Abstract
Developing sympathetic neurons depend on NGF for survival. When sympathetic neurons are deprived of NGF in vitro, a well documented series of events, including c-Jun N-terminal kinase (JNK) pathway activation, release of cytochrome c from the mitochondria, and caspase activation, culminates in the death of the neuron by apoptosis within 24–48 h. This process requires de novo gene expression, suggesting that increased expression of specific genes activates the cell death program. Using rat gene microarrays, we found that NGF withdrawal induces the expression of many genes, including mkp1, which encodes a MAPK phosphatase that can dephosphorylate JNKs. The increase in mkp1 mRNA level requires the MLK-JNK-c-Jun pathway, and we show that Mkp1 is an important regulator of JNK-dependent apoptosis in sympathetic neurons. In microinjection experiments, Mkp1 overexpression can inhibit JNK-mediated phosphorylation of c-Jun and protect sympathetic neurons from apoptosis, while Mkp1 knockdown accelerates NGF withdrawal-induced death. Accordingly, the number of superior cervical ganglion (SCG) neurons is reduced in mkp1−/− mice at P1 during the period of developmental sympathetic neuron death. We also show that c-Jun and ATF2 bind to two conserved ATF binding sites in the mkp1 promoter in vitro and in chromatin. Both of these ATF sites contribute to basal promoter activity and are required for mkp1 promoter induction after NGF withdrawal. These results demonstrate that Mkp1 is part of a negative feedback loop induced by the MLK-JNK-c-Jun signaling pathway that modulates JNK activity and the rate of neuronal death in rat sympathetic neurons following NGF withdrawal.
Introduction
Programmed cell death (apoptosis) has important functions in the developing mammalian nervous system and is essential for adjusting the size of neuronal populations as well as ensuring that appropriate connections are formed between neurons and their targets (Oppenheim, 1991; Jacobson et al., 1997; Yuan and Yankner, 2000). The size of neuronal populations is determined by the limited availability of survival factors secreted by their target tissues. During late embryonic and early postnatal development, sympathetic neurons depend on the prototypical neurotrophin, nerve growth factor (NGF), for survival. When sympathetic neurons are deprived of NGF in vitro, they die by apoptosis, and this death involves the mitochondrial (intrinsic) pathway of caspase activation (Deckwerth and Johnson, 1993; Edwards and Tolkovsky, 1994; Deshmukh and Johnson, 1998; Neame et al., 1998; Wright et al., 2007). However, inhibitors of transcription or protein synthesis can protect the neurons from NGF withdrawal-induced apoptosis, suggesting that new gene expression is required for cell death to occur (Martin et al., 1988). Therefore, sympathetic neurons have proved to be a useful model for studies of neuronal apoptosis and for identifying new genes that are regulated in response to NGF withdrawal.
In mammalian cells, the c-Jun N-terminal kinases (JNKs) are one of the major subfamilies of the MAPK (mitogen-activated protein kinase) superfamily, which also includes the extracellular signal-regulated kinases (ERKs) and the p38 MAP kinases. Following NGF withdrawal in sympathetic neurons, the JNK pathway is activated and required for cell death (Estus et al., 1994; Ham et al., 1995; Virdee et al., 1997; Eilers et al., 1998). JNKs are activated by reversible dual phosphorylation on the threonine and tyrosine residues of the Thr-Pro-Tyr motif in the catalytic domain by the upstream JNK kinases MKK4 and MKK7 (Davis, 2000). Activated JNKs phosphorylate a variety of downstream targets, such as the AP-1 transcription factors c-Jun and ATF2. This increases their ability to activate the transcription of their target genes (Gupta et al., 1995; Karin, 1995; van Dam et al., 1995; Hazzalin and Mahadevan, 2002), which include members of the dual specificity phosphatase (DUSP) family (Hayakawa et al., 2004; Breitwieser et al., 2007).
MAPK phosphatases (MKPs) are a family of DUSPs that inactivate MAP kinases by dephosphorylating both phospho-threonine and phospho-tyrosine residues located in the activation loop. This family includes Mkp1 (DUSP1, 3CH134), which was the first family member to be identified as a phosphatase and was originally cloned as CL100 by subtractive hybridization using RNA isolated from hydrogen peroxide-treated cells (Keyse and Emslie, 1992). Mkp1 functions to negatively regulate MAPK signaling (Sun et al., 1993; Keyse, 2000); however, little is known about the function and regulation of Mkp1 in the mammalian nervous system, and nothing is known about the relationship between Mkp1 and the JNK signaling pathway in sympathetic neurons deprived of NGF.
Here, we show that the mkp1 gene is a direct transcriptional target of the MLK-JNK-c-Jun pathway and that Mkp1 plays a crucial role in the negative regulation of JNK signaling in sympathetic neurons after NGF withdrawal.
Materials and Methods
Cell culture.
Sympathetic neurons were isolated from the superior cervical ganglia (SCG) of 1-d-old Sprague Dawley rats (Biological Services Unit, University College London, London, UK) and cultured as described previously (Whitfield et al., 2004). Animal experiments were performed according to the Animals (Scientific Procedures) Act 1986 under a license reviewed and approved by the Biological Services Unit at University College London. Neurons were cultured in DMEM (Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS), 2 mm glutamine (Invitrogen), and penicillin-streptomycin (SCG medium). The antimitotic agents fluorodeoxyuridine (20 μm) and uridine (20 μm) were added to limit the proliferation of non-neuronal cells and when required, NGF (Cedarlane Laboratories) was added at 50 ng/ml. Neurons were plated on 13 mm diameter glass coverslips coated with poly-l-lysine and laminin and cultured in 3.5 cm diameter dishes containing 2 ml of SCG medium and NGF for 5–7 d. For NGF withdrawal experiments, neurons were washed twice in SCG medium lacking NGF and then refed with SCG medium supplemented with a neutralizing anti-NGF antibody at 100 ng/ml (Millipore Bioscience Research Reagents Europe). The MLK3 inhibitor CEP-11004 (Cephalon) was dissolved in DMSO and used at a final concentration of 400 nm.
The PC6-3 subline of PC12 pheochromocytoma cells (Pittman et al., 1993) was plated on collagen-coated 9 cm dishes and cultured in PC6-3 complete medium consisting of RPMI 1640 (Invitrogen), 10% horse serum, 5% FCS, 2 mm glutamine, and penicillin/streptomycin. Cells were maintained in a humidified 5% CO2 incubator at 37°C and passaged every 5–7 d. To induce differentiation, 1 × 106 PC6-3 cells per 9 cm dish were maintained for 7 d in medium containing RPMI 1640, 2% horse serum, 1% FCS, 2 mm glutamine, penicillin/streptomycin, and NGF at 100 ng/ml (Promega). For NGF withdrawal experiments, differentiated PC6-3 cells were gently rinsed twice with medium lacking NGF and then refed with medium containing NGF or anti-NGF antibody.
Animals.
Mkp1−/− mice (Dorfman et al., 1996) were provided by Bristol-Myers Squibb with a mixed C57BL/6–129/Sv genetic background. To generate mkp1+/+ and mkp1−/− lines with uniform C57BL/6 genetic background, the original line was back-crossed against C57BL/6 for 10 generations, wild-type and interrupted mkp1 alleles being detected by PCR screening.
Analysis of mkp1−/− mice.
For morphometric analyses, SCGs were removed from mkp1+/+ or mkp1−/− mice at P1 and were immersion fixed in 10% formalin. After embedding in paraffin, serial sections 7 μm wide were cut through the ganglia, and every section was collected on Surgipath X-tra adhesive slides (Leica). After Nissl staining, ImageJ image analysis software was used to count all neuronal profiles containing nucleoli on every third section. The number obtained was multiplied by 3, as described by Jacobs et al. (2005) and Coggeshall et al. (1984). This method does not correct for split nucleoli. Statistical results were expressed as the mean ± SEM and were tested for significance by a one-tailed Student's t test. Alternate sections were immunostained for neuron-specific MAP2 (Clone HM-2, Sigma). TUNEL staining was performed using an in situ cell death detection kit (Millipore) according to the manufacturer's protocol. ImageJ image analysis software was used to count all TUNEL-positive neurons in every third section, and this number was multiplied by three according to the method of Coggeshall et al. (1984) to obtain the total number of apoptotic cells per ganglion. Sections were also immunostained with Ki67 (Clone MIB-1, DAKO) as a marker of cellular proliferation.
Plasmid constructs.
Sympathetic neurons from 1-d-old Sprague Dawley rats were cultured for 6 d in vitro, and RNA was extracted and used to generate cDNA by reverse transcription as described below. To construct pcDMkp1, a DNA fragment containing the rat Mkp1 coding sequences (1–1104 bp) was generated by PCR amplification using the primers 5′-ATGGTGATGGAGGTGGGCAT-3′ and 5′-TCAGCAGCTCGGAGAGGTTG-3′ followed by cloning into HindIII and EcoRI-restricted pcDNA1.0 (Invitrogen). The expression vectors for JunΔ169, the JIP-1 JNK binding domain (JBD), wtMLK3, and kdMLK3 have been previously described (Ham et al., 1995; Eilers et al., 2001; Mota et al., 2001). The mkp1-LUC reporter plasmid was constructed as follows. Genomic DNA was extracted from 1-d-old Sprague Dawley rat brains using the QIAGEN DNA Blood and Tissue Kit. An mkp1 promoter fragment was amplified by PCR using the sense oligonucleotide: 5′-GCGACGACAACGTGCTTGAC-3′ and the antisense oligonucleotide: 5′-GGCGAAGAAGGAGCGACAATCC-3′. The genomic DNA was denatured at 94°C for 5 min. Amplification was performed at 94°C for 30 s, 63°C for 45 s, and 72°C for 140 s for 35 cycles; and then at 72°C for 7 min. The resulting mkp1 promoter fragment (−1010 to +1) was then cloned into the HindIII and NcoI sites of the pGL3 basic vector (Promega) upstream of the luciferase gene. Fragment orientation and positioning were confirmed by restriction enzyme analysis and DNA sequencing. The mkp1-LUC 2xATF mutant construct was generated using the following complementary primer pairs: ATF site 1 5′-CGCTCCCAGGCCGACGAGTTATTTGCTTTTGGCTTTG-3′ and 5′-CAAAGCCAAAAGCAAATAACTCGTCGGCCTGGGAGCG-3′; ATF site 2 5′-GCAGGGCGGGCGAGTTCCCCACCCGGTCAC-3′ and 5′-GTGACCGGGTGGGGAACTCGCCCGCCCTGC-3′. The altered bases create 4 point mutations in each of the ATF/CRE sites (the mutated bases are underlined). Mutagenesis was performed using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene). DNA sequencing confirmed that the mutations had been incorporated correctly.
Real-time quantitative PCR.
Total RNA was extracted from sympathetic neurons using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. Briefly, total RNA was eluted in 30 μl of RNase-free water and 1 μg of total RNA was reverse transcribed into cDNA using 200 units of Moloney murine leukemia virus (M-MLV) reverse transcriptase in the presence of 2.5 μm N6 random primers, 0.5 mm dNTPs, and amplification grade DNase I (all Invitrogen). Five nanograms of the resulting cDNA were used for real-time PCR using the ABI-Prism 7900HT fast Sequence Detection System and Taqman PCR Master Mix (Applied Biosystems). Primers were designed with PrimerExpress software v2.0 (Applied Biosystems) and used at the following concentrations: for c-jun, 300 nm forward primer (5′-TCCACGGCCAACATGCT-3′), 900 nm reverse primer (5′-CCACTGTTAACGTGGTTCATGAC-3′), and 175 nm probe (5′-FAM-AGGGAACAGGTGGCACAGCTTAACAGA-TAMRA-3′); for bim, 300 nm forward primer (5′-CCAGGCCTTCAACCATTATCTC-3′), 900 nm reverse primer (5′-GCGCAGATCTTCAGGTTCCT-3′), and 175 nm probe (5′-FAM-TGCAATGGCTTCCATAAGGCAGTCTCA-TAMRA-3′); for mkp1, 300 nm forward primer (5′-GGACAACCACAAGGCAGACA-3′), 300 nm reverse primer (5′-CAGCATCCTTGATGGAGTCTATAAAG-3′), and 175 nm probe (5′-FAM-TAGCTCCTGGTTCAAGGAGGCGATTG-TAMRA-3′). The levels of the c-jun, bim, and mkp1 mRNAs were normalized to the level of the Gapdh (forward 5′-CTGAGAATGGGAAGCTGGTC-3′ and reverse 5′-ACTGTGGTCATGAGCCCTTC-3′) or Hprt1 (forward 5′-AGTCCCAGCGTCGTGATTA-3′ and reverse 5′-CCCGTTGACTGGTCATTACA-3′) mRNA. The 2−ΔΔCT relative quantitation method was used to determine the relative expression.
Immunoblotting.
Sympathetic neurons were harvested in 1 ml of ice-cold PBS, spun down, and lysed in sample buffer (2% SDS, 2 mm β-mercaptoethanol, 60 mm Tris, pH 6.8, 0.01% bromophenol blue) for 10 min at 100°C. Proteins were separated on 12% SDS polyacrylamide gels and transferred to Immobilon-P (Millipore). After blocking for 45 min with 5% nonfat milk in TBS supplemented with 0.5% Tween 20, the membrane was incubated with different primary antibodies overnight at 4°C. The following primary antibodies were used: mouse monoclonal c-Jun antibody (610327, BD Transduction Laboratories), mouse monoclonal phospho-c-Jun (ser63) antibody (sc-822; Santa Cruz Biotechnology), rabbit polyclonal Bim antibody (#2819, Cell Signaling Technology), and rabbit polyclonal Mkp1 antibody (M-18 and V-15, Santa Cruz Biotechnology). Equivalent protein loading was confirmed by stripping membranes and reprobing with a rabbit polyclonal ERK 1/2 antibody (Cell Signaling Technology).
PC6-3 cells were harvested in 1 ml of ice-cold PBS, spun down, and lysed in SDS lysis buffer (10 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.5 mm EDTA, 0.5 mm EGTA, 1% SDS) containing a protease inhibitor cocktail (Sigma) and 1 mm phenylmethylsulfonyl fluoride (Ham et al., 1995) and heated for 20 min at 90°C. The lysate was spun for 20 min at 4°C and the resulting supernatant quantified using the Bio-Rad protein assay (Bio-Rad) according to the manufacturer's instructions. Typically, 20 μg of protein from PC6-3 cells was loaded per lane.
Immunofluorescence.
Sympathetic neurons cultured on poly-l-lysine/laminin-coated glass coverslips were fixed using 4% paraformaldehyde at room temperature for 20 min, washed three times with PBS, and then permeabilized with 0.5% Triton X-100 in PBS at room temperature for 5 min. Neurons were then incubated in 50% normal goat serum in 1% BSA in PBS for 30 min at room temperature. After washing, neurons were incubated with primary antibody for 1 h at room temperature, followed by a 45 min incubation with secondary antibody at room temperature. The primary antibodies against Mkp1 and c-Jun were diluted 1:5000 and 1:1000, respectively. Fluorescein-conjugated goat anti-rabbit or anti-mouse secondary antibodies were typically used at a dilution of 1:250. Neurons were rinsed in PBS and nuclei stained with DAPI dye in Antifade (DAKO) or Hoechst dye and mounted on glass slides. Slides were viewed on a Zeiss Axioplan 2 microscope using a Plan-Apochromat 63x/1.40 oil objective. Images were captured at room temperature (20°C) using a Quantix digital camera (Photometrics) and SmartCapture VP software. Images were then saved as TIFF files and viewed using Adobe Photoshop CS4.
siRNA and electroporation.
For electroporation experiments, confluent 9 cm dishes of PC6-3 cells were harvested in PC6-3 complete medium, adjusted to 1 × 107 cells/ml, and 300 μl was transferred to a polycarbonate 4 mm electroporation cuvette (Thermo Fisher Scientific). Mkp1 siRNAs (Dharmacon) were added to the cells at a final concentration of 3 μm and were electroporated by applying a short electrical pulse (400–1200 V) for 1–10 ms using an Electroporation Pulse Generator EPI 2500. Electroporated cells were returned to culture for 24 h and knockdown was assessed by immunoblotting as described above. Four mkp1 siRNA oligonucleotides with target sequences UGGAGCAUAUCGUGCCGAA (Dharmacon accession number J-096434-09), AAGAUAUGCUCGACGCCUU (J-096434-10), UGAUCAACGUCUCGGCCAA (J-096434-11), and GUGAAGCAGAGGCGGAGUA (J-096434-12) and nonspecific control siRNA oligonucleotides (Dharmacon accession numbers D-001820-03-05 and D-001810-03-05) were purchased from Dharmacon.
Microinjection.
Microinjection of sympathetic neurons was performed as described previously (Whitfield et al., 2001; Towers et al., 2009). Briefly, injection mixes containing the DNA being tested (at the indicated concentrations) in 0.5× PBS (−Ca2+, −Mg2+) were injected into the nuclei of sympathetic neurons. At least three independent experiments were performed with 120 and 140 neurons injected in each experiment. Typically, >90% of the cells survived injection. To assess the effect of different expression vectors on neuronal survival after NGF withdrawal, neurons were cultured for 5–7 d before injection. As a marker, neutral 70,000 MW Texas Red dextran (Invitrogen) was added to the injection mix at a final concentration of 5 μg/μl together with each expression vector at 50 ng/μl. After NGF withdrawal, the number of viable Texas Red dextran-positive neurons was determined at 0, 24, 48, 72, and 96 h in a blinded fashion. For immunofluorescence analysis, purified guinea pig IgG was added to the injection mix as a marker at a final concentration of 5 μg/μl. After NGF withdrawal, the injected neurons were identified by staining with a rhodamine-conjugated donkey anti-guinea pig IgG antibody (Jackson ImmunoResearch Laboratories) diluted 1:100.
Mkp1 siRNAs and controls were microinjected as described previously (Aalto et al., 2007). Typically, 3 μm of the individual siRNA or pools (Dharmacon) were microinjected into the nucleus together with Texas Red Dextran (5 μg/μl) or guinea pig IgG (2.5 μg/μl) as a marker. Microinjected cells were returned to culture for 24 h to allow the preexisting Mkp1 protein to degrade. Sixteen hours after NGF withdrawal, either neurons were fixed and stained for immunofluorescence analysis or the number of Texas Red dextran-positive cells was determined in a survival assay.
In reporter gene assays, sympathetic neurons were microinjected with mkp1-LUC or the ATF site mutant at 10 ng/μl together with the control Renilla luciferase construct pRL-TK at 5 ng/μl.
For antibody coinjection experiments, rabbit polyclonal antibodies against c-Jun (H-79X; Santa Cruz Biotechnology) and ATF2 (C-19X; Santa Cruz Biotechnology) were diluted in PBS (−Ca2+, −Mg2+) and centrifuged for 3 h at 4°C in Microcon YM3 centrifugal filters (Millipore) to remove sodium azide. The final antibody concentration was adjusted to 2 μg/μl in PBS (−Ca2+, −Mg2+). Purified rabbit IgG (Jackson ImmunoResearch Laboratories) was used as a control. Neurons were microinjected with the mkp1-LUC reporter construct (20 ng/μl), pRL-TK (10 ng/μl) and antibody (1 μg/μl). A few hours after injection, NGF was withdrawn, and 16 h later a dual luciferase assay was performed.
Dual luciferase assay.
Injected neurons were harvested in 1 ml of ice-cold PBS, spun down, and then lysed in 25 μl of passive lysis buffer. Luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega), and the luciferase assay was performed using a Lumat LB 9507 luminometer according to the manufacturer's instructions. Output for firefly was normalized to the Renilla luciferase output. Each experiment was performed at least three times and the SEM was calculated.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as described previously (Towers et al., 2009). PC6-3 cells were plated at a density of 1 × 106 cells per 9 cm dish and differentiated for 7 d. Cells were then rinsed twice and cultured in medium containing NGF or anti-NGF antibody. After 16 h, proteins and DNA were crosslinked by adding formaldehyde (1% final concentration) and incubating at room temperature for 3 h. For preclearing and for recovery of the immune complexes 50% protein A/G-agarose beads (Santa Cruz Biotechnology) in ChIP dilution buffer containing BSA were used. ChIP samples were analyzed by PCR using TaqDNA polymerase with CoralLoad PCR buffer and Q-solution (Qiagen). To detect binding of c-Jun and ATF2 to the region of the rat mkp1 promoter that contains the two conserved ATF sites, the primers described by Ryser et al. (2004) were used: 5′-CGGAGCCAGCGCTCAAAG-3′ and 5′-GATCCTAATCTGGCTTCACCGCGCG-3′. PCR conditions were as follows: 94°C for 5 min, then 30–35 cycles of 94°C for 30 s, 60°C for 45 s, 72°C for 1 min, followed by a final incubation at 72°C for 10 min. To study binding of c-Jun and ATF2 to the TREs in the rat c-jun promoter the primers described by Towers et al. (2009) were used: 5′-TGGAGAAAGAAGGGCCCAACTGTAG-3′ and 5′-GTGCAACTCTGAGTCCTTATC-3′. The PCR conditions were 94°C for 5 min followed by 30–35 cycles of 20 s at 94°C, 45 s at 52°C, 1 min at 72°C followed by 10 min at 72°C. The PCR products were run on non-denaturing 8% polyacrylamide/1×TBE gels and then stained with SYBR green 1 (Sigma-Aldrich). Images were captured using a UVIdoc gel documentation system (UVItec).
Electrophoretic mobility shift assay.
Double stranded oligonucleotides were labeled with [α-32P]dCTP (3000 Ci/mmol; PerkinElmer) using Klenow polymerase (Roche Diagnostics) to fill in 5′ overhangs. The following pairs of oligonucleotides were used (ATF binding sites are underlined): mkp1 ATF site 2, 5′-CTAGCGGGTGACGTCACCAC-3′ and 5′-GATCGTGGTGACGTCACCCG-3′; mkp1 ATF site mutant, 5′-CTAGCGGGCGAGTTCCCCAC-3′ and 5′-GATCGTGGGGAACTCGCCCG-3′.
Sympathetic neuron extracts for EMSA experiments were prepared as described previously (Towers et al., 2009) using whole-cell extract buffer (0.1% NP-40, 250 mm KCl, 50 mm HEPES pH 7.9, 10% glycerol, 0.2 mm EDTA, 0.2 mm EGTA, containing the following inhibitors added just before use, 4 mm NaF, 4 mm Na3VO4, 1 mm DTT, 0.5 mm PMSF, 2% v/v Sigma mammalian protease inhibitor cocktail). Protein concentration was determined using the Bio-Rad protein assay.
Binding reactions were prepared with 40 mm KCl, 20 mm HEPES, pH 7.9, 5 mm MgCl2, 1 mm EGTA, 0.5 mm DTT, 10% glycerol, 0.5 μg/μl BSA, 1 μg of poly(dI-dC), and 4 μl of whole-cell extract buffer in a volume of 24 μl. Four micrograms of whole-cell extract were used per binding reaction, and for supershift assays 2–4 μl of antibody was added. The antibodies were c-Jun (H-79) X, ATF2 (C-19) X, phospho-c-Jun (KM-1) X (all from Santa Cruz Biotechnology), phospho-ATF2 (Thr71) (Cell Signaling Technology), and, as a control, Bim AB17003 (Millipore Bioscience Research Reagents). Binding reactions were for 2 h at 4°C, after which 0.4 ng of the 32P-labeled double-stranded oligo was added. The samples were incubated for 15 min at room temperature, and then electrophoresed on a 5% polyacrylamide, 0.25× TBE gel. Following electrophoresis at 180 V for ∼2 h at room temperature, the gel was fixed for 15 min in 10% acetic acid, 10% methanol and dried at 80°C under vacuum. The bands were visualized by exposing the dried gel to Kodak MXB x-ray film (Kodak). Exposed x-ray films were scanned using an Epson photo scanner (model 4990) and images saved as TIFF files.
Statistical analysis.
Data were analyzed by paired Student's t test (for two-tailed distributions), and significance is expressed as follows: #p < 0.001, *p < 0.01, +p < 0.1, and NS (not significant, p > 0.1). For all graphs, error bars represent mean ± SEM.
Results
Inhibition of the MLK-JNK-c-Jun pathway reduces the increase in mkp1 mRNA level after NGF withdrawal
To identify mRNAs upregulated after NGF withdrawal in sympathetic neurons, we performed a gene microarray analysis using Affymetrix Exon arrays and RNA isolated from sympathetic neurons that had been cultured for 16 h in the presence of NGF, absence of NGF or absence of NGF but with the mixed-lineage kinase (MLK) inhibitor CEP-11004 added to the medium (M. Kristiansen, F. Menghi, and J. Ham, unpublished data). Sixteen hours corresponds to the transcriptional commitment point for NGF withdrawal-induced death. The level of expression of genes already known to be induced by NGF deprivation, such as c-jun, bim, dp5, eglN3, and atf3, is high at this time point. In our microarray experiment, these genes behaved as predicted and their transcripts increased in level following NGF withdrawal (Fig. 1A). The MLK-JNK-c-Jun pathway is activated after NGF deprivation (Virdee et al., 1997; Eilers et al., 1998; Mota et al., 2001; Xu et al., 2001). To identify which of the genes induced after NGF withdrawal are potential targets of the MLK-JNK-c-Jun pathway, we used a small molecule MLK inhibitor, CEP-11004, which is related to the compound CEP-1347. MLKs are upstream activators of the JNK-c-Jun pathway and are inhibited by CEP-11004 and CEP-1347 in NGF-deprived sympathetic neurons (Maroney et al., 2001; Mota et al., 2001; Wang et al., 2005). With the exception of eglN3, the induction of the known targets of the MLK-JNK-c-Jun pathway (c-jun, bim, dp5, and atf3) was significantly reduced by CEP-11004 (Fig. 1A). One of the genes most highly upregulated after NGF withdrawal in our microarray experiment was the MAPK phosphatase, mkp1 (Fig. 1A). The mkp1 mRNA was previously shown to be induced as early as 5 h after NGF withdrawal (Estus et al., 1994). Interestingly, we found that an increase of 4.51-fold in the level of the mkp1 mRNA was reduced to 1.42-fold in the presence of CEP-11004. This suggests that mkp1 is a potential target of the MLK-JNK-c-Jun pathway.
Figure 1.
Mkp1 mRNA levels increase in sympathetic neurons after NGF withdrawal, and this requires MLK activity. Sympathetic neurons from 1-d-old rats were cultured for 6 d in vitro and then rinsed and refed with medium containing NGF (+NGF), or lacking NGF (−NGF), or lacking NGF but containing 400 nm CEP-11004 (−NGF+CEP-11004) for 16 h. RNA was then isolated using an RNeasy mini kit. A, Affymetrix Rat Exon 1.0ST microarray analysis of gene expression in sympathetic neurons at the whole gene level. c-jun, bim, dp5, and atf3 are induced after NGF withdrawal, and this is reduced by CEP-11004 validating them as known targets of the MLK-JNK-c-Jun pathway. mkp1 induction after NGF withdrawal (4.51-fold) is reduced by CEP-11004 to 1.42-fold, suggesting that it too may be a transcriptional target of the JNK pathway. B, c-jun, bim, and mkp1 mRNA levels were measured by real-time PCR and normalized to the housekeeping genes gapdh and hprt1. Similar results were obtained in both cases. The data shown represents the average of three independent experiments ± SEM. Statistical comparisons were made between +NGF and −NGF or −NGF and −NGF+CEP for each gene. #p < 0.001; *p < 0.01; +p = 0.02.
To validate these results, we cultured sympathetic neurons for 6 d in the presence of NGF and then for a further 16 h in the presence of NGF, anti-NGF antibody, or CEP-11004. The level of mkp1 mRNA, and as a control, c-jun and bim mRNA, was then measured by quantitative PCR (Fig. 1B). After NGF withdrawal, the level of the mkp1 mRNA increased by 4.93-fold, but this was reduced to 1.91-fold in the presence of CEP-11004 when normalized to gapdh mRNA levels, or from 4.42-fold to 1.39-fold when normalized to hprt1 mRNA levels (Fig. 1B). Similarly, levels of c-jun and bim mRNA also mirrored the patterns from the microarray analysis (Fig. 1B). A similar increase in mkp1 mRNA was also observed in neuronally differentiated PC6-3 cells after NGF withdrawal (supplemental Fig. S1, available at www.jneurosci.org as supplemental material). These data confirm the validity of the microarray experiment and reveal that mkp1 is a potential MLK-JNK-c-Jun target gene.
Induction of Mkp1 protein following NGF withdrawal
The primary function of Mkp1 is to dephosphorylate JNKs (Slack et al., 2001), and it is known that JNK activity rapidly increases after NGF withdrawal (by 4 h) but starts to return to basal level at later time points (Xia et al., 1995; Virdee et al., 1997; Eilers et al., 1998). We examined the effect of NGF withdrawal on the level of Mkp1 protein and its localization (Fig. 2). In immunoblotting experiments, we observed that by 4 h after NGF withdrawal, there was a small increase in the level of Mkp1 protein in sympathetic neurons. However, by 8 and 16 h, levels of Mkp1 protein had significantly increased. In contrast, when sympathetic neurons were deprived of NGF in the presence of 400 nm CEP-11004 for 16 h, there was no significant increase in Mkp1 protein level (Fig. 2A). The addition of CEP-11004 had no effect on the level of Mkp1 in the presence of NGF (Fig. 2A). Levels of Mkp1 protein and its subcellular localization were also studied by immunofluorescence (Fig. 2B). In the presence of NGF, the Mkp1 protein was distributed throughout the cell, albeit at low levels (Fig. 2B, 0 h). However, by 8 h after NGF withdrawal, the level of Mkp1 protein had increased, and by 16 h, there was a significant amount of Mkp1 protein present throughout the cell, including the nucleus. This increase was reversed by the addition of CEP-11004 (Fig. 2B). These results suggest that the Mkp1 protein increases in level in a time-dependent manner after NGF withdrawal and that CEP-11004 prevents this increase.
Figure 2.
The Mkp1 protein increases in level in sympathetic neurons following NGF withdrawal. A, Immunoblotting analysis of Mkp1 protein levels using extracts prepared from sympathetic neurons at 0, 4, 8, and 16 h after NGF withdrawal. Neurons were maintained in the absence of NGF for the indicated times and were then harvested, lysed in 2× SDS sample buffer, and immunoblotted with an anti-Mkp1 antibody. Four independent experiments were performed and a representative immunoblot is shown. ERK levels are shown as a loading control. B, Analysis of Mkp1 protein levels and subcellular localization by immunocytochemistry. Sympathetic neurons were treated as indicated and fixed at 0, 4, 8, or 16 h after NGF withdrawal ± CEP-11004. The cells were then stained with an anti-Mkp1 antibody followed by a rhodamine-conjugated secondary antibody, and Hoechst dye to label the nuclear DNA. By 16 hours after NGF withdrawal, Mkp1 protein levels had significantly increased, and Mkp1 was localized in both the nucleus and the cytosol. The scale bar represents 25 μm.
Overexpression of Mkp1 prevents c-Jun N-terminal phosphorylation and protects against NGF withdrawal-induced death
c-Jun N-terminal phosphorylation increases after NGF withdrawal due to an increase in JNK activity and JNK activity is required for NGF withdrawal-induced death (Xia et al., 1995; Virdee et al., 1997; Eilers et al., 1998, 2001; Harding et al., 2001). Since we demonstrated a time-dependent increase in the level of Mkp1 protein after NGF withdrawal (Fig. 2) and since it is known that Mkp1 can dephosphorylate JNKs, we investigated the effect of Mkp1 overexpression on NGF withdrawal-induced death in sympathetic neurons. We constructed an Mkp1 expression vector, pcDMkp1, by subcloning the rat mkp1 coding sequence into pcDNA1. Sympathetic neurons were then microinjected with the pcDMkp1 expression construct together with guinea pig IgG as a marker and maintained in the presence of NGF for 16 h. Neurons microinjected with the pcDMkp1 construct expressed much higher levels of Mkp1 protein compared to neurons injected with the empty vector pcDNA1 (data not shown). We then studied the effect of Mkp1 overexpression on the phosphorylation of c-Jun, a well characterized JNK substrate, by microinjecting sympathetic neurons with the pcDMkp1 expression vector or the empty vector, pcDNA1. In the presence of NGF, <2% of neurons injected with the empty vector pcDNA1 expressed a detectable level of phospho-c-Jun (supplemental Fig. S2B, available at www.jneurosci.org as supplemental material). After NGF withdrawal, phosphorylation of c-Jun at serine 63 was evident by strong immunostaining in almost all of the neurons (92%) injected with pcDNA1 (Fig. 3A; supplemental Fig. S2B, available at www.jneurosci.org as supplemental material). However, when pcDMkp1 was injected only 5% of the neurons stained for phospho-c-Jun after NGF withdrawal (Fig. 3A; supplemental Fig. S2A,B, available at www.jneurosci.org as supplemental material). This suggests that the dephosphorylation of JNKs by Mkp1 prevented the subsequent phosphorylation of c-Jun. Since JNK activity and c-Jun phosphorylation are required for NGF withdrawal-induced death, we investigated the effect of pcDMkp1 expression on the survival of sympathetic neurons after NGF withdrawal (Fig. 3B). The survival of sympathetic neurons injected with the pcDMkp1 expression construct was compared directly with either empty vectors (pcDNA1 or pcDNA3), a pcDNA1 expression construct encoding JunΔ169 (a dominant negative c-Jun mutant lacking the transactivation domain) or a pcDNA3 expression construct encoding the JBD (a direct JNK inhibitor protein), together with Texas Red dextran as a marker. A few hours after injection, cells were deprived of NGF for a total period of 72 h and the percentage of viable injected cells was calculated (Fig. 3B). As a positive control, the viability of neurons injected with the empty vector, pcDNA1 was also determined in the presence of NGF at 72 h. Expression of both JunΔ169 and the JBD delays NGF withdrawal-induced death (Ham et al., 1995; Eilers et al., 2001). Twenty-four hours after NGF withdrawal, there was an increased rate of cell death in sympathetic neurons injected with the empty vectors compared to neurons injected with pcDJunΔ169, pcDJBD, and pcDMkp1 (data not shown). By 72 h, the percentage of viable cells injected with pcDMkp1 (57%) was significantly higher when compared to neurons injected with either pcDNA1 (30%) or pcDNA3 (24%) (Fig. 3B). Furthermore, the viability of sympathetic neurons injected with pcDMkp1 (57%) was similar to that of neurons injected with pcDJunΔ169 (62%, p = 0.24) or pcDJBD (68%, p = 0.422) (Fig. 3B), demonstrating that overexpression of Mkp1 delays NGF withdrawal-induced death in sympathetic neurons.
Figure 3.

Overexpression of Mkp1 protects sympathetic neurons against NGF withdrawal-induced death. A, Overexpressing Mkp1 prevents the phosphorylation of c-Jun. Sympathetic neurons were cultured for 6 d in vitro and then microinjected with pcDNA1 (empty vector) or pcDMkp1 at 10 ng/μl, together with guinea pig IgG as a marker. After injection, neurons were rinsed and refed with medium lacking NGF for 16 h before being fixed and stained with a phospho-c-Jun antibody. Arrows indicate injected neurons. The scale bar represents 25 μm. B, Neurons were microinjected with Texas Red dextran (5 μg/μl) and the following expression constructs at 50 ng/μl: the empty expression vector pcDNA1 or pcDNA3 (negative controls), pcDJunΔ169, pcDJBD (a direct JNK inhibitor protein), or pcDMkp1. The cells were then rinsed and refed with −NGF or +NGF medium and counted on an inverted microscope. After 72 h, the percentage of viable injected (Texas Red positive) neurons that remained was determined. pcDMkp1 delayed neuronal cell death after NGF withdrawal in a similar manner to pcDJunΔ169 and pcDJBD (when compared to the empty vectors pcDNA1 or pcDNA3). The average of three experiments ± SEM is shown. #p < 0.001, *p < 0.01, Student's t test.
Knocking down Mkp1 accelerates NGF withdrawal-induced death in sympathetic neurons
Since overexpression of Mkp1 delayed NGF withdrawal-induced death in sympathetic neurons, we next investigated the effect of knocking down Mkp1 on neuronal viability. We tested four commercially available siRNAs (Dharmacon) that directly target different regions of the rat mkp1 gene. To study the effect of the siRNAs on Mkp1 protein levels in immunoblotting experiments, we used neuronally differentiated PC6-3 cells rather than sympathetic neurons. PC6-3 cells were electroporated with siRNAs against mkp1 either individually or as a combination and immediately returned to culture. Nontargeting siRNAs were also electroporated as a control for non-sequence-specific effects. Unstimulated PC6-3 cells that had not been electroporated were also used as controls to monitor the effects of electroporation on the viability of PC6-3 cells. After 24 h to allow for degradation of preexisting endogenous Mkp1, cells were harvested, and immunoblotting performed with an Mkp1 antibody to assess the effect of the siRNAs (Fig. 4A). PC6-3 cells that were electroporated with a nontargeting control siRNA had a similar level of Mkp1 protein to the unelectroporated control cells. Likewise, electroporation of a control siRNA against a nonessential gene (cyclophilin B) also had no effect on Mkp1 protein levels. Individual siRNAs that target different exons of the mkp1 gene significantly knocked down the Mkp1 protein by >80% when compared to controls. Pooling combinations of siRNAs showed even greater target specificity for mkp1 and appeared to have an additive effect by knocking down Mkp1 protein levels by >95% (Fig. 4A). ERK protein levels were not altered by any of the siRNAs. Next we investigated whether the siRNAs were capable of knocking down Mkp1 in sympathetic neurons. We therefore microinjected individual or pooled mkp1 siRNAs into the nucleus together with guinea pig IgG as a marker. Injected neurons were left for 24 h after injection and then were deprived of NGF for 16 h, fixed, and then immunostained for Mkp1. Neurons injected with individual siRNAs showed a significantly reduced level of Mkp1 protein (by ∼80%) by immunostaining compared with those neurons injected with the nontargeting control (Fig. 4B). Knockdown of Mkp1 was enhanced further (to ∼90%) by pooling various combinations of siRNAs (Fig. 4B). Over 85% of cells injected with siRNAs against mkp1 showed a significant reduction in Mkp1 levels.
Figure 4.

siRNAs against mkp1 knock down Mkp1 protein levels and increase NGF withdrawal-induced death. A, PC6-3 cells were electroporated for 10 ms with siRNAs against mkp1 (3 μm) or controls as indicated and returned to culture. After 24 h, electroporated cells were harvested and Mkp1 protein levels were assessed by immunoblotting. siRNAs were effective individually at knocking down Mkp1, while pooling at least two siRNAs gave almost complete knockdown of endogenous Mkp1. As a control, ERK protein levels were measured in the same cell extracts. B, Sympathetic neurons were cultured in the presence of NGF for 6 d and were then microinjected with guinea pig IgG as a marker (red) together with a control siRNA or mkp1 siRNA (3 μm) either alone or pooled as indicated. After 24 h, the neurons were rinsed and refed with medium lacking NGF for 16 h. After this, the neurons were fixed and stained with an antibody against Mkp1. Neurons injected with siRNAs against mkp1 showed little or no staining for Mkp1 (green). The scale bar represents 25 μm. C, Knocking down Mkp1 accelerates neuronal cell death after NGF withdrawal. Sympathetic neurons were cultured in vitro for 6 d and then microinjected with Texas Red dextran together with 10 ng/μl of either pcDNA1, pcDJunΔ169, pcDJBD, or pcDMkp1 as indicated, or mkp1 siRNAs. After injection, neurons were rinsed and refed with medium lacking NGF. The percentage of viable injected neurons at 72 h after NGF withdrawal was calculated. Individual and pooled siRNAs accelerate neuronal cell death when compared to nontargeting control (NTC) siRNA. The average of three experiments ± SEM is shown. #p < 0.001, *p < 0.01, Student's t test.
To test the effect of knocking down Mkp1 on NGF withdrawal-induced death, we microinjected sympathetic neurons with siRNAs against mkp1 or the nontargeting control siRNA together with Texas Red dextran as a marker and measured neuronal viability after NGF withdrawal over a period of 72 h. The viability of sympathetic neurons injected with the mkp1 siRNAs was compared directly to the siRNA nontargeting control (siRNA NTC) and empty vector. We also microinjected the expression vectors pcDMkp1, pcDJunΔ169, and pcDJBD for comparison. Microinjection of siRNAs against mkp1 increased the rate of neuronal death. At 72 h after NGF withdrawal, only 23% of neurons injected with siRNA 1 and 20% of neurons injected with siRNA 2 were viable compared to the siRNA NTC (36%) and the empty vector pcDNA1 (35%). When the two siRNAs were coinjected together there was increased cell death and only 15% of the cells were viable. When compared to the empty vector pcDNA1, the expression vectors pcDMkp1, pcDJunΔ169, and pcDJBD significantly delayed neuronal cell death, as previously demonstrated, while knocking down Mkp1 significantly accelerated neuronal cell death, suggesting that in sympathetic neurons Mkp1 plays an important role in regulating the rate of NGF withdrawal-induced death.
c-Jun and ATF2 bind to two conserved ATF sites in the mkp1 promoter
After NGF withdrawal in sympathetic neurons, the AP-1 transcription factors c-Jun and, to a lesser extent, ATF2 are phosphorylated by JNKs, and this increases their transcriptional activity (Eilers et al., 1998, 2001; Towers et al., 2009). c-Jun and ATF2 bind to specific palindromic sequences in promoters, the AP-1 site/TPA responsive element (TRE) (5′-TGA G/C TCA-3′) or the ATF site/cAMP response element (CRE) (5′-TGA CG TCA-3′), by forming c-Jun/ATF2 heterodimers or ATF2 homodimers. We investigated whether c-Jun and ATF2 could bind to the mkp1 promoter. Initially, we aligned the mkp1 promoter sequences of four species and searched for consensus transcription factor binding sites. We found two conserved, potential ATF binding sites in the mkp1 promoter, the first of which (ATF site 1) is located at position −172 to −165 in relation to the transcriptional start site and is one base different from the ATF/CRE consensus site. The second site (ATF site 2), located at position −124 to −117, is an exact match for the ATF/CRE consensus site (Fig. 5).
Figure 5.
Alignment of the promoter sequences for the rat, human, mouse, and cow mkp1 genes. Shaded regions indicate two conserved ATF sites, an E box and a TATA box. Bases conserved across all four species are shown by an asterisk below the alignment. The transcriptional start site of the mkp1 gene in each species is indicated by a square box.
We investigated whether c-Jun and ATF2 can bind to the conserved ATF sites in the mkp1 promoter in living cells by performing chromatin immunoprecipitation (ChIP) assays using antibodies against c-Jun and ATF2 and also Bim as a negative control. For ChIP experiments, we used neuronally differentiated PC6-3 cells, rather than primary sympathetic neurons, since a large number of cells are required for conventional ChIP assays. PC6-3 cells were differentiated in the presence of NGF for 7 d and then either maintained in the presence or absence of NGF for 16 h. The binding of c-Jun and ATF2 to the mkp1 promoter was studied by PCR using primers that flank mkp1 ATF site 1 and site 2 and that amplify a 259 bp region (Fig. 6A). The control antibody (Bim) immunoprecipitated a low level of chromatin containing the mkp1 promoter (Fig. 6A, top panel, lanes 3–4). However, both the c-Jun and ATF2 antibodies precipitated a significantly increased proportion of the region of the mkp1 promoter containing the two ATF sites (Fig. 6A, top panel, lanes 5–8). At 16 hours after NGF withdrawal, the amount of c-Jun and ATF2 bound to the mkp1 promoter had not increased. One possible explanation for this is that the mkp1 promoter ATF sites are already bound by c-Jun- and ATF2-containing complexes in the presence of NGF. We then tested the effect of NGF withdrawal on the amount of phospho-c-Jun and phospho-ATF2 bound to the mkp1 promoter. Again, there was a low background level of mkp1 promoter chromatin immunoprecipitated using the Bim control antibody (Fig. 6A, bottom panel, lanes 3–4). There was only a slight increase above background level when the region was immunoprecipitated with the phospho-c-Jun (serine 63) antibody in the presence of NGF (Fig. 6A, bottom panel, lane 5). However, this was significantly enriched following NGF withdrawal (Fig. 6A, bottom panel, lane 6). We found only a minor increase in the amount of phosphorylated ATF2 (threonine 71) after NGF withdrawal (Fig. 6A, bottom panel, lanes 7–8). As a control, we also studied the binding of c-Jun and ATF2 to the c-jun promoter using primers that flank the jun1 and jun2 TREs (supplemental Fig. S3, available at www.jneurosci.org as supplemental material). We found that both c-Jun and ATF2 were bound to the c-jun promoter in the presence of NGF and that the amount of c-Jun and ATF2 bound to the jun1 and jun2 TREs did not increase after NGF withdrawal. However, the level of c-Jun phosphorylation at serine 63 did increase after NGF deprivation (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), as observed for the mkp1 promoter (Fig. 6A). These results are consistent with previous results in ChIP assays studying the binding of c-Jun and ATF2 to a conserved ATF site in the dp5 promoter (Towers et al., 2009).
Figure 6.

c-Jun and ATF2 bind to two conserved ATF sites in the mkp1 promoter. A, Neuronally differentiated PC12 cells were cultured in the presence (+N) or absence (−N) of NGF for 16 h. The cells were then crosslinked, chromatin prepared and ChIP assays performed with antibodies against Bim (a negative control), c-Jun, ATF2, phospho-c-Jun (serine 63), or phospho-ATF2 (threonine 71). The mkp1 promoter contains two conserved ATF sites (Fig. 5), and the ChIP samples (as indicated) and 1% of the input chromatin samples were analyzed by PCR using primers that flank the region containing both of the ATF sites. As a negative control, a PCR without chromatin was also tested (H2O). B, Sympathetic neurons were cultured for 7 d in vitro and then refed with medium containing NGF (+N) or anti-NGF antibody (−N). Whole-cell extracts were prepared 16 h later. EMSA experiments were performed using oligonucleotides containing either the wild-type mkp1 ATF site 2 or mutant site, in which 4 nt have been changed. Four micrograms of +N or −N whole-cell extract were used per binding reaction as indicated. A control antibody against Bim or c-Jun, ATF2, phospho-c-Jun (ser63), or phospho-ATF2 (Thr71) antibodies were added as shown. The binding reactions were separated on a 5% polyacrylamide gel. Free probe and the AP-1 complexes are indicated. The four point mutations inhibited the binding of AP-1 proteins to the mkp1 ATF site. The c-Jun, ATF2, phospho-c-Jun, and phospho-ATF2 antibodies all caused a supershift of the AP-1 complexes, and these are indicated (SS).
We then investigated whether c-Jun and ATF2 in sympathetic neuron extracts can bind to mkp1 ATF site 2 in vitro by performing an EMSA experiment (Fig. 6B). Whole-cell extracts were prepared from neurons cultured in the presence of NGF for 7 d and then either maintained in the presence or absence of NGF for 16 h. Extracts from neurons maintained in the presence of NGF contained proteins that bound to mkp1 ATF site 2 (Fig. 6B, lane 2), and this binding activity increased slightly after NGF withdrawal (Fig. 6B, lane 3). We also tested a mutant ATF/CRE site in which four bases had been altered (mkp1Mut). Binding of the AP-1 proteins (marked AP-1) was abolished by introducing these point mutations (Fig. 6B, lanes 5 and 6). To determine whether the specific protein complexes contained c-Jun or ATF2, antibodies specific for c-Jun or ATF2 were added to the binding reactions. The Bim antibody was added to binding reactions as a negative control (lane 7), but there was no difference in the binding pattern when compared with the −NGF extract (lane 3). Both the c-Jun and ATF2 antibodies (lanes 8 and 9) supershifted a significant proportion of the AP-1 complexes toward the top of each lane (marked SS). A significant proportion of the AP-1 proteins bound to the mkp1 ATF site was also supershifted when an antibody against phospho-c-Jun (serine 63) was added and, to a lesser extent, when a phospho-ATF2 (threonine 71) antibody was used. These antibodies gave supershifts that were just above the AP-1 complex (Fig. 6B, lanes 10–11). These results demonstrate that after NGF withdrawal the c-Jun in sympathetic neuron extracts can bind to mkp1 ATF site 2 in vitro and is phosphorylated at serine 63, while the amount of ATF2 (phosphorylated at threonine 71) bound to mkp1 ATF site 2 is lower.
mkp1 promoter activation requires the two ATF sites, JNK, c-Jun, and ATF2
To study how mkp1 expression is regulated, we constructed a luciferase reporter plasmid (mkp1-LUC) containing 1 kb of the mkp1 promoter sequence. Sympathetic neurons were cultured for 6 d in the presence of NGF and then microinjected with mkp1-LUC together with the control Renilla luciferase construct pRL-TK. After injection, the cells were maintained for 16 h in the presence or the absence of NGF, after which time relative luciferase activity was determined by dual luciferase assay. The mkp1-LUC reporter construct is significantly activated by 1.53-fold (p = 0.001) following withdrawal of NGF (Fig. 7A). To test whether the two conserved ATF sites are necessary for activation of the mkp1 promoter after NGF withdrawal, we introduced four point mutations into both ATF sites to abolish AP-1 binding, in the construct mkp1-LUC (2xATF mutant). Mutation of the ATF sites resulted in a 37% decrease in basal promoter activity in the presence of NGF when compared to wild type (Fig. 7A). After NGF withdrawal, activation of the 2xATF mutant was completely abolished (Fig. 7A). This demonstrates that mkp1 promoter activation is mediated through the two ATF sites.
Figure 7.
The two conserved ATF sites in the mkp1 promoter contribute to basal promoter activity and are required for promoter induction after NGF withdrawal in sympathetic neurons. A, A 1 kb DNA fragment containing the rat mkp1 promoter was cloned upstream of the Firefly luciferase gene in pGL3-basic. The two conserved ATF sites were then mutated using the QuikChange IIXL Site-Directed Mutagenesis Kit. The wild-type and mutant mkp1-LUC constructs (10 ng/μl) were microinjected into sympathetic neurons together with the Renilla luciferase construct pRL-TK (5 ng/μl). The injected cells were then refed with medium containing or lacking NGF, as indicated. After 16 h, a dual luciferase assay was performed and normalized activity was calculated relative to +NGF (gray bars). Statistical significance was calculated relative to +NGF (gray bars) for each construct. The data shown represents the mean of 4 experiments ± SEM. B, Sympathetic neurons were microinjected with wild-type or the 2xATF mutant mkp1-LUC (20 ng/μl), pRL-TK(5 ng/μl), and wild-type or kinase-dead MLK3 expression vectors (20 ng/μl) or pcDNA3 as a control. Cells were maintained in medium containing NGF for 16 h and then luciferase activity was measured. Luciferase activity was calculated relative to the level for mkp1-LUC coinjected with pcDNA3 (control), which was set as 1. Statistical significance was calculated relative to pcDNA3 (control) for each construct. The mean of three independent experiments ± SEM is shown. C, Sympathetic neurons were comicroinjected with mkp1-LUC (20 ng/μl) and pRL-TK(5 ng/μl) together with wtMLK3 or kdMLK3 (20 ng/μl) in the presence and absence of mkp1 siRNA (3 μm). Cells were maintained in medium containing NGF for 16 h and then luciferase activity was measured. Luciferase activity was calculated relative to the level for mkp1-LUC coinjected with the kdMLK3 construct and NTC siRNA. The mean of three independent experiments ± SEM is shown. D, Expression of the JIP-1 JBD reduces induction of an mkp1 reporter construct after NGF withdrawal. Sympathetic neurons were microinjected with mkp1-LUC (20 ng/μl), pRL-TK (5 ng/μl), and pcDNA3 or pcDJBD (50 ng/μl). Cells were maintained +NGF or −NGF for 16 h and then luciferase activity was measured. Luciferase activity was calculated relative to the level for mkp1-LUC injected with pcDNA3 +NGF, which was set as 1. Statistical significance was calculated relative to +NGF (gray bars) for each construct. The mean of three independent experiments ± SEM is shown. E, Sympathetic neurons were cultured for 6 d in vitro and then microinjected with mkp1-LUC (20 ng/μl), pRL-TK (5 ng/μl), and the c-Jun (H-79) or ATF2 (C-19) antibodies or rabbit Ig as a control (1 μg/μl) as indicated. Following injection, the cells were maintained in medium containing NGF (+) or anti-NGF antibody (−) for 16 h before luciferase activity was measured. For each experiment, the level of normalized luciferase activity for control IgG +NGF was set as 1 and other values were calculated relative to this. Statistical significance was calculated relative to +NGF (gray bars) for each antibody coinjection. The mean ± SEM. for five independent experiments is shown. Student's t test was used to determine whether inductions were significant: #p < 0.001; *p < 0.01; +p < 0.1, NS (not significant) p > 0.1.
Mixed-lineage kinase 3 (MLK3) belongs to a family of MAP kinase kinase kinases that can activate the JNK pathway. MLK3 is activated after NGF withdrawal and overexpression of this upstream activator can promote neuronal death, while kinase-dead MLK3 mutants can block apoptosis as well as c-Jun phosphorylation induced by NGF withdrawal (Mota et al., 2001). We investigated whether overexpression of MLK3 was sufficient to activate the mkp1 promoter in the presence of NGF. Sympathetic neurons were microinjected with wild-type mkp1-LUC or the 2xATF mutant together with an expression vector for wild-type MLK3 (wtMLK3), kinase-dead MLK3 (kdMLK3), or the empty vector pcDNA3. Neurons were cultured for 16 h in the presence of NGF, after which luciferase activity was determined. When mkp1-LUC and the kdMLK3 were coinjected, there was a small, insignificant increase in relative luciferase activity compared to empty vector. However, when mkp1-LUC and wtMLK3 were coinjected there was a significant increase in luciferase activity (3.1-fold). This induction was greatly reduced (to 1.19-fold) when wtMLK3 was coinjected with the 2xATF mutant. These results suggest that overexpression of MLK3 is sufficient to activate the mkp1 promoter, and this requires the two ATF sites. To test whether wtMLK3-driven JNK activity can also increase endogenous mkp1 expression, we coinjected sympathetic neurons with mkp1-LUC together with wtMLK3 or kdMLK3 in the presence and absence of mkp1 siRNA (Fig. 7C). Neurons were cultured for 16 h in the presence of NGF, after which luciferase activity was determined. When mkp1-LUC and kdMLK3 were coinjected in the presence of mkp1 siRNA there was a small, insignificant increase in luciferase activity (1.2-fold) when compared to neurons coinjected in the presence of NTC siRNA. However, when mkp1-LUC and wtMLK3 where coinjected in the presence of mkp1 siRNA, there was a significant increase in luciferase activity (9-fold). This demonstrates that the wtMLK3 can induce the endogenous mkp1, which in turn would limit the activation of the mkp1-LUC reporter construct by wtMLK3.
We next investigated the effect of directly inhibiting JNK activity by expressing the JNK binding domain (JBD) of the scaffold protein JNK interacting protein 1 (JIP-1), a selective JNK inhibitor (Davis, 2000). Expression of the JBD in sympathetic neurons inhibits JNK activity and can delay NGF withdrawal-induced death (Eilers et al., 2001; Harding et al., 2001) (Fig. 3B). To investigate whether JNK activity was necessary for induction of the mkp1 promoter, we microinjected an expression vector for the JBD or the empty vector pcDNA3 into sympathetic neurons together with mkp1-LUC and measured luciferase activity 16 h after NGF withdrawal (Fig. 7D). When the empty vector, pcDNA3 was injected, there was a significant 1.51-fold induction of the mkp1-LUC reporter construct after NGF deprivation. Expression of the JBD completely abolished the induction of the mkp1-LUC reporter construct after NGF withdrawal. In addition, there was a 62% decrease in basal promoter activity when the mkp1-LUC basal levels with the pcDNA3 and pcDJBD expression vectors were compared. These results suggest that JNK activity is required for the normal induction of the mkp1 reporter construct following NGF withdrawal.
It has been shown previously that the NGF withdrawal-induced death of sympathetic neurons can be inhibited by microinjecting antibodies against c-Jun, but not Jun B or Jun D antibodies or control IgG (Estus et al., 1994). To investigate the individual roles of c-Jun and ATF2 in the activation of mkp1-LUC after NGF withdrawal, we performed an antibody coinjection experiment (Fig. 7E). Sympathetic neurons were injected with the mkp1-LUC reporter construct together with the control rabbit Ig or the c-Jun or ATF2 antibodies. Following injection with the control antibody, the mkp1-LUC reporter construct was activated 2.31-fold after NGF withdrawal. However, in comparison, activation of the mkp1-LUC reporter construct was significantly reduced following NGF withdrawal when antibodies against c-Jun (1.26-fold) or ATF2 (1.47-fold) were coinjected. The basal promoter activity in the presence of NGF was not affected. These results suggest that following NGF withdrawal both c-Jun and ATF2 are important for the activation of the mkp1 promoter in sympathetic neurons.
Knock-out of mkp1 results in increased sympathetic neuron apoptosis during early postnatal development
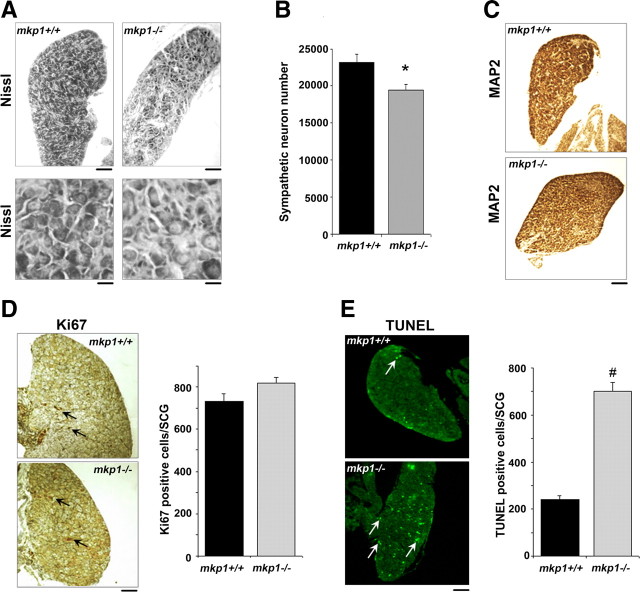
Finally, we studied the role of mkp1 in sympathetic neurons in vivo by comparing the number of sympathetic neurons in the superior cervical ganglia of 1-d-old wild-type (mkp1+/+) and mkp1−/− mice (Fig. 8). At this time point of naturally occurring cell death, there were almost 20% fewer sympathetic neurons in the mkp1−/− SCG than in the mkp1+/+ SCG (Fig. 8A,B). To confirm that the Nissl-stained cell bodies counted were indeed sympathetic neurons, sections were immunostained with an antibody to the neuronal marker MAP2 (Fig. 8C).
Figure 8.
The number of sympathetic neurons is reduced during early postnatal development in mkp1−/− SCG compared to mkp1+/+ SCG. A, Brightfield images showing sections of mkp1+/+ and mkp1−/− P1 SCGs stained with Nissl. The top panel shows sections at low magnification (scale bar, 100 μm). The bottom panel shows a higher magnification (scale bar, 30 μm). B, Sympathetic neuron numbers in P1 SCGs isolated from mkp1+/+ and mkp1−/− mice were quantified in three animals of each genotype. Error bars represent SEM. *p < 0.02, Student's t test. C, Sections from mkp1+/+ and mkp1−/− P1 SCGs were also stained with an antibody to the neuronal marker MAP2. Scale bar, 100 μm. D, Sections from mkp1+/+ and mkp1−/− P1 SCGs were stained with the proliferation marker Ki67. Arrows indicate Ki67-positive cells. Scale bar, 100 μm. The number of Ki67-positive cells was determined in the mkp1+/+ and mkp1−/− P1 SCGs. Error bars represent SEM. p = 0.12. E, Sections from mkp1+/+ and mkp1−/− P1 SCGs were analyzed for apoptosis using TUNEL. Arrows indicate TUNEL-positive cells. Scale bar, 100 μm. The number of TUNEL-positive cells was determined in the mkp1+/+ and mkp1−/− P1 SCGs. Error bars represent SEM. p = 0.0004.
To determine whether the difference in sympathetic neuron number at this time point was due to an increase in neuronal apoptosis or a decrease in the level of proliferation of sympathetic neuroblasts, we used Ki67 as a marker of proliferating cells and TUNEL to measure the total number of apoptotic cells in the SCG at P1 (Fig. 8D,E). Immunostaining of SCG sections with Ki67 showed that there was no significant difference in the number of proliferating cells (Fig. 8D). However, at P1, the mkp1−/− SCG had approximately three times more apoptotic cells than the mkp1+/+ SCG (Fig. 8E). These data suggest that there is a decrease in sympathetic neuron number in the SCG of mkp1−/− mice that is attributable to an increase in neuronal death rather than a decrease in cellular proliferation. During naturally occurring sympathetic neuron death, mkp1 therefore acts as an inhibitor of apoptosis.
Discussion
Inhibitors of transcription and protein synthesis can block NGF withdrawal-induced apoptosis in sympathetic neurons, suggesting that new gene expression is required for cell death (Martin et al., 1988). However, only a limited number of genes induced by NGF withdrawal have been identified so far. Using microarray technology (Affymetrix Exon arrays) and RNA isolated from rat sympathetic neurons, we identified a MAPK phosphatase, mkp1, as a potential transcriptional target of the MLK-JNK pathway. Mkp1 is an immediate early gene and therefore many factors can increase mkp1 mRNA levels in different cell types, including heat shock and oxidative stress (Keyse and Emslie, 1992; Franklin et al., 1998), UV light (Franklin et al., 1998), and survival factor withdrawal (Fig. 1). We found not only that the mkp1 RNA significantly increased in level after NGF withdrawal in sympathetic neurons, as previously described (Estus et al., 1994), but also that this increase is strongly reduced by a MLK inhibitor (CEP-11004). Interestingly, although we could detect the expression of seven other MAPK phosphatases (MKPs/DUSPs) in sympathetic neurons (Fig. 9), only the mkp1 mRNA increased in level after NGF withdrawal. One of the other DUSPs, DUSP6/MKP3, significantly decreased in level after NGF deprivation, in agreement with the previous observation that the expression of MKP3 is induced by the addition of NGF and that this depends on ERK activity (Camps et al., 1998), which falls after NGF withdrawal (Xia et al., 1995). At the protein level, Mkp1 is rapidly induced in response to NGF withdrawal peaking at 16 h; however, we found that this increase is prevented by CEP-11004 (Fig. 2). Given that the increase in mkp1 mRNA and Mkp1 protein level occurs after JNK activation and depends on the MLK-JNK pathway, we studied the function and regulation of mkp1.
Figure 9.

A gene microarray heat map of DUSP genes and selected control genes in sympathetic neurons. RNA was isolated from sympathetic neurons cultured in medium containing NGF (+NGF), lacking NGF (−NGF), or lacking NGF but containing 400 nm CEP-11004 (−NGF+CEP-11004) and analyzed using the Affymetrix Rat Exon 1.0ST microarray. A heat map generated from the microarray data reflecting log2 normalized gene expression values is shown for all DUSP family members represented on the chip and a selection of control genes. Only dusp1/mkp1 increased in expression after NGF withdrawal. Green represents lower gene expression and red represents higher gene expression. Each condition is represented in triplicate.
After NGF withdrawal in sympathetic neurons, JNKs phosphorylate the AP-1 transcription factor c-Jun, which ultimately promotes neuronal death. In microinjection experiments, we found that overexpressing Mkp1 in sympathetic neurons prevented the phosphorylation of c-Jun at serine 63 after NGF withdrawal, suggesting that the dephosphorylation of JNKs by Mkp1 prevented the subsequent phosphorylation of c-Jun. By performing survival assays, we found that Mkp1 overexpression significantly delayed apoptosis in sympathetic neurons after NGF withdrawal. This delay was similar to that seen with expression constructs encoding a dominant negative c-Jun mutant (JunΔ169) or the JBD (a direct JNK inhibitor protein), both of which are known to protect against NGF withdrawal-induced death. We then microinjected siRNAs targeting different regions of the mkp1 gene into sympathetic neurons. In survival assays, knockdown of Mkp1 using individual siRNAs accelerated the death of sympathetic neurons and this effect could be enhanced when pools of siRNAs were injected. These observations suggest that Mkp1 plays an important role in regulating the rate of NGF withdrawal-induced death. In agreement with this, we found that at P1 during the period of naturally occurring sympathetic neuron apoptosis, the number of SCG neurons in mkp1−/− mice was reduced by 20% compared to wild type (Fig. 8B) and the number of TUNEL-positive cells/SCG was increased from 240 in wild-type mice to 700 in mkp1−/− mice (Fig. 8E).
We identified two conserved ATF-binding sites in the rat mkp1 promoter and showed that these sites bind c-Jun and ATF2 in the chromatin of neuronally differentiated PC12 cells and in sympathetic neuron extracts in vitro. One of these sites (TGACGTCA) was shown to bind ATF2 in ChIP assays with chromatin from mouse embryonic fibroblasts (Breitwieser et al., 2007). To study the role of the ATF sites in the mkp1 promoter, we constructed an mkp1 reporter plasmid in which both ATF binding sites had been mutated. This abolished the binding of AP-1 proteins and the induction of the reporter plasmid after NGF withdrawal. It also significantly reduced the basal promoter activity, suggesting that these sites are important mkp1 promoter elements. In sympathetic neurons cultured in the presence of NGF, the mkp1 ATF sites may contribute to basal promoter activity by binding c-Jun/ATF heterodimers. After NGF withdrawal, these heterodimers would be targeted by nuclear JNKs for phosphorylation, which would lead to an increase in their transcriptional activity and thereby contribute to the induction of mkp1 transcription. In agreement with this hypothesis, we found that JNK activity and c-Jun and ATF2 are required for the induction of mkp1-LUC after NGF withdrawal. We obtained similar results for the two ATF binding sites in the c-jun promoter, the jun1 and jun2 TREs (supplemental Fig. S3, available at www.jneurosci.org as supplemental material) (Eilers et al., 1998, 2001). These observations are consistent with ChIP data published by Hayakawa et al. (2004), who found that activation of the JNK pathway by cisplatinum in a breast carcinoma cell line did not increase the amount of c-Jun and ATF2 bound to the promoters of c-jun, c-fos, and other c-Jun target genes but did increase the amount of phosphorylated c-Jun on the same promoters.
MAPK phosphatases, such as mkp1, belong to a larger DUSP subgroup that all have the capability of dephosphorylating MAPKs. MAPK signaling cascades regulate important cellular processes, including gene expression, cell proliferation, cell survival, and death. Mkp1 functions to negatively regulate MAPK signaling (Sun et al., 1993; Keyse, 2000) and has been implicated in metabolic control (Wu et al., 2006), immune regulation [for review, see Abraham and Clark (2006), Jeffrey et al. (2007), and Liu et al. (2007)], and cancer (Keyse, 2008). Several studies have shown that in cancer cell models, spontaneous or conditional expression of Mkp1 can protect against apoptosis by its ability to dephosphorylate MAP kinases (Magi-Galluzzi et al., 1997; Franklin et al., 1998; Srikanth et al., 1999; Sánchez-Pérez et al., 2000). Interestingly, mutations in the Drosophila mkp1 homolog, puckered, cause hyperactivation of DJNK, resulting in cytoskeletal defects that lead to failure in dorsal closure, while a knock-out of puckered can trigger apoptosis in the developing Drosophila embryo (Martín-Blanco et al., 1998; McEwen and Peifer, 2005). Conversely, overexpression of puckered can mimic basket (Drosophila JNK homolog) mutant phenotypes and therefore inactivate signaling through JNK/basket (Martín-Blanco et al., 1998).
In this study, we compared the number of SCG neurons at P1 in wild-type mice and a conventional mkp1−/− knock-out line. In the future, it would be interesting to study a neuron-specific conditional knock-out of mkp1 to control for any compensatory changes that may have occurred earlier during development. Since the JNK kinases MKK4 and MKK7 bind to the same region of JNKs as Mkp1 (the CD domain) (Tanoue et al., 2000), it would also be interesting to count the number of SCG neurons in a neuron-specific conditional double knock-out of mkk4 and mkk7. This would be predicted to reduce JNK activity and therefore increase the number of SCG neurons at P1.
Mkp1 can be phosphorylated by ERKs, which increases its stability and therefore reinforces its negative feedback function (Brondello et al., 1999). After NGF withdrawal in sympathetic neurons, the balance of MAPK activation is markedly shifted in favor of the JNK pathway, while ERK activity falls (Xia et al., 1995). Thus, although the mkp1 mRNA is rapidly induced after NGF deprivation, the decrease in ERK activity may delay or reduce the magnitude of the increase in Mkp1 protein level, which in turn may lead to a more protracted activation of JNKs. After NGF deprivation, JNKs phosphorylate c-Jun and ATF2, which increases their transcriptional activity and c-Jun expression (Fig. 10). c-Jun binds to two ATF binding sites present in the promoter of c-jun itself (supplemental Fig. S3, available at www.jneurosci.org as supplemental material), to AP-1 sites in cell death genes, such as dp5 and bim (Towers et al., 2009), and also to two ATF binding sites in the mkp1 promoter (Fig. 6). After NGF withdrawal in sympathetic neurons, JNK kinase activity increases twofold after 4 h but by 16 h starts to return to basal levels (Eilers et al., 1998). The time at which JNK activity is highest (4 h after NGF withdrawal) slightly precedes the time at which c-Jun levels and c-Jun N-terminal phosphorylation start to increase (Ham et al., 1995; Eilers et al., 1998), which in turn slightly precedes the time at which Mkp1 starts to increase in level (∼8 h). The strong induction of mkp1 after 16 h of NGF withdrawal could explain why at this time point JNK activity in sympathetic neurons returns to basal levels (Eilers et al., 1998). Thus, by dephosphorylating JNKs, Mkp1 would form part of a negative feedback loop (Fig. 10).
Figure 10.
Hypothetical model showing how Mkp1 negatively regulates the MLK-JNK-c-Jun pathway after NGF withdrawal. After NGF withdrawal, JNK activity increases leading to phosphorylation of the AP-1 transcription factors c-Jun and ATF2. This increases their ability to activate the transcription of their target genes. c-Jun binds to ATF binding sites present in the promoters of c-jun itself (the jun1 and jun2 TREs) to enhance transcription of c-jun in a positive feedback manner. c-Jun can also bind to AP-1 sites in cell death genes, such as dp5, bim, and puma and also to two conserved ATF binding sites in the mkp1 promoter. This leads to increased expression of Mkp1, which in turn can dephosphorylate JNKs in a negative feedback manner. Induction of the mkp1 promoter after NGF withdrawal can be reduced by mutating both of the ATF sites, by expressing JunΔ169 or the JIP1-JBD, or by using the MLK inhibitor CEP-11004. Mkp1 acts as a negative regulator of the JNK pathway and thereby modulates the rate of neuronal death after NGF withdrawal.
We conclude that mkp1 is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons and is the first NGF withdrawal-induced gene to be described that inhibits apoptosis in this system. Studying the role of Mkp1 in sympathetic neurons has contributed to our understanding of the gene network activated by the MLK-JNK-c-Jun pathway after NGF withdrawal. Mkp1 is part of a negative feedback loop induced by the MLK-JNK pathway that modulates JNK activity. Since the level of Mkp1 activity determines the rate of cell death after NGF withdrawal, signals that increase the expression or activity of Mkp1, such as cAMP (Zhang et al., 2008), would be predicted to promote neuronal survival.
Footnotes
This project was supported by the Wellcome Trust. We thank Francesca Menghi for help with the analysis of microarray data, Jasper de Boer for advice on electroporation, and Cephalon, Inc., for CEP-11004. We are grateful to Mike Hubank for critical reading of the manuscript. We are also grateful to Bristol-Myers Squibb for providing the mkp1−/− mouse strain. We would also like to thank Victoria Brown, Nouruja Rahman, Olga Boruc, and Mino Medghalchi for expert advice and technical help.
References
- Aalto AP, Sarin LP, van Dijk AA, Saarma M, Poranen MM, Arumäe U, Bamford DH. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase. RNA. 2007;13:422–429. doi: 10.1261/rna.348307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham SM, Clark AR. Dual-specificity phosphatase 1: a critical regulator of innate immune responses. Biochem Soc Trans. 2006;34:1018–1023. doi: 10.1042/BST0341018. [DOI] [PubMed] [Google Scholar]
- Breitwieser W, Lyons S, Flenniken AM, Ashton G, Bruder G, Willington M, Lacaud G, Kouskoff V, Jones N. Feedback regulation of p38 activity via ATF2 is essential for survival of embryonic liver cells. Genes Dev. 2007;21:2069–2082. doi: 10.1101/gad.430207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello JM, Pouysségur J, McKenzie FR. Reduced MAP kinase phosphatase-1 degradation after p42/p44MAPK-dependent phosphorylation. Science. 1999;286:2514–2517. doi: 10.1126/science.286.5449.2514. [DOI] [PubMed] [Google Scholar]
- Camps M, Chabert C, Muda M, Boschert U, Gillieron C, Arkinstall S. Induction of the mitogen-activated protein kinase phosphatase MKP3 by nerve growth factor in differentiating PC12. FEBS Lett. 1998;425:271–276. doi: 10.1016/s0014-5793(98)00250-6. [DOI] [PubMed] [Google Scholar]
- Coggeshall RE, Chung K, Greenwood D, Hulsebosch CE. An empirical method for converting nucleolar counts to neuronal numbers. J Neurosci Methods. 1984;12:125–132. doi: 10.1016/0165-0270(84)90011-6. [DOI] [PubMed] [Google Scholar]
- Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- Deckwerth TL, Johnson EM., Jr Temporal analysis of events associated with programmed cell death (apoptosis) of sympathetic neurons deprived of nerve growth factor. J Cell Biol. 1993;123:1207–1222. doi: 10.1083/jcb.123.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh M, Johnson EM., Jr Evidence of a novel event during neuronal death: development of competence-to-die in response to cytoplasmic cytochrome c. Neuron. 1998;21:695–705. doi: 10.1016/s0896-6273(00)80587-5. [DOI] [PubMed] [Google Scholar]
- Dorfman K, Carrasco D, Gruda M, Ryan C, Lira SA, Bravo R. Disruption of the erp/mkp-1 gene does not affect mouse development: normal MAP kinase activity in ERP/MKP-1-deficient fibroblasts. Oncogene. 1996;13:925–931. [PubMed] [Google Scholar]
- Edwards SN, Tolkovsky AM. Characterisation of apoptosis in cultured rat sympathetic neurons after nerve growth factor withdrawal. J Cell Biol. 1994;124:537–546. doi: 10.1083/jcb.124.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers A, Whitfield J, Babij C, Rubin LL, Ham J. Role of the Jun kinase pathway in the regulation of c-Jun expression and apoptosis in sympathetic neurons. J Neurosci. 1998;18:1713–1724. doi: 10.1523/JNEUROSCI.18-05-01713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilers A, Whitfield J, Shah B, Spadoni C, Desmond H, Ham J. Direct inhibition of c-Jun N-terminal kinase in sympathetic neurones prevents c-jun promoter activation and NGF withdrawal-induced death. J Neurochem. 2001;76:1439–1454. doi: 10.1046/j.1471-4159.2001.00150.x. [DOI] [PubMed] [Google Scholar]
- Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr Altered gene expression in neurons during programmed cell death: identification of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin CC, Srikanth S, Kraft AS. Conditional expression of mitogen activated protein kinase phosphatase-1, MKP-1, is cytoprotective against UV-induced apoptosis. Proc Natl Acad Sci U S A. 1998;95:3014–3019. doi: 10.1073/pnas.95.6.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Campbell D, Dérijard B, Davis RJ. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- Ham J, Babij C, Whitfield J, Pfarr CM, Lallemand D, Yaniv M, Rubin LL. A c-Jun dominant negative mutant protects sympathetic neurons against programmed cell death. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Harding TC, Xue L, Bienemann A, Haywood D, Dickens M, Tolkovsky AM, Uney JB. Inhibition of JNK by overexpression of the JNK binding domain of JIP-1 prevents apoptosis in sympathetic neurons. J Biol Chem. 2001;276:4531–4534. doi: 10.1074/jbc.C000815200. [DOI] [PubMed] [Google Scholar]
- Hayakawa J, Mittal S, Wang Y, Korkmaz KS, Adamson E, English C, Ohmichi M, McClelland M, Mercola D. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol Cell. 2004;16:521–535. doi: 10.1016/j.molcel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Hazzalin CA, Mahadevan LC. MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol. 2002;3:30–40. doi: 10.1038/nrm715. [DOI] [PubMed] [Google Scholar]
- Jacobs WB, Govoni G, Ho D, Atwal JK, Barnabe-Heider F, Keyes WM, Mills AA, Miller FD, Kaplan DR. P63 is an essential proapoptotic protein during neural development. Neuron. 2005;48:743–756. doi: 10.1016/j.neuron.2005.10.027. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–16486. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- Keyse SM. Protein phosphatases and the regulation of mitogen-activated protein kinase signalling. Curr Opin Cell Biol. 2000;12:186–192. doi: 10.1016/s0955-0674(99)00075-7. [DOI] [PubMed] [Google Scholar]
- Keyse SM. Dual-specificity MAP kinase phosphatases (MKPs) and cancer. Cancer Metastasis Rev. 2008;27:253–261. doi: 10.1007/s10555-008-9123-1. [DOI] [PubMed] [Google Scholar]
- Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359:644–647. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- Liu Y, Shepherd EG, Nelin LD. MAPK phosphatases—regulating the immune response. Nat Rev Immunol. 2007;7:202–212. doi: 10.1038/nri2035. [DOI] [PubMed] [Google Scholar]
- Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, Wishnow K, Kaplan I, Stork PJ, Loda M. Mitogen-activated protein kinase phosphatase 1 is overexpressed in prostate cancers and is inversely related to apoptosis. Lab Invest. 1997;76:37–51. [PubMed] [Google Scholar]
- Maroney AC, Finn JP, Connors TJ, Durkin JT, Angeles T, Gessner G, Xu Z, Meyer SL, Savage MJ, Greene LA, Scott RW, Vaught JL. Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family. J Biol Chem. 2001;276:25302–25308. doi: 10.1074/jbc.M011601200. [DOI] [PubMed] [Google Scholar]
- Martin DP, Schmidt RE, DiStefano PS, Lowry OH, Carter JG, Johnson EM., Jr Inhibitors of protein synthesis and RNA synthesis prevent neuronal death caused by nerve growth factor deprivation. J Cell Biol. 1988;106:829–844. doi: 10.1083/jcb.106.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, Martinez-Arias A. Puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12:557–570. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen DG, Peifer M. Puckered, a Drosophila MAPK phosphatase, ensures cell viability by antagonizing JNK-induced apoptosis. Development. 2005;132:3935–3946. doi: 10.1242/dev.01949. [DOI] [PubMed] [Google Scholar]
- Mota M, Reeder M, Chernoff J, Bazenet CE. Evidence for a role of mixed lineage kinases in neuronal apoptosis. J Neurosci. 2001;21:4949–4957. doi: 10.1523/JNEUROSCI.21-14-04949.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neame SJ, Rubin LL, Philpott KL. Blocking cytochrome c activity within intact neurons inhibits apoptosis. J Cell Biol. 1998;142:1583–1593. doi: 10.1083/jcb.142.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- Pittman RN, Wang S, DiBenedetto AJ, Mills JC. A system for characterizing cellular and molecular events in programmed neuronal cell death. J Neurosci. 1993;13:3669–3680. doi: 10.1523/JNEUROSCI.13-09-03669.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser S, Massiha A, Piuz I, Schlegel W. Stimulated initiation of mitogen activated protein kinase phosphatase-1 (MKP-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem J. 2004;378:473–484. doi: 10.1042/BJ20031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Pérez I, Martínez-Gomariz M, Williams D, Keyse SM, Perona R. CL100/MKP-1 modulates JNK activation and apoptosis in response to cisplatin. Oncogene. 2000;19:5142–5152. doi: 10.1038/sj.onc.1203887. [DOI] [PubMed] [Google Scholar]
- Slack DN, Seternes OM, Gabrielsen M, Keyse SM. Distinct binding determinants for ERK2/p38alpha and JNK map kinases mediate catalytic activation and substrate selectivity of MAP kinase phosphatase-1. J Biol Chem. 2001;276:16491–16500. doi: 10.1074/jbc.M010966200. [DOI] [PubMed] [Google Scholar]
- Srikanth S, Franklin CC, Duke RC, Kraft RS. Human DU145 prostate cancer cells overexpressing mitogen-activated protein kinase phosphatase-1 are resistant to Fas ligand-induced mitochondrial perturbations and cellular apoptosis. Mol Cell Biochem. 1999;199:169–178. doi: 10.1023/a:1006980326855. [DOI] [PubMed] [Google Scholar]
- Sun H, Charles CH, Lau LF, Tonks NK. MKP-1 (3CH134), an immediate early gene product, is a dual specificity phosphatase that dephosphorylates MAP kinase in vivo. Cell. 1993;75:487–493. doi: 10.1016/0092-8674(93)90383-2. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- Towers E, Gilley J, Randall R, Hughes R, Kristiansen M, Ham J. The proapoptotic dp5 gene is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons. Nucleic Acids Res. 2009;37:3044–3060. doi: 10.1093/nar/gkp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dam H, Wilhelm D, Herr I, Steffen A, Herrlich P, Angel P. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 1995;14:1798–1811. doi: 10.1002/j.1460-2075.1995.tb07168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virdee K, Bannister AJ, Hunt SP, Tolkovsky AM. Comparison between the timing of JNK activation, c-Jun phosphorylation, and onset of death commitment in sympathetic neurones. J Neurochem. 1997;69:550–561. doi: 10.1046/j.1471-4159.1997.69020550.x. [DOI] [PubMed] [Google Scholar]
- Wang LH, Paden AJ, Johnson EM., Jr Mixed-lineage kinase inhibitors require the activation of Trk receptors to maintain long-term neuronal trophism and survival. J Pharmacol Exp Ther. 2005;312:1007–1019. doi: 10.1124/jpet.104.077800. [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- Whitfield J, Neame SJ, Ham J. Methods for culturing primary sympathetic neurons and for determining neuronal viability. Methods Mol Biol. 2004;282:157–168. doi: 10.1385/1-59259-812-9:157. [DOI] [PubMed] [Google Scholar]
- Wright KM, Vaughn AE, Deshmukh M. Apoptosome dependent caspase-3 activation pathway is non-redundant and necessary for apoptosis in sympathetic neurons. Cell Death Differ. 2007;14:625–633. doi: 10.1038/sj.cdd.4402024. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Roth RJ, Anderson EJ, Hong EG, Lee MK, Choi CS, Neufer PD, Shulman GI, Kim JK, Bennett AM. Mice lacking MAP kinase phosphatase-1 have enhanced MAP kinase activity and resistance to diet-induced obesity. Cell Metab. 2006;4:61–73. doi: 10.1016/j.cmet.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- Xu Z, Maroney AC, Dobrzanski P, Kukekov NV, Greene LA. The MLK family mediates c-Jun N-terminal kinase activation in neuronal apoptosis. Mol Cell Biol. 2001;21:4713–4724. doi: 10.1128/MCB.21.14.4713-4724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang Q, Zhu N, Yu M, Shen B, Xiang J, Lin A. Cyclic AMP inhibits JNK activation by CREB-mediated induction of c-FLIP(L) and MKP-1, thereby antagonizing UV-induced apoptosis. Cell Death Differ. 2008;15:1654–1662. doi: 10.1038/cdd.2008.87. [DOI] [PubMed] [Google Scholar]