdU (1:50, AdB Serotec), mouse anti-ki67 (1:1000, BD Pharmingen). The rabbit anti-LHX6 (1:600–1:1500) antibody used in this study has been described previously (Lavdas et al., 1999). Secondary antibodies used were Alexa Fluor 488-conjugated, Alexa Fluor 568-conjugated, and Alexa Fluor 647-conjugated goat anti-rabbit IgG or goat anti-rat IgG or goat anti-mouse IgG (all used at 1:750; Invitrogen).
Interneuron birth dating with BrdU.
To label adequate numbers of interneurons, E13 and E16 pregnant mothers were given five doses 2 h apart of 2 mg of BrdU (20 mg/ml in PBS) (starting at 10:00 A.M.). The animals were killed 30 d after birth and brain tissue was processed for immunohistochemistry as described above. Following detection of the Venus signal, the floating sections were transferred onto slides and dried before being fixed first with 70% (v/v) ethanol/20% (v/v) acetic acid for 10 min at room temperature followed by 20 min at −20°C in 70% ice-cold ethanol. Sections were next incubated for 15 min at room temperature in 1% (v/v) Triton X-100 in PBS and then in 6 m HCl/1% Triton X-100 in PBS. Following three washes in PBS for 5 min each, anti-BrdU antibody was applied in block solution at 4°C overnight. Washing, secondary antibody incubation and mounting was performed as described above.
Quantification.
For all quantification experiments a minimum of three mice were used for each genotype. The extent of colocalization between GFP/YFP/Venus and the various interneuron markers in each of the transgenic mice (Zic4-CreTg/R26R-YFP, Lhx6-CreTg/Dlx1-Venusfl and Dlx1-Venusfl) was determined as previously described (Fogarty et al., 2007). In all experiments quantification was performed in a defined area of the cortex (1250 μm width and 30 μm depth) spanning the pial–white matter extent of the cortex. In some cases this was subdivided into 5 equal bins and the numbers of cells in each bin was determined.
Results
Zic4-Cre transgenic mice label the septal germinal zone
The Zic family of transcription factors consists of five highly related zinc finger protein-encoding genes (Zic1–5) (Aruga, 2004). Zic1, Zic2 and Zic3 have overlapping patterns of expression and are essential for development of forebrain midline structures during embryogenesis (Nagai et al., 1997; Aruga, 2004; Inoue et al., 2007, 2008; Maurus and Harris, 2009). Zic4 expression has also been reported in the mouse forebrain (Gaston-Massuet et al., 2005). We characterized the detailed expression pattern of Zic4 in the forebrain and generated transgenic mice carrying a genomic BAC that was modified to express Cre under control of Zic4 (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). To fate-map Zic4 precursors and identify their neuronal progeny in the cortex we crossed these mice to the Cre reporter Rosa26-lox-STOP-lox-YFP (hereafter referred to as R26R-YFP) (Srinivas et al., 2001).
Cre activity was observed at E10.5 in midline regions of the telencephalon in Zic4-CreTg/R26R-YFP+/− transgenic embryos (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Analysis of the expression of the endogenous Zic4 gene and the YFP reporter at E12.5 in wild type and transgenic embryos, respectively, showed Zic4 gene activity in the telencephalic midline (Fig. 1A–J). Within the septal ventricular zone all proliferating neuroepithelial cells expressing Ki67, coexpressed YFP (supplemental Fig. 3, available at www.jneurosci.org as supplemental material), indicating that the Zic4-Cre mice can be used to fate-map the septal germinal zone. Expression of Zic4 and YFP partly overlapped with Nkx2.1 (Fig. 1K,L″), which is expressed only in the posterior ventral septum (Flames et al., 2007). In dorsal regions, endogenous Zic4 expression was observed in the cortical hem and the meninges (Fig. 1A–D) as previously reported for Zic1 and Zic2 (Inoue et al., 2008) but activation of the YFP reporter was not detected (Figs. 1F–J,M–O, 2B) suggesting that the Zic4-Cre transgene does not drive expression of Cre in these regions. YFP expression in Zic4-CreTg/R26R-YFP+/− transgenic embryos was restricted to the subpallium and was excluded from Emx1-expressing pallial septal neuroepithelial cells (Fig. 1P,Q).
Figure 1.
Expression of Zic4 and activation of the YFP reporter in wild type and Zic4-CreTg/R26R-YFP+/− embryos. A–E, Expression of Zic4 transcripts in wild type embryos at different rostro-caudal levels of the forebrain at E12.5. Arrows in A point to expression in the meninges. F–J, Expression of YFP in E12.5 Zic4-CreTg/R26R-YFP+/− embryos. The red boxes indicate the areas shown in M–O. K–K″, Coexpression of Zic4 and Nkx2.1 transcripts in the posterior septum in wild type embryos. L–L″, Immunolabeling for YFP and NKX2.1 shows activation of the YFP reporter within the NKX2.1-expressing region of the septum in Zic4-CreTg/R26R-YFP+/− embryos. P, Q, Expression of YFP and Emx1 at E11.5 in Zic4-CreTg/R26R-YFP+/− embryos. Se, Septum; Cx, cortex; hem, cortical hem. Scale bars: A–L″, 300 μm; M–O, 100 μm; P, Q, 150 μm.
Figure 2.
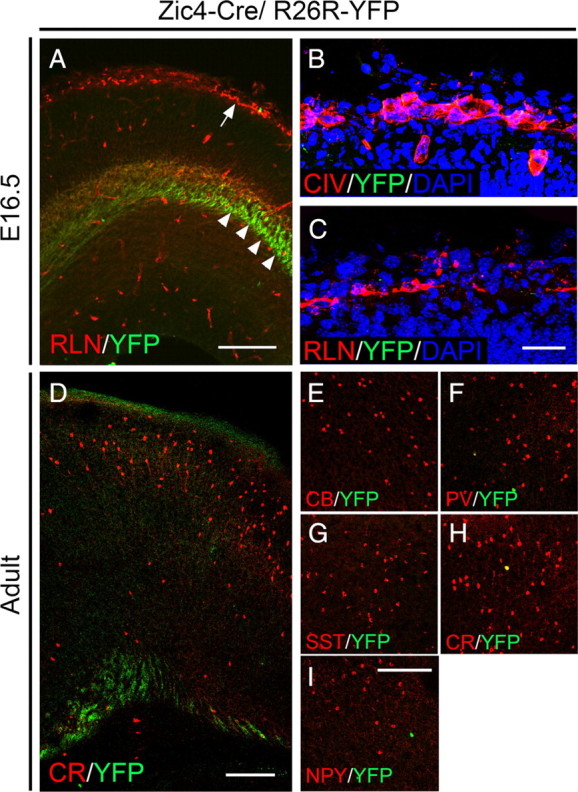
Absence of colocalization between YFP and molecular markers that label the meninges, C–R cells and interneurons in the neocortex in Zic4-CreTg/R26R-YFP+/− transgenic mice. A, E16.5 cortex showing expression of YFP and the C–R cell marker RLN. Some YFP-labeled thalamocortical fibers can be observed (arrowheads). The arrow points to C–R cells in the marginal zone where virtually no expression of YFP is observed. B, C, Absence of coexpression between YFP and Collagen IV (B) or RLN (C). D, Section through the adult cortex showing sparse, if any, YFP-expressing cells. Some YFP-labeled fibers can be observed. E–I, Immunolabeling for YFP and the interneuron markers CB, PV, SST, CR and NPY shows no or very little colocalization with all markers examined. Scale bars: A, 150 μm; B, C, 50 μm; D, 200 μm; E–I, 200 μm.
Subpallial septal precursors do not generate Cajal–Retzius cells or GABAergic interneurons for the cortex
To identify septal-derived cells in the cortex we examined transgenic embryos at E16.5. The cortex was largely devoid of YFP-expressing cells at this stage when cortical interneurons migrating in from the subpallium are normally abundant. Some fibers immunopositive for YFP were detected in the cortex (Fig. 2A, arrowheads). These presumably correspond to fibers originating in the thalamus where Cre recombination is abundant (Fig. 1H–J). There was no expression of YFP within the Collagen IV (CIV)-expressing zone confirming the absence of Cre activity within the meningeal region (Fig. 2B). In addition, none of the Reelin (RLN)-expressing Cajal–Retzius (C–R) cells located in the cortical marginal zone (MZ) underneath the meninges coexpressed YFP (Fig. 2A,C), consistent with recent findings that septal-derived C–R cells have a pallial septal origin (Bielle et al., 2005; Zimmer et al., 2010). Immunolabeling of the adult cortex of Zic4-CreTg/R26R-YFP+/− transgenic animals with a range of markers that detect known cortical interneuron subtypes in mice showed a near complete absence of colocalization at all anterior–posterior levels of the cortex (Fig. 2D–I; supplemental Fig. 4, available at www.jneurosci.org as supplemental material). The occasional YFP+ cells that were observed in the cortex coexpressed LHX6 (98.6% n = 216 cells) (supplemental Fig. 5, available at www.jneurosci.org as supplemental material) suggesting that they are derived from adjacent MGE-territories where sparse Cre activity was sometimes observed (data not shown). Altogether our data indicate that the septum does not generate interneurons for the cortex.
Genetic strategy for subtractive labeling of cell progeny from the LGE/dCGE germinal zone
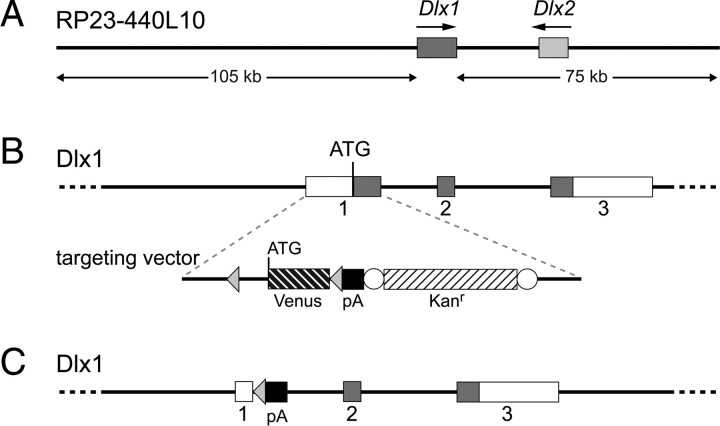
To label the LGE and the dCGE and identify cortical interneurons derived from these regions we adopted a subtractive genetic labeling strategy. The Dlx family of homeobox genes includes six family members in mice, four of which (Dlx1, Dlx2, Dlx5 and Dlx6) are expressed in largely overlapping patterns in the developing forebrain (Bulfone et al., 1993; Simeone et al., 1994; Eisenstat et al., 1999; Liu et al., 2009). At early stages, expression is restricted to the subcortical telencephalon and more caudally in defined diencephalic domains (Bulfone et al., 1993). Later on, interneuron populations migrating from the subpallium to the cortex maintain Dlx expression and all four Dlx genes (Dlx1, Dlx2, Dlx5 and Dlx6) are essential for this migration (Anderson et al., 1997, 1999; Cobos et al., 2005, 2007; Le et al., 2007; Wang et al., 2010). We made use of the expression pattern of Dlx1, to label migrating cortical interneurons as soon as they emerge from the subpallial germinal zones. Previous work using BAC transgenic mice expressing green fluorescent protein (GFP) under control of Dlx1 had shown that BAC RP23–440L10 can drive correct expression of GFP (Cobos et al., 2006) (http://www.gensat.org/). We modified the same BAC by inserting the Venus variant of yellow fluorescent protein into the Dlx1 gene. To allow in vivo subtractive labeling of interneuron subpopulations we flanked Venus with loxP sites (Fig. 3).
Figure 3.
Genetic strategy for subtractive labeling of the LGE/dCGE. A, The unmodified BAC containing the Dlx1 and Dlx2 genes. B, Strategy for modification of the genomic BAC by insertion of Venus flanked by loxP sites. The loxP sites are indicated by arrowheads. A bacterial kanamycin resistance cassette (Kanr) was included in the targeting vector. This was flanked by FRT sites (open circles) and was removed before microinjection of the modified BAC into fertilized eggs. C, Structure of the Dlx1-Venusfl transgene following Cre recombination in transgenic mice.
Dlx1-lox-Venus-loxTg (hereafter referred to as Dlx1-Venusfl) transgenic embryos show high levels of Venus expression in the subcortical telencephalon including the septum, the MGE, LGE and CGE, the POA, the anterior entopeduncular area (AEP) and the amygdala, as previously described for the endogenous Dlx1 gene (Bulfone et al., 1993) (Figs. 4A, 5A–F). Venus is detected in all LHX6-expressing migrating GABAergic interneurons (Fig. 4B–B″) at E15.5 but is excluded from oligodendrocyte lineage cells (Fig. 4C–C″), consistent with its proposed role in regulating neuronal versus oligodendroglial cell fate (Petryniak et al., 2007). Coimmunolabeling of adult cortex from Dlx1-Venusfl mice for Venus and various interneuron markers showed colocalization of Venus with all markers examined [calbindin (CB), parvalbumin (PV), somatostatin (SST), calretinin (CR), neuropeptide Y (NPY), RLN and vasoactive intestinal peptide (VIP)] (Fig. 4D–J′). Unlike the endogenous Dlx gene which is maintained in only a subset of cortical GABAergic interneurons in the adult brain (Cobos et al., 2006), expression of the Dlx1-Venusfl transgene was found to be maintained in the entire cortical GABAergic interneuron population (Fig. 4K,L) as previously reported for this BAC (Cobos et al., 2006). The Dlx1-Venusfl transgenic mouse therefore allows us to follow the development of cortical GABAergic interneurons at all embryonic and adult stages.
Figure 4.

Expression of Venus in Dlx1-Venusfl transgenic mice. A, Widespread expression of Venus can be observed in the subcortical telencephalon at E13.5. B–B″, All LHX6-expressing GABAergic interneurons found in the cortex coexpress Venus. Arrows indicate cells expressing the two markers. C–C″ Absence of colocalization between Venus and PDGFRa in the oligodendrocyte lineage. Arrowheads indicate PDGFRa+ cells. D–J′, Coexpression of Venus with CB, PV, SST, CR, NPY, VIP and RLN. K, The extent of colocalization between Venus and each of the seven markers was quantified, and the data are presented as percentage of the total number of cells expressing each of the markers (% contribution). L, The number of Venus-positive cells coexpressing CB, PV, SST, CR, NPY, VIP or RLN are presented as a percentage of the total number of Venus-positive cells. Data in K and L are presented for the motor and somatosensory cortex. Error bars indicate SD. Scale bars: A, 250 μm; B–C″, 25 μm; D–J′, 60 μm.
Figure 5.
Subtractive genetic labeling of LGE/dCGE-derived cell progeny. A–F, Expression of Venus in Dlx1-Venusfl transgenic embryos at E12.5. Venus can be detected in Dlx1-expressing regions of the forebrain including the septum, the MGE, LGE, CGE, POA, AEP and subdivisions of the diencephalon. G–L, Expression of Venus and β-gal in Nkx2.1-CreTg/Dlx1-Venusfl/R26R-LacZ transgenic embryos at E12.5. Boxes in H indicate the areas shown in M– N″. There is no colocalization between Venus and β-galactosidase confirming efficient removal of Venus from the entire Nkx2.1-Cre lineage. Arrowheads point to β-gal+ cells. Scale bars: A–L, 500 μm; M–N″, 25 μm.
To subtract the Venus tag from MGE and POA-derived cells and allow visualization of interneurons originating outside these regions we crossed the Dlx1-Venusfl mice to ones expressing Cre under control of Nkx2.1 (Fig. 5G–L) (Kessaris et al., 2006; Fogarty et al., 2007). In the presence of the Rosa26-lox-STOP-lox-LacZ (R26R-LacZ) reporter allele we observed efficient removal of the Venus reporter in all Nkx2.1-Cre-expressing cells and their descendants [identified by expression of β-galactosidase (β-gal)] (Fig. 5G–L). These included the posterior septum, the MGE, the POA and the AEP (Fogarty et al., 2007) (Fig. 5H–J). Although Venus expression can be observed throughout the mantle of the telencephalon, it is excluded from all β-gal+ cells confirming the efficient removal of the reporter from the Nkx2.1 lineage (Fig. 5M–N″). Venus-positive cells in the mantle of areas such as the MGE presumably represent migrating cells from Nkx2.1-negative germinal zones.
Our data demonstrate that our Dlx1-Venusfl transgenic mice in combination with Nkx2.1-CreTg can be used to trace cortical interneurons generated outside the MGE. Given that (1) the septum does not generate interneurons for the cortex and (2) Nkx2.1-CreTg recombines in the MGE, AEP and POA and subtracts the label from these germinal zones and their progeny, we can infer that any remaining Venus-positive interneurons migrating to the cortex in Nkx2.1-CreTg/Dlx1-Venusfl embryos are likely to be generated from the LGE/dCGE, with a small contribution from the dMGE where the Nkx2.1-Cre transgene recombines inefficiently (Fogarty et al., 2007).
Interneurons generated from the LGE/dCGE enter the cortex late and migrate initially through the subventricular zone but not the MZ
We compared the distribution of MGE/POA-derived cells in the cortex of Nkx2.1-CreTg/R26R-GFP+/− transgenic embryos, with the distribution of Venus-labeled cells in Nkx2.1-CreTg/Dlx1-Venusfl embryos (Fig. 6). At E13.5 when large numbers of MGE-derived interneurons were present in the cortex and were advanced as far dorsally as the developing motor cortex, only a few LGE/dCGE-derived neurons were present (Fig. 6A–E,K,P). These were all negative for LHX6, even at anterior levels of the cortex, confirming their LGE/dCGE origin (supplemental Fig. 6, available at www.jneurosci.org as supplemental material). At E13.5 there are no POA-derived interneurons in the cortex (Gelman et al., 2009). From E14.5 onwards LGE/dCGE-derived interneurons were abundant at all anterior–posterior regions of the cortex (Fig. 6F–J). Unlike MGE-derived interneurons which enter the cortex initially through the (MZ) and subventricular zone/intermediate zone (SVZ/IZ) (Lavdas et al., 1999) (Fig. 6P,Q), and POA-derived interneurons which invade the cortex through the MZ and subplate (Gelman et al., 2009), LGE/dCGE-derived cells invaded the cortex initially through the SVZ avoiding the cortical plate and the MZ (Fig. 6F–J,L,M,L′,M′). By E16.5, LGE/dCGE-derived interneurons began to invade the cortical plate at all anterior–posterior regions of the cortex (Fig. 6N,N′). Increasing numbers of Venus-positive cells appeared in the MZ from E16.5 onwards (Fig. 6N,N′) and by E17.5 large numbers of these cells were observed in the superficial layers of the developing cortex (Fig. 6O,O′). LGE/dCGE interneurons entered the hippocampal anlage at E14.5-E15.5 and accumulated in abundance by E17.5 (Fig. 6O). This is consistent with previous suggestions that the CGE may be a major source of interneurons for the hippocampus (Nery et al., 2002; Yozu et al., 2005). Altogether our data show that LGE/dCGE-derived interneurons enter the cortex 1–2 d later than the MGE-derived ones. They migrate initially within the deeper SVZ/IZ route and only later join the superficial MZ stream.
Figure 6.
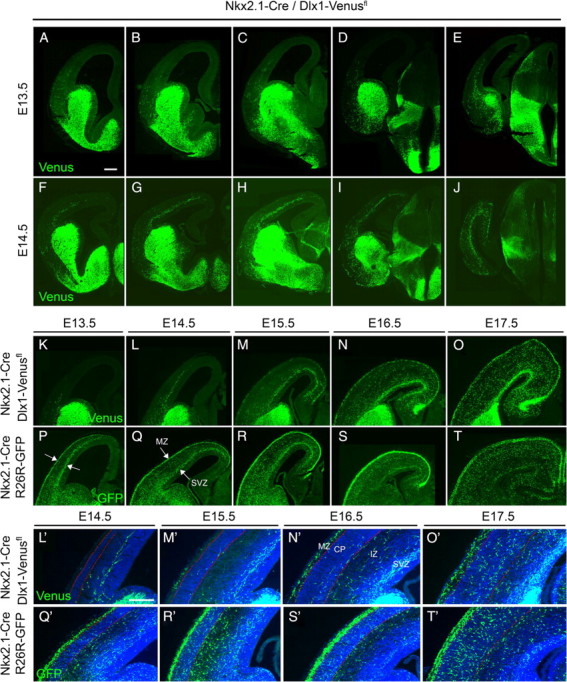
Migration of LGE/dCGE- and MGE-derived interneurons into the cortex during embryogenesis. Migration of LGE/dCGE-derived cells in Nkx2.1-CreTg/Dlx1-Venusfl transgenic mice at E13.5 (A–E) and E14.5 (F–J) at different anterior–posterior levels of the cortex. K–O, Progressive accumulation of LGE/dCGE-derived interneurons in the cortex during embryogenesis. Significant numbers of Venus-positive cells can be detected from E14.5 onwards. Migration initially occurs only within the SVZ/IZ. By E15.5-E16.5 migration within the MZ can be observed. P–T, MGE-derived interneurons in Nkx2.1-CreTg/R26R-YFP embryos migrate into the cortex earlier than LGE/dCGE-derived ones and enter the cortex initially in two migration streams (arrows in P, Q). Sections shown in K–T were taken at the level of the POA. L′–T′, Timing of cortical plate invasion for LGE/dCGE- and MGE-derived cells. Red lines in L′–T′ delineate the cortical plate. CP, cortical plate. Scale bars: A–T, 100 μm; L′–T′, 85 μm.
Distribution and identity of LGE/dCGE-derived interneurons in the postnatal cortex
We examined the postnatal distribution of LGE/dCGE-derived interneurons within the cortex and their identity in the adult animals. For this we made use of a mouse that expresses Cre under control of the Lhx6 gene (Fogarty et al., 2007), which acts downstream of Nkx2.1 in postmitotic neurons (Du et al., 2008). Lhx6-Cre recombines efficiently in all MGE-derived interneurons (Fogarty et al., 2007). It was not possible, however, to use these mice for embryonic studies because we could observe remaining Venus fluorescence in β-gal+ migrating interneurons in Lhx6-CreTg/Dlx1-Venusfl/R26R-LacZ+/− transgenic embryos (data not shown). This was presumably due to expression of Lhx6-Cre in postmitotic neurons, as opposed to VZ precursors, and consequent carry-over of Venus protein inside the cells. Although Lhx6-CreTg does not subtract Venus from Nkx5.1-derived interneurons generated in the POA, the contribution of this region to interneurons of the cortex is small (Gelman et al., 2009). Therefore, the distribution of Venus-positive cells in the cortex of Lhx6-CreTg/Dlx1-Venusfl pups largely reflects that of LGE/dCGE-cells. At P1, the distribution of MGE and LGE/dCGE-derived interneurons within the cortex was similar (Fig. 7A,B) (for quantification see Fig. 7G). The majority of interneurons at this stage were found within the superficial layers. At all embryonic as well as postnatal stages the MGE-derived interneuron population was clearly larger than the LGE/dCGE-derived one (Figs. 6, 7). The distribution of LGE/dCGE-derived interneurons changed little postnatally with the majority remaining within the superficial cortex where they ended up in the adult mouse (Fig. 7A,C,E,G–I). In contrast, MGE-derived cells appeared to shift positions extensively at postnatal stages to gradually distribute themselves across the cortical layers with the majority residing within the middle layers of the cortex (Fig. 7B,D,F,G–I).
Figure 7.

Distribution of LGE/dCGE- and MGE-derived interneurons within the cortex at postnatal stages. A, C, E, LGE/dCGE-derived interneurons in Lhx6-CreTg/Dlx1-venusfl transgenic mice at P1, P3 and in the adult cortex. B, D, F, MGE-derived interneurons in Lhx6-CreTg/R26R-YFP transgenic mice at P1, P3 and in the adult cortex. G–I, Quantification of LGE/dCGE- and MGE-derived interneurons within the somatosensory cortex at P1, P3 and in the adult. The cortex was subdivided into 5 bins along the pial surface-white matter axis (1–5, pial to white matter) and the number of labeled cells in each bin was determined. Data for each bin are presented as a proportion of the total number of labeled cells. Error bars indicate SD. J, Quantification in the adult cortex of the distribution of LGE/dCGE-derived interneurons born at E13 and E16. **p < 0.01, two-way ANOVA test. Error bars indicate SEM. Str, striatum. Scale bar: 150 μm.
Although most LGE/dCGE-derived interneurons were settled in superficial layers of the cortex, some were also present in deeper layers. To determine whether laminar fate is dependent on birthdate, we examined the distribution in the adult cortex of LGE/dCGE-derived interneurons born at E13 and E16. Proportionally more cells were located in the upper layers of the cortex regardless of their birthdate (Fig. 7J). The LGE/dCGE germinal zone generated more interneurons for the cortex at E13 compared with E16 [% of Venus+BrdU+ cells out of the total number of Venus-positive cells was 22.4 ± 0.2 SEM when females were injected with BrdU at E13 and 13.8 ± 4.5 SEM when females were injected with BrdU at E16].
We also examined the identity of Venus-positive cells in the adult cortex of Lhx6-CreTg/Dlx1-Venusfl mice. Venus-positive cells coexpressed NEUN and Gad67 (% of Venus-positive cells coexpressing Gad67 mRNA was 93.7 ± 3.2 SD) but not OLIG2, PDGFRa or GFAP, confirming their GABAergic neuronal identity (supplemental Fig. 7, available at www.jneurosci.org as supplemental material). Colocalization was observed between Venus and the cortical interneuron markers CR, NPY, VIP and RLN, but not PV or SST and very little with CB, which have been shown to specifically label MGE-derived interneurons (Fig. 8A–G) (Wonders and Anderson, 2006; Fogarty et al., 2007; Miyoshi et al., 2007; Q. Xu et al., 2008; Sousa et al., 2009). Similar analysis performed in the CA1 region of the hippocampus showed colocalization between Venus and CR, NPY, VIP and nNOS but not PV or SST (data not shown). This is consistent with an MGE-origin of the latter two populations (Fogarty et al., 2007) and a dual MGE/CGE origin of CR+, NPY+ and nNOS+ cells (Fogarty et al., 2007; Tricoire et al., 2010).
Figure 8.
LGE/dCGE/dCGE-derived interneuron subtypes in the adult cortex. A–G, Immunolabeling for Venus and the interneuron markers CB, PV, SST, RLN, CR, NPY and VIP in sections from the adult cortex of Lhx6-CreTg/Dlx1-Venusfl transgenic mice. Arrowheads indicate cells that show absence of colocalization. Arrows point to cells expressing Venus and the interneuron marker indicated. H, The extent of colocalization between Venus and each of the markers examined was quantified, and the data are presented as percentage of the total number of cells expressing each of the markers. I, The number of Venus-positive cells coexpressing CB, PV, SST, CR, NPY, VIP or RLN are presented as a percentage of the total number of Venus-positive cells. J, The distribution of RLN+SST− and VIP+ interneurons in the adult motor cortex is shown. The cortex was subdivided into 5 bins along the pial-white matter axis (1–5, pia to white matter) and the number of labeled cells in each bin was determined. Data for each bin are presented as a proportion of the total number of marker-positive cells. K, Comparison of the number and distribution of Venus- or YFP-expressing cells in the adult motor cortex of Lhx6-CreTg/Dlx1-Venusfl, Lhx6-CreTg/R26R-YFP+/− and Dlx1-Venusfl transgenic mice which label the LGE/dCGE, the MGE and the MGE+LGE/dCGE interneuron populations, respectively. The cortex was subdivided into 5 bins along the pial-white matter axis and the number of labeled cells in each bin was determined. Data for each bin are presented as numbers of cells in each bin. Error bars indicate SD. Scale bar: 75 μm.
We quantified the extent of colocalization between Venus and the interneuron markers examined. Nearly all of the RLN+SST− and the VIP-positive cells as well as the majority of CR+ and NPY+ cells coexpressed Venus indicating that they are generated from LGE/dCGE-precursors (Fig. 8H). A small proportion of cells expressing NPY (but not PV, CR, SST or VIP) in our Lhx6-CreTg/Dlx1-Venusfl mice correspond to Nkx5.1-derived POA neurons. VIP+ and RLN+SST− cells are thought to be nonoverlapping cortical interneuron populations (Miyoshi et al., 2010) that are distinct from the MGE-derived PV+ and SST+RLN+ interneurons (Fogarty et al., 2007; X. Xu et al., 2010). We find that VIP+ and RLN+SST− cells account for the majority of interneurons generated from the LGE/dCGE (∼35% and ∼26%, respectively, in the somatosensory cortex) (Fig. 8I). Most RLN+SST− cells were located in the superficial cortex whereas VIP+ cells were distributed more across cortical layers with a bias toward the superficial cortex (Fig. 8J).
To compare the contribution of LGE/dCGE- and MGE-derived cells to the total population of interneurons in the adult cortex we counted the total number of Venus-positive cells in Lhx6-CreTg/Dlx1-Venusfl mice and compared it to equivalent regions in Lhx6-CreTg/R26R-YFP and Dlx1-Venusfl mice (Fig. 8K). In total, across all layers, LGE/dCGE/-derived interneurons represented ∼30–40% of the total interneuron population (∼28% in the somatosensory cortex and ∼39% in the motor cortex). A large number of these interneurons were located in the superficial layers of the cortex whereas higher densities of MGE-derived neurons were found in middle layers (Fig. 8K).
Discussion
We examined the contribution of subpallial septal and LGE/dCGE germinal zones to neurons of the cortex. We demonstrate that the septum does not generate cortical interneurons whereas the LGE/dCGE together with the POA generate ∼30–40% of the total cortical GABAergic population. LGE/dCGE-derived interneurons enter the cortex later than MGE-derived ones and migrate initially in one stream. At early postnatal stages the distribution of MGE and LGE/dCGE-derived cells within the cortex is similar with most immature interneurons being located in the marginal zone. The LGE/dCGE-derived population redistributes little at postnatal stages and settles mostly within the upper cortical layers whereas the MGE-derived cells move extensively to occupy middle and deeper layers. LGE/dCGE-derived neurons do not express PV or SST, which identify MGE neurons, confirming that spatially and molecularly distinct germinal zones generate different GABAergic interneurons for the cortex. This reinforces the view that genetic restriction of neuronal potential at the precursor stage contributes to the generation of interneuron diversity in the adult cortex.
Neurons derived from septal precursor domains do not migrate to the cortex
Evidence that the septum may generate GABAergic interneurons for the cortex came from mice lacking the transcription factor VAX1. They have a small reduction in the size of the MGE but a complete loss of septal structures (Taglialatela et al., 2004). Interneuron numbers in the cortex are severely reduced and interneurons migrating through the septum are missing from mutant embryos suggesting that the septum may be a major source of interneurons for the cortex (Taglialatela et al., 2004). Our work shows that there are no interneurons in the cortex that have their origins in the septal neuroepithelium. The defect in the numbers of interneurons in the cortex of Vax1 mutants may be attributed to the severe loss of Dlx expression in the MGE (Taglialatela et al., 2004), which is essential for cortical interneuron migration (Anderson et al., 1997).
Migration of LGE/dCGE-derived neurons to the cortex
We observed an apparent difference between the migration of MGE- and LGE/dCGE-derived interneurons to the cortex. While the early MGE interneurons migrate into the cortex in two streams (the MZ and the SVZ/IZ) (Lavdas et al., 1999), LGE/dCGE-derived ones enter the cortex initially within the SVZ/IZ and only later appear in the MZ. Where did the MZ stream appear from and why was it not there at earlier stages? At E14.5 when LGE/dCGE cells enter the cortex there is already a well developed cortical plate. In order for neurons generated from the ganglionic eminences to reach the MZ they would have to first cross the cortical plate. Interneurons entering the cortex tangentially do not invade the cortical plate immediately but instead ‘wait’ for 24–48 h during which time they migrate and disperse within the cortex (Ang et al., 2003; Tanaka et al., 2003, 2006, 2009; López-Bendito et al., 2008). Consistent with these observations, the MZ stream of LGE/dCGE cells appeared within ∼24–48 h of the SVZ/IZ stream coinciding with the time when cortical plate invasion could be observed. The stream of LGE/dCGE-derived interneurons that appears in the MZ may thus consist entirely or partly of interneurons originating in the SVZ/IZ stream. Recent evidence has shown that later-born MGE interneurons entering the cortex at E14.5 do so also within the SVZ/IZ. They later transiently enter the MZ before descending into the cortical plate (Tanaka et al., 2009). This may suggest that all ganglionic eminence-derived interneurons entering the cortex late follow the SVZ stream until they are able to cross the cortical plate.
Movement of interneurons from the SVZ to the MZ and vice versa has been reported in vitro and in vivo (Tanaka et al., 2003, 2009). Cells moving from one stream to another would presumably have to reenter the cortical plate to take their final position. It is unknown why they change streams or why they do not remain within the cortical plate when they first invade it. It is possible that signals from pyramidal neurons are initially absent thus allowing interneurons to disperse further before reentering the cortical plate and ending their journey. Radial migration of interneurons continues until the early postnatal stages when these cells acquire their final layer positions. Although the overall distribution LGE/dCGE-derived interneurons changed little at postnatal stages, it was impossible to determine from static images of the entire population whether and how much movement was taking place. LGE/dCGE-derived interneurons may be moving postnatally but without changing the overall distribution. Our Lhx6-CreTg/Dlx1-Venusfl mice will allow us for the first time to examine the movement of LGE/dCGE-derived interneurons separately from their MGE-derived counterparts.
Are MGE- and LGE/dCGE-derived interneurons guided into the cortex by different signaling systems? Signaling through Semaphorin/Neuropillin, Neuregulin 1/ErbB4 and Cxcl12/Cxcr4/7 have all been implicated in cortical interneuron guidance (Stumm et al., 2003; Flames et al., 2004; Tiveron et al., 2006; Li et al., 2008; López-Bendito et al., 2008). Whether the same signaling systems guide both the MGE and the LGE/dCGE interneurons is unknown. The fact that the two populations follow the same migration routes to reach and navigate through the cortex, albeit at different times, strongly suggests that they follow the same cues. Recent transcriptional profiling experiments suggest that LHX6 may regulate interneuron migration by promoting the expression of the ErbB4, Cxcr4 and Cxcr7 receptor genes (Zhao et al., 2008). If true, and if MGE and LGE/dCGE-derived interneurons follow the same signals to enter the cortex, then different mechanisms must have evolved to regulate expression of the same receptors in the two lineages, given that expression of LHX6 is restricted to the MGE. Unlike MGE and LGE/dCGE interneurons, POA-derived ones enter the cortex initially through the MZ and subplate, avoiding the SVZ (Gelman et al., 2009). This may suggest that POA interneurons follow unique cues to enter the cortex. Comparative transcriptional profiling of migrating cortical interneuron cohorts with different embryonic origins will allow us to examine the differences among them and potentially identify differential regulators of migration and cell fate.
Subtype identity of interneurons generated from the LGE/dCGE
30–40% of all cortical GABAergic interneurons are generated from the LGE/dCGE and include subtypes expressing the neurochemical markers CR, NPY, VIP and RLN. VIP+ and RLN+SST− interneurons are generated exclusively from germinal zones outside the MGE and represent the majority of LGE/dCGE-derived interneurons. Our data are consistent with recent inducible fate-mapping studies of CGE-derived cells whereby the neurochemical and physiological identity of these cells has been characterized. Other markers identifying CGE interneurons include the transcription factor COUPTFII (expressed preferentially but not exclusively in CGE cells) (Kanatani et al., 2008; Miyoshi et al., 2010) and the Serotonin 3A (5-HT3A) receptor gene (Vucurovic et al., 2010). The role of these genes in CGE interneuron development is unknown although COUPTFII is likely to be involved in the caudal migration (Kanatani et al., 2008). Other molecular determinants of dCGE cell fate and development remain to be identified.
The embryonic origin of cortical interneurons and the generation of diversity
We can narrow down the embryonic sources of cortical GABAergic interneurons in rodents to two major regions: the MGE and the LGE/dCGE. We were unable to distinguish between the LGE and the dCGE in our mice. Transplantation studies of E13.5 LGE failed to show significant numbers of interneurons migrating to the cortex (Wichterle et al., 2001). The possibility that at later stages the LGE generates cortical interneurons remains unexplored although recent data argue against this (Miyoshi et al., 2007). In addition to these sources, the POA is the only other germinal zone known to generate interneurons for the cortex. To date only a subdomain of the POA has been fate-mapped therefore the full contribution of this region to cortical interneurons remains to be determined (Gelman et al., 2009).
Although the MGE, the LGE/dCGE and the POA generate distinct cortical interneurons, they cannot account for the full extent of interneuron heterogeneity observed in the adult cortex. Other mechanisms must exist that contribute to the generation of diversity. For example, smaller subdivisions of the neuroepithelium and biases in subtype generation have been identified in the MGE (Flames et al., 2007; Fogarty et al., 2007; Wonders et al., 2008). Temporal shifts in cell fate provide an additional mechanism for producing multiple cell types from restricted progenitor pools (Miyoshi et al., 2007). Finally, some aspects of interneuron identity may be determined postmitotically either during migration or once the immature neurons reach their destination. This has been shown for cortical projection neurons (Shoemaker and Arlotta, 2010). Whether it occurs for cortical interneurons remains unknown.
Our subtractive labeling approach has enabled us to tag cell progeny from germinal zones that lacked unique expression of molecular markers. Similar subtractive and combinatorial fate-mapping work will allow further refinement of subcortical VZ subdivisions to determine the full extent of the contribution of germinal zone specialization to cortical interneuron diversity.
Footnotes
Financial support for the work was provided to N.K. by a United Kingdom Medical Research Council New Investigator Award (grant reference G0501173 ID: 75618) and a European Research Council (ERC) Starting Grant under the European Community's Seventh Framework Programme (FP7/2007-2013)/ERC Grant agreement no. 207807. A.N.R., M.P.H., C.K.P.C., and S.F.R. are supported by PhD studentships from the Wellcome Trust, the United Kingdom Biotechnology and Biological Sciences Research Council, the United Kingdom Medical Research Council, and the Portuguese Fundação para a Ciência e Tecnologia, respectively. We thank our colleagues at the Wolfson Institute for Biomedical Research (University College London) for helpful comments and discussions. Ulla Dennehy and Matthew Grist provided outstanding transgenic and technical support. We thank the following for providing reagents: Neal Copeland (recombineering reagents), Andre Goffinet (anti-reelin antibody), Shioko Kimura (Nkx2.1 cDNA), Vassilis Pachnis (anti-Lhx6 antibody), Rolf Sprengel (iCre plasmid), and Shankar Srinivas (R26R-YFP mice). We thank Gord Fishell for sharing data before publication.
References
- Anderson S, Mione M, Yun K, Rubenstein JL. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9:646–654. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Ang ES, Jr, Haydar TF, Gluncic V, Rakic P. Four-dimensional migratory coordinates of GABAergic interneurons in the developing mouse cortex. J Neurosci. 2003;23:5805–5815. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol Cell Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Batista-Brito R, Fishell G. The developmental integration of cortical interneurons into a functional network. Curr Top Dev Biol. 2009;87:81–118. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Nême N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Bulfone A, Puelles L, Porteus MH, Frohman MA, Martin GR, Rubenstein JL. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005;8:1059–1068. doi: 10.1038/nn1499. [DOI] [PubMed] [Google Scholar]
- Cobos I, Long JE, Thwin MT, Rubenstein JL. Cellular patterns of transcription factor expression in developing cortical interneurons. Cereb Cortex. 2006;16(Suppl 1):i82–i88. doi: 10.1093/cercor/bhk003. [DOI] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Xu Q, Ocbina PJ, Anderson SA. NKX2.1 specifies cortical interneuron fate by activating Lhx6. Development. 2008;135:1559–1567. doi: 10.1242/dev.015123. [DOI] [PubMed] [Google Scholar]
- Eisenstat DD, Liu JK, Mione M, Zhong W, Yu G, Anderson SA, Ghattas I, Puelles L, Rubenstein JL. DLX-1, DLX-2, and DLX-5 expression define distinct stages of basal forebrain differentiation. J Comp Neurol. 1999;414:217–237. doi: 10.1002/(sici)1096-9861(19991115)414:2<217::aid-cne6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marín O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marín O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marín O, Pachnis V, Kessaris N. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston-Massuet C, Henderson DJ, Greene ND, Copp AJ. Zic4, a zinc-finger transcription factor, is expressed in the developing mouse nervous system. Dev Dyn. 2005;233:1110–1115. doi: 10.1002/dvdy.20417. [DOI] [PubMed] [Google Scholar]
- Gelman DM, Martini FJ, Nóbrega-Pereira S, Pierani A, Kessaris N, Marín O. The embryonic preoptic area is a novel source of cortical GABAergic interneurons. J Neurosci. 2009;29:9380–9389. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Ota M, Ogawa M, Mikoshiba K, Aruga J. Zic1 and Zic3 regulate medial forebrain development through expansion of neuronal progenitors. J Neurosci. 2007;27:5461–5473. doi: 10.1523/JNEUROSCI.4046-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Ogawa M, Mikoshiba K, Aruga J. Zic deficiency in the cortical marginal zone and meninges results in cortical lamination defects resembling those in type II lissencephaly. J Neurosci. 2008;28:4712–4725. doi: 10.1523/JNEUROSCI.5735-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatani S, Yozu M, Tabata H, Nakajima K. COUP-TFII is preferentially expressed in the caudal ganglionic eminence and is involved in the caudal migratory stream. J Neurosci. 2008;28:13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TN, Du G, Fonseca M, Zhou QP, Wigle JT, Eisenstat DD. DLX homeobox genes promote cortical interneuron migration from the basal forebrain by direct repression of the semaphorin receptor neuropilin-2. J Biol Chem. 2007;282:19071–19081. doi: 10.1074/jbc.M607486200. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Li G, Adesnik H, Li J, Long J, Nicoll RA, Rubenstein JL, Pleasure SJ. Regional distribution of cortical interneurons and development of inhibitory tone are regulated by Cxcl12/Cxcr4 signaling. J Neurosci. 2008;28:1085–1098. doi: 10.1523/JNEUROSCI.4602-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Novosedlik N, Wang A, Hudson ML, Cohen IL, Chudley AE, Forster-Gibson CJ, Lewis SM, Holden JJ. The DLX1and DLX2 genes and susceptibility to autism spectrum disorders. Eur J Hum Genet. 2009;17:228–235. doi: 10.1038/ejhg.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Sánchez-Alcañiz JA, Pla R, Borrell V, Picó E, Valdeolmillos M, Marín O. Chemokine signaling controls intracortical migration and final distribution of GABAergic interneurons. J Neurosci. 2008;28:1613–1624. doi: 10.1523/JNEUROSCI.4651-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by Cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Maurus D, Harris WA. Zic-associated holoprosencephaly: zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, Hedgehog, and retinoic acid signaling. Genes Dev. 2009;23:1461–1473. doi: 10.1101/gad.517009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Alifragis P, Wong RO, Parnavelas JG. Ventricle-directed migration in the developing cerebral cortex. Nat Neurosci. 2002;5:218–224. doi: 10.1038/nn813. [DOI] [PubMed] [Google Scholar]
- Nagai T, Aruga J, Takada S, Günther T, Spörle R, Schughart K, Mikoshiba K. The expression of the mouse Zic1, Zic2, and Zic3 gene suggests an essential role for Zic genes in body pattern formation. Dev Biol. 1997;182:299–313. doi: 10.1006/dbio.1996.8449. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Shoemaker LD, Arlotta P. Untangling the cortex: Advances in understanding specification and differentiation of corticospinal motor neurons. Bioessays. 2010;32:197–206. doi: 10.1002/bies.200900114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Pannese M, D'Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K. Cloning and characterization of two members of the vertebrate Dlx gene family. Proc Natl Acad Sci U S A. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6–2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19(Suppl 1):i1–i10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanco A, Szekeres C, Patel N, Rao S, Campbell K, Kreidberg JA, Polleux F, Anton ES. Netrin-1-alpha3beta1 integrin interactions regulate the migration of interneurons through the cortical marginal zone. Proc Natl Acad Sci U S A. 2009;106:7595–7600. doi: 10.1073/pnas.0811343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumm RK, Zhou C, Ara T, Lazarini F, Dubois-Dalcq M, Nagasawa T, Höllt V, Schulz S. CXCR4 regulates interneuron migration in the developing neocortex. J Neurosci. 2003;23:5123–5130. doi: 10.1523/JNEUROSCI.23-12-05123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela P, Soria JM, Caironi V, Moiana A, Bertuzzi S. Compromised generation of GABAergic interneurons in the brains of Vax1−/− mice. Development. 2004;131:4239–4249. doi: 10.1242/dev.01299. [DOI] [PubMed] [Google Scholar]
- Tanaka D, Nakaya Y, Yanagawa Y, Obata K, Murakami F. Multimodal tangential migration of neocortical GABAergic neurons independent of GPI-anchored proteins. Development. 2003;130:5803–5813. doi: 10.1242/dev.00825. [DOI] [PubMed] [Google Scholar]
- Tanaka DH, Maekawa K, Yanagawa Y, Obata K, Murakami F. Multidirectional and multizonal tangential migration of GABAergic interneurons in the developing cerebral cortex. Development. 2006;133:2167–2176. doi: 10.1242/dev.02382. [DOI] [PubMed] [Google Scholar]
- Tanaka DH, Yanagida M, Zhu Y, Mikami S, Nagasawa T, Miyazaki J, Yanagawa Y, Obata K, Murakami F. Random walk behavior of migrating cortical interneurons in the marginal zone: time-lapse analysis in flat-mount cortex. J Neurosci. 2009;29:1300–1311. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiveron MC, Rossel M, Moepps B, Zhang YL, Seidenfaden R, Favor J, König N, Cremer H. Molecular interaction between projection neuron precursors and invading interneurons via stromal-derived factor 1 (CXCL12)/CXCR4 signaling in the cortical subventricular zone/intermediate zone. J Neurosci. 2006;26:13273–13278. doi: 10.1523/JNEUROSCI.4162-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricoire L, Pelkey KA, Daw MI, Sousa VH, Miyoshi G, Jeffries B, Cauli B, Fishell G, McBain CJ. Common origins of hippocampal Ivy and nitric oxide synthase expressing neurogliaform cells. J Neurosci. 2010;30:2165–2176. doi: 10.1523/JNEUROSCI.5123-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucurovic K, Gallopin T, Ferezou I, Rancillac A, Chameau P, van Hooft JA, Geoffroy H, Monyer H, Rossier J, Vitalis T. Serotonin 3A receptor subtype as an early and protracted marker of cortical interneuron subpopulations. Cereb Cortex. 2010 doi: 10.1093/cercor/bhp310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dye CA, Sohal V, Long JE, Estrada RC, Roztocil T, Lufkin T, Deisseroth K, Baraban SC, Rubenstein JL. Dlx5 and Dlx6 regulate the development of parvalbumin-expressing cortical interneurons. J Neurosci. 2010;30:5334–5345. doi: 10.1523/JNEUROSCI.5963-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Taylor L, Welagen J, Mbata IC, Xiang JZ, Anderson SA. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev Biol. 2008;314:127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Gashghaei HT, Han C, Watson H, Campbell KJ, Anton ES. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS One. 2007;2:e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yozu M, Tabata H, Nakajima K. The caudal migratory stream: a novel migratory stream of interneurons derived from the caudal ganglionic eminence in the developing mouse forebrain. J Neurosci. 2005;25:7268–7277. doi: 10.1523/JNEUROSCI.2072-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Flandin P, Long JE, Cuesta MD, Westphal H, Rubenstein JL. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J Comp Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Lee J, Griveau A, Arber S, Pierani A, Garel S, Guillemot F. Role of Fgf8 signalling in the specification of rostral Cajal-Retzius cells. Development. 2010;137:293–302. doi: 10.1242/dev.041178. [DOI] [PMC free article] [PubMed] [Google Scholar]