Abstract
Over the last 30 years the process of in vitro fertilisation (IVF) has evolved considerably, yet the efficiency of this treatment remains relatively poor. The principal challenge faced by doctors and embryologists is the identification of the embryo with the greatest potential for producing a child. Current methods of embryo viability assessment provide only a rough guide to potential. In order to improve the odds of a successful pregnancy it is typical to transfer more than one embryo to the uterus. However, this often results in multiple pregnancies (twins, triplets, etc), which are associated with significantly elevated risks of serious complications. If embryo viability could be assessed more accurately, it would be possible to transfer fewer embryos without negatively impacting IVF pregnancy rates. In order to assist with the identification of viable embryos, several scoring systems based on morphological criteria have been developed. However, these mostly rely on a subjective visual analysis. Automated assessment of morphological features offers the possibility of more accurate quantification of key embryo characteristics and elimination of inter- and intra-observer variation. In this paper, we describe the main embryo scoring systems currently in use and review related works on embryo image analysis that could lead to an automatic and precise grading of embryo quality. We summarise achievements, discuss challenges ahead, and point to some possible future directions in this research field.
Keywords: embryo grading, image analysis, in vitro fertilisation.
INTRODUCTION
According to the 2006 American report on Assisted Reproductive Technology (ART) Success Rates [1], the dream of having a child is not easily realised for many people who want to start a family. One couple in six cannot conceive naturally and approximately 12% of women of childbearing age in the United States have used an infertility service.
In vitro fertilisation (IVF) is one of the principal methods employed for the treatment of infertility. It is estimated that there are more than one million IVF treatments carried out worldwide each year. Utilisation is particularly high in developed countries, where IVF and related treatments now account for 1-4% of all births. The process of IVF has evolved considerably since the first successful treatment three decades ago [2]. However, the efficiency of the treatment remains relatively poor, mainly due to the low probability of an individual embryo successfully implanting in the uterus and producing a child. For this reason, IVF clinics generally transfer more than one embryo per cycle, in the hope of increasing the probability of success. While this approach has helped to maintain IVF pregnancy rates at an acceptable level, it has also led to an explosion in the number of multiple pregnancies.
Currently, over 40% of deliveries following IVF in the USA consist of twins, triplets or even higher numbers of births [3]. High-order multiple pregnancies, such as those frequently produced by IVF, are associated with significantly elevated risks of serious complications. Mothers carrying twins or triplets have an increased incidence of preeclampsia, maternal haemorrhage, operative delivery, uterine rupture, and preterm labour [4]. Infants from multiple pregnancies face increased perinatal mortality, 4.5 fold and 9 fold for twins and triplets respectively. The incidence of cerebral palsy displays similar increases [5]. Preterm deliveries resulting from multiple IVF pregnancies also have financial implications for health services and insurers, costing an estimated $890 million in the USA annually [6].
Multiple pregnancies can easily be prevented by transferring fewer embryos to the mother's uterus each cycle, the ideal strategy being single embryo transfer. However, restricting the number of embryos transferred has a negative impact on the likelihood of a patient becoming pregnant each cycle. The reason for this is that the embryos produced in a typical IVF cycle are extremely variable in terms of their ability to form a viable pregnancy. In cases of single embryo transfer (SET) it is therefore essential that the embryo chosen for transfer is that having the greatest potential for forming a pregnancy and producing a healthy child.
Currently, the decision of which embryo to transfer is made on the basis of morphological assessments conducted in the IVF laboratory. However, despite a large number of published studies, there is no consensus about the most accurate method for embryo quality assessment. Additionally, the available grading systems rely mostly on visual information obtained by the embryologist and are thus subject to interobserver (and to some extent intraobserver) variance.
In a study of morphological grading carried out for day-3 embryos (i.e. three days after fertilisation), Bendus et al. [7] confirmed the existence of a moderate intraobserver variability and demonstrated substantial differences between embryologists in the morphological scores given to a group of embryos. It is inevitable that in some circumstances this variability will affect the decision of which embryo(s) is (are) selected for transfer and will, as a result, have a direct impact on the probability of IVF success.
Automated image analysis may add objectivity to the process of embryo selection and, consequently, lead to an improvement of the IVF process. However, the automatic assessment of embryo's features may often be problematic due to microscope image quality, differences in the embryo morphology in different development stages, volume of data to be analysed, position and transparency of the embryo, etc. In this paper, we briefly describe some of the most widely used embryo grading systems, review the published works toward automation of these methods, and discuss challenges and perspectives.
This paper is organised as follows: in the introduction section, we describe the main issues on in vitro fertilisation. In the embryo grading systems section, we briefly describe the main grading systems currently in use. In the section automatic image analysis, we review the works published on automatic human embryo image analysis that could lead to automatic grading. Finally, in the discussion section, we comment on the relationship among the published works, progress made, and challenges ahead.
EMBRYO GRADING SYSTEMS
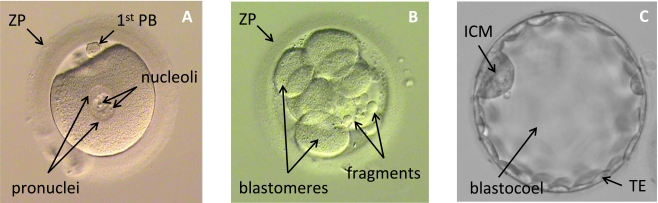
There are several embryo grading systems currently in use. They vary according to the embryo stage of development and rely on visual analysis of the embryo morphology. Currently, there is no consensus about which grading system is the most accurate. In Fig. (1) are shown examples of embryo images on day 1, day 2, and day 5, of development and their structures assessed for grading are indicated.
Fig. (1).
Examples of embryo image: (A) fertilised oocyte; (B) cleaving embryo; (C) blastocyst; ZP - zona pellucida; 1st PB - first polar body; ICM - inner cell mass; TE - trophectoderm.
The key morphological features of relevance to embryo viability are:
Cell number and degree of symmetry: if all cells are similar in size and an appropriate number of cells are present this indicates that the embryo has a good chance of being viable.
Fragmentation of cells: a low proportion of embryo volume composed of cell fragments is an indicator of high viability, while an embryo containing many fragmented cells is considered to have reduced potential.
Characteristics of the zona pellucida (ZP): embryos with a thinner ZP and higher variation in ZP thickness have a greater likelihood of producing a pregnancy.
ZONA PELLUCIDA THICKNESS VARIATION
The ZP, indicated in Fig. (1a), is a glycoprotein membrane encapsulating the oocyte and early embryo. The study presented by Gabrielsen et al. [8] has shown that the ZP thickness is a reliable indicator of IVF success rate and can be used as a criterion for embryo selection.
In most cases, the variation of ZP thickness is measured at three points, as suggested by Cohen et al. [9], and the following equations are used:
where: ZPTn (n = 1, 2, 3) is the Zona Pellucida Thickness at the point n; ZPTV is the Zona Pellucida Thickness Variation; ZPTmax is maximum Zona Pellucida Thickness; ZPTmean is the mean Zona Pellucida Thickness.
The measurement of the ZP is well documented and can be used to select embryos for transfer on days 2 and 3 after fertilisation when the ZP is at its most prominent and it can be easily visualised and measured. The thickness of the ZP is inversely correlated with embryo viability. This criterion also seems to be highly relevant on occasions where only suboptimal morphology embryos are available for transfer. It may also be useful to identify cases where the embryos may benefit from assisted hatching, a process in which the ZP is deliberately breached (usually with a laser) to aid the embryo's escape from the membrane (hatching) [9]. Hatching usually occurs naturally on day-5 or day-6 post-fertilisation, but may be impeded if the ZP is too thick or hardened. Viable embryos tend to have thinner ZP and higher ZP variation.
PRONUCLEAR MORPHOLOGY
Scott and Smith [10] have developed a method for embryo grading based on analysis of pronuclear morphology. The pronuclei, indicated in Fig. (1a), are spherical bodies that appear in the cytoplasm of the fertilised oocyte after insemination. Normal fertilisation results in the generation of two pronuclei, one containing the genetic contribution of the sperm, the other the contribution derived from the oocyte. The embryo is assessed at 16-18 hours post-insemination, with scores assigned for: (a) position of the pronuclei in the cytoplasm, (b) positioning of nucleoli (small spherical bodies inside each pronucleus), and (c) appearance of the cytoplasm.
The position of the pronuclei is evaluated and scored 5 or 1. A score of 5 is given if the pronuclei are close or aligned, while a score of 1 is given if the pronuclei are well separated.
The positioning of nucleoli is evaluated and scored 5, 4, or 3. If the nucleoli are aligned in a row at the pronuclear junction a score of 5 is given for this feature. If the nucleoli are beginning to align a score of 4 is awarded, while scattered nucleoli result in a score of 3.
The appearance of the cytoplasm is evaluated and scored 5 or 3. If the cytoplasm is heterogeneous in appearance with a clear halo around the edges, occasionally with a clear area in the centre around the pronuclei, and a darkened ring in the middle, then this characteristic scores 5. If the cytoplasm is clear homogeneous, or pitted and/or darkened, then a score of 3 is given.
The final grade of the embryo is then the sum of the three obtained scores. Additionally, at 24-26 hours the embryo is examined and if it presents breakdown of the nuclear membrane or cleavage to two-cell stage (both signs that the embryo is progressing on schedule) an additional score of 10 is added to the final embryo grade. Thus, the final score range is from 7 to 25 and a higher score is an indication of higher viability.
FRAGMENTATION ANALYSIS AND DEVELOPMENTAL SPEED
Some studies have shown that the amount of anucleate cellular fragments and the rate of development of a given embryo can be good indicators of its chances of establishing pregnancy [11-14]. These two aspects of morphology are the most widely used for embryo evaluation, being routinely scored in all IVF laboratories.
Steer et al. [12] have proposed the following scoring system: grade 4 for embryos with blastomeres (divided cells in Fig. (1b)) equal sized and symmetrical; grade 3 for an embryo with uneven blastomeres and less than 10% of the embryo volume occupied by fragments (Fig. (1b)); grade 2 for an embryo with 10% to 50% of fragmentation; and grade 1 for an embryo with more than 50% of fragmentation or pronucleate single cell embryos. Then, the grade of embryo is multiplied by the number of blastomeres to define the final grade of the embryo. This grading system is applied two days after the oocyte recovery. Cell number and degree of fragmentation are also routinely scored on day-3 post-fertilisation.
BLASTOCYST ANALYSIS
In recent years there has been increasing interest in culturing IVF embryos to the blastocyst stage. This stage is characterised by the formation of a fluid filled cavity (the blastocoel) in the middle of the embryo, surrounded by a single layer of cells called the trophectoderm (TE) (Fig. (1c)). A small protuberance of cells called the inner cell mass (ICM), which will eventually form the fetus, is also visible at this time. The blastocyst stage is usually reached five days after fertilisation of the oocyte. Traditional IVF embryo culture methods only extend for three days post-fertilisation.
Some nonviable embryos or embryos carrying chromosomal abnormalities may spontaneously stop growing during culture from day-3 to day-5 and be eliminated. Thus the transfer of embryos at the blastocyst stage assists embryologists in the selection of viable embryos [15]. However, successfully reaching the blastocyst stage is still no guarantee of viability. Even among blastocysts there remains a great deal of variability in potential, with fewer than half having the capacity to produce a pregnancy.
The most widely used blastocyst grading systems are those proposed by Dokras et al. [16] and Gardner et al. [17]. Balaban et al. [15] have shown that clinical pregnancy was higher when transferred embryos were selected using Gardner's grading system compared with those using the Dokras system, despite a similar number of embryos transferred.
Briefly, in the Gardner's grading systems, the blastocyst is graded from one to six according to the following criteria shown in Table 1:
Table 1.
Expansion Grading of Blastocyst
| Expansion grade | Blastocyst development and stage status |
|---|---|
| 1 | Early blastocyst: the blastocoel is less than half the volume of the embryo |
| 2 | Blastocyst: the blastocoel is greater than or equal to half of the volume of the embryo |
| 3 | Full blastocyst: the blastocoel completely fills the embryo |
| 4 | Expanded blastocyst: the blastocoel volume is larger than that of the early embryo and the zona pellucida is thinning. |
| 5 | Hatching blastocyst: the trophectoderm has started to herniate through zona pellucida |
| 6 | Hatched blastocyst: the blastocyst has completely escaped from the zona pellucida |
If the blastocyst is graded as 3 to 6, then the development of the ICM and TE is also graded. The ICM an TE grades are shown in Tables 2 and 3, respectively:
The TE grading is as shown in Table 3:
Despite the costs of longer culturing embryos, the study of Gardner et al. [17] has shown that high ongoing pregnancy rates (60.9%) can be achieved with the selection and transfer of a single blastocyst in good prognosis patients.
AUTOMATIC IMAGE ANALYSIS
The development of algorithms for biomedical image analysis is not an easy task. Usually, the variability in the quality of the images (due to illumination, artefacts, noise, etc.) does not permit application of straightforward segmentation techniques, such as thresholding, and it is difficult to design feature descriptors that could reliably express the information we want to obtain from the images under analysis.
Due to rapid development of the bioinformatics field, much effort has been focused toward automatic microscope image analysis of C.elegans embryos [18-20], fly embryos [21], and several other non-human embryos. An extensive recent review of this biological literature is presented by Peng [22]. We focus here on the literature concerning human embryo image analysis.
As IVF becomes more and more popular, some groups in the image analysis community have started to work on developing algorithms for automatic human embryo image analysis.
MODELLING BLASTOMERES AND ZP
Pedersen et al. [23] developed an approach, based on a multiphase variational level set [24], for modelling 3D shape and relative position of blastomeres of the embryos at day 2 after fertilisation. Using a Hoffman Modulation Contrast (HMC) light microscope [25], they detected contour locations and then found the outlines of blastomeres and the ZP. In a study of 4 HMC-image focus sequences (each sequence consisting of 17 images focused 5 micrometers apart), the results of the proposed approach visually appear good but were not quantified.
COMPUTER-CONTROLLED MULTILEVEL MORPHOMETRIC ANALYSIS
Hnida et al. [26] al have shown that even semi-automatic image segmentation software can be very useful for embryo morphology analysis. Using the FertiMorph system [27], sequences of 26 images per embryo were recorded, and blastomere size and nuclear structures of 232 human embryos were analysed semi-automatically. The results have shown that highly fragmented embryos presented a 43 to 46% reduction in blastomere volume compared with embryos with no fragmentation. Multinucleated blastomeres were on average, 51.5%, 67.8%, and 73.1% larger than non-multinucleated sibling blastomeres at the 2-, 3-, and 4-cell stages of development, respectively. In conclusion, the study suggested that the average blastomere size is affected by the level of fragmentation and multinuclearity, and that computer assisted multilevel analysis of blastomere size may be used as indicator of embryo viability.
Agerholm et al. [28], also using sequences of 26 images per embryo, investigated whether nuclear size and number are indicative of aberrant chromosome content in blastomeres and embryos. Analysing blastomere size and nuclear structures in a semi-automatic manner they concluded that mean nuclear size is not a marker for chromosome content in mononucleated embryos but it seems that the nuclei size can be related to multinucleation, which is frequently associated with abnormal chromosome content.
Giusti et al. [29] presented a method for segmentation of day-2 embryos using sets of images taken in different focus planes that were subsequently converted into polar coordinate images with an energy value associated to each pixel. A graph was then constructed and the minimal energy path of the graph led to a curve that estimated the contour of each blastomere. Using images of 53 embryos at the 4-cell stage, 71% of them had all four blastomeres correctly detected.
Beuchat et al. [30] developed a method to analyse day-1 embryo images semi-automatically. Using thresholding and ellipse fitting, the proposed method provided measurements of 24 variables (such as pronuclei major axis, minor axis, axis ratio, centering, etc.) useful for assessment of embryo viability. Then, classification in to one of two classes (low and high implantation probability) was performed using linear discriminant analysis. The performance of the proposed method was evaluated in terms of whether it could accurately predict embryo implantation. The average classification error when subjective grading was used was 0.39 and the average classification error of the proposed method was 0.36.
AUTOMATIC SEGMENTATION OF ZP
Karlsson et al. [31] have proposed a variational segmentation method based on an image model for the ZP which takes advantage of the characteristic appearance of HMC microscopy images.
In that work [31], the segmentation of ZP was divided in two parts: (a) first the segmentation of the embryo from its background, which assists in finding the outer boundary of the ZP, and (b) the segmentation of the inner boundary of ZP from the rest of the embryo. Both parts of this algorithm were based on variational analysis. The method appeared to work well on illustrative examples but has not undergone extensive evaluation to our knowledge.
Morales et al. [32], also, developed an algorithm for automatic ZP segmentation. Their algorithm, unlike that of Karlsson et al. [31] was based on an active contour model which identifies the external and internal boundaries of ZP. As a pre-processing step, this proposed approach enhanced the input images by high-pass-Gaussian convolution filter in combination with differential hysteresis processing [33] leading to improved clarity into the ZP on the images. The validation of the algorithm was done on 76 images through comparison with images manually segmented by human experts, yielding 91.65% accuracy in localisation of the boundaries.
BLASTOCYST IMAGES ANALYSIS
With the coming of more physiological culture media and advances in embryo culture systems and laboratory practices, culture of human embryos to the blastocyst phase has become increasingly routine [34]. However, automatic analysis of blastocyst images has not yet attracted the attention of the image analysis community.
Kock et al. [35] have shown that blastocysts can be better selected if objective digital image analysis and morphometric assessments can be done prior to blastocyst transfer. Their study showed the correlation between morphometric measurements and blastocyst implantation. In this prospective study they measured blastocyst diameter, ICM diameter, ZP thickness, blastocyst surface area, and ICM surface area, in images acquired over a period of one year. Their study demonstrated that the blastocysts that resulted in live birth presented on average: ICM ratio 12% larger, ICM area 14% larger, ICM diameter 11% larger, blastocyst area 21% larger, blastocyst diameter 10% larger, and ZP thickness 27% thinner. During Kock's study the gathering of morphometric data was assisted by embryologists, features for analysis being selected and defined using digital image analysis software. Clearly, full automation of this process would be advantageous.
DECISION SUPPORT SYSTEMS
It is well known that decision support systems have potential to greatly reduce medical errors and improve patient care [36, 37]. Due to the many variables that influence the IVF outcome, decision support systems may be a useful tool in helping clinicians during treatment planning.
Patrizi et al. [38] developed an algorithm, called TRACE, which classified embryos in two classes: those suitable for procreation and those not suitable. The algorithm used training sets to establish centres of classes and classification was then performed based upon a measure of distance from the classes centre. The feature vectors that represented each embryo were comprised of a set of moments calculated based on the embryo image histogram. In tests on 165 images, the proposed algorithm presented mean precision up to 0.85. However, its binary classification does not indicate which embryo is the best one.
Jurisica et al. [39] proposed a decision support system called TA3IVF to assist IVF practitioners in dealing with the complexity of the procedure during the planning of an IVF treatment. TA3IVF used case-based reasoning (CBR) [40] and relied on the notion of relevance assessment for the search for useful information in an available knowledge base. A modified nearest-neighbour matching algorithm [41] was used for data retrieval. The proposed system achieved an accuracy of 71.2% over 20 random trials in predicting pregnancy outcome.
Morales et al. [42] developed a decision support system for prediction of the outcome of a batch of embryos to be transferred to a woman's uterus. The approach was based on a Bayesian classifier that took into account variables based on morphology (thickness of ZP, number of cells, level of fragmentation of blastomeres) and on the clinical data of the patients. In tests on 63 cases, using different types of Bayesian classifiers (such as naive Bays [43], selective naive Bays [44], semi naive Bays [45], tree augmented naive Bays [46], and k-dependence Baysian classifier [47]), the accuracy of the systems ranged from 63.49% to 71.43%.
DISCUSSION
Most of the grading systems currently used for assessing the viability of IVF embryos are very subjective and rely on visual inspection of morphological characteristics of the embryos that are only qualitatively evaluated. For example, in the analysis of pronuclear morphology, as presented above, the guidelines for grading the alignment of the pronuclei rely on qualitative evaluations like close aligned, well separated, or, in the case of cytoplasm evaluation, heterogeneous or homogenous. Grading based only on these qualitative criteria may be one of the main causes of the interobserver variance reported by Bendus et al. [7], since terms like close, well, and very are imprecise.
Image analysis may be a way to overcome the subjectivity of the existing grading systems if it could provide quantitative evaluation of key aspects of the embryo morphology. Ultimately, an automatic system might simulate different grading systems and then combine the grades obtained from them in a final single grade. One possible method to combine them would be through the use of an AdaBoost [48] algorithm where each grading system could be regarded as a weak classifier that would be used to compose a strong classifier. Combination of classifiers seems to be a possible way to improve the performance of medical image analysis systems [49-51].
Also, image analysis techniques would allow storage of accurate measurements of several embryo characteristics that could be used for improvement of our knowledge about early embryo development and lead to further refinements of morphological grading systems. For instance, based on measurements of ICM, Richter [52] showed that Blastocysts with relatively large and/or slightly oval (roundness index 1.1 -1.2) ICMs are more likely to implant than other blastocysts. With a full automatic segmentation and quantification of embryos morphological features, more findings of this kind will hopefully be found.
However, automatic embryo image analysis poses some challenges to the image analysis community. Firstly, many of the morphological features analysed are not always well perceived immediately, requiring some adjustment of the microscope focus. The embryo is three dimensional and some features may be clearly observed in one focus plane, but invisible in another. Accurate analysis would require an automatic system to analyse several images of the same embryo taken through different planes of focus in order to quantify the features needed. This may greatly increase the processing time of an automatic system whereas the embryologist can perform an embryo evaluation in a matter of seconds. An automatic system would have to be at least as good as an embryologist in terms of its ability to select viable embryos. It would be validated by assessing the proportion of selected embryos that produce a pregnancy. There are also other issues like the differences in image quality, magnification, contrast method used, timing of image acquisition, that should be taken into consideration in the design of a system that aims to standardise embryo grading.
The promising works described above show that some advances have been made toward automatic image analysis of human embryo. The embryo model proposed by Pedersen et al. [23] may be compatible with automation of grading systems based on ZP morphology and counting of blastomeres.
The works of Karlsson et al. [31] and Morales et al. [35], are significant steps toward a fully automatic grading system, since they would provide input data for decision support system like the one proposed by Morales et al. [41]. However, much more refinement and validation is necessary prior to clinical application.
There is little published work concerning automatic image analysis of human embryos in general. However, there is a particular paucity of publications concerning efforts to automate evaluation at the blastocyst stage. This may be due to the fact that embryo culture to this later stage of development has only recently become available. Thus, it is expected that, in the future, increasing effort will be made toward automating blastocyst image analysis, especially as the transfer of embryos at this stage is growing in popularity and associated with high pregnancy rates.
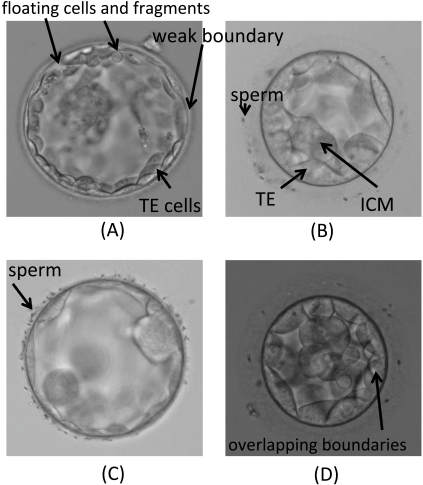
In Fig. (1) we show examples of typical challenges presented by blastocyst images. In Fig. (2a) we can observe a region of weak boundary and the presence of fragments and floating cells inside the blastocoel which can be an obstacle for a segmentation algorithm based on curve evolution trying to evolve from the image centre toward the TE inner boundary. The overlapping of TE cell boundaries, due to cell transparency, shown in Fig. (2d), also adds difficulties. Features of this type can often cause the evolving curves to remain partially stuck to neighbouring cell boundaries. Sperm attached to the ZP, as shown in Fig. (2b) and Fig. (2c), are likely to disturb the segmentation of outer TE boundaries.
Fig. (2).
Examples of problematic images of blastocyst.
Automated image analysis may increase the objectivity of the process of embryo selection and, consequently, lead to an improvement of the IVF process. Automation also has the advantage that embryo observation and scoring could be carried out at more time points than currently possible, including during the night when IVF laboratories are not typically staffed. There is growing data to support the notion that a cumulative morphological score, derived from multiple independent evaluations, may provide the best guide to embryo potential [53]. The new generation of combined incubator-microscope systems, such as the Biostation (Nikon) or the Primo-Vision (Cryoinnovation), offers the potential for morphological evaluation to be carried out without the need to remove embryos from the optimised gas and temperature conditions inside the incubator. This should reduce environmental stress experienced by the embryo and may lead to improvements in embryo viability and pregnancy rates as a result. Automated morphological evaluation could be adapted for use with such systems.
It is likely that the use of automated systems will save the embryology staff a significant amount of time and the availability of twenty four hour observation may allow IVF procedures to be conducted in a more flexible manner throughout the day, rather than at specific time points defined by the need to conduct observations a set number of hours later. Ultimately, it is hoped that computer assisted embryo evaluation will improve and streamline the IVF procedure, reducing costs and increasing the ability of embryologists to indentify the embryo most likely to produce a child. Advances of this type will be vital if efficient single embryo transfer and the elimination of high-order multiple pregnancies are to be achieved.
Table 2.
ICM Grading of Blastocyst
| ICM grade | ICM quality |
|---|---|
| A | Tightly packed, many cells |
| B | Loosely grouped, several cells |
| C | Very few cells |
Table 3.
TE Grading of Blastocyst
| TE grade | TE quality |
|---|---|
| a | Many cells forming a tightly knit epithelium |
| b | Few cells |
| c | Very few cells forming a loose epithelium |
ACKNOWLEDGEMENTS
This work was supported by the Oxford Biomedical Research Centre. We thank Tracey Griffiths and Gerri Emerson for providing the images of embryos used in this paper.
REFERENCES
- 1."2006 Assisted reproductive technology success rates: national summary and fertility clinic report".[Online] Available:www.cdc.gov/ART/ART2006/508PDF/2006ART.pdf . 2006. [23 March 2009]. [DOI] [PubMed]
- 2.Wang J, Sauer M V. “In vitro fertilization (IVF): a review of 3 decades of clinical innovation and technological advancement ". Ther. Clin. Risk Manag. 2006;2:355–364. doi: 10.2147/tcrm.2006.2.4.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3."SART, Society for Assisted Reproductive Technology". [Online] Available: http://www.sart.org . [23 March 2009].
- 4.Crosignani PG. (ESHRE Capri Workshop Group), "Multiple gestation pregnancy". Hum. Reprod. 2000;15:1856–1864. [PubMed] [Google Scholar]
- 5.Petterson B, Stanley F, Henderson D. " Cerebral palsy in multiple births in Western Australia". Am. J. Med. Genet. 1990;37:346–351. doi: 10.1002/ajmg.1320370311. [DOI] [PubMed] [Google Scholar]
- 6.Bromer JG, Seli E. "Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics". Curr. Opin. Obstet. Gynecol. 2008;20:234–241. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 7.Bendus AEB, Mayer JF, Shipley SK, Catherino WH. “ Interobserver and intraobserver variation in day 3 embryo grading". Fertil. Steril. 2006; 86:1608–1615. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 8.Gabrielsen A, Bhatnager PR, Petersen K, Lindenberg S. "Influence of zona pellucida thickness of human embryos on clinical pregnancy outcome following in vitro fertilization treatment". J. Assis. Reprod. Genet. 2000;17(6):335–343. doi: 10.1023/A:1009453011321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J, Inge K L, Suzman M, Wiker SR, Wright G. "Videocinematography of fresh and cryopreserved embryos: a retrospective analysis of embryonic morphology and implantation". Fertil. Steril. 1989;51(3):820–827. doi: 10.1016/s0015-0282(16)60673-8. [DOI] [PubMed] [Google Scholar]
- 10.Scott L A, Smith S. “The successful use of pronuclear embryo transfers the day following oocyte retrieval". Hum. Reprod. 1998;13(4):1003–1013. doi: 10.1093/humrep/13.4.1003. [DOI] [PubMed] [Google Scholar]
- 11.Puissant F, Van Rysselberg M, Barlow P, Deweze J, Leroy F. “Embryo scoring as a prognostic tool in ivf treatment". Hum. Reprod. 1987;2:705–708. doi: 10.1093/oxfordjournals.humrep.a136618. [DOI] [PubMed] [Google Scholar]
- 12.Steer C V, Mills C L, Tan S L, Campbell S, Edwards R G. "The cumulative embryo score: a predictive embryo scoring technique to select the optimal number of embryos to transfer in an in-vitro fertilization and embryo transfer programme". Hum. Reprod. 1992;7:117–119. doi: 10.1093/oxfordjournals.humrep.a137542. [DOI] [PubMed] [Google Scholar]
- 13.Sakkas D, Shoukir Y, Chardonnens D, Bianchi P G, Campana A. "Early cleavage of human embryos to the two-cell stage after intracytoplasmic sperm injection as an indicator of embryo viability". Hum. Reprod. 1998;13:182–187. doi: 10.1093/humrep/13.1.182. [DOI] [PubMed] [Google Scholar]
- 14.Loi K, Prasath E B, Huang Z W, Loh S F, Loh S K E. "A cumulative embryo scoring system for the prediction of pregnancy outcome following intracytoplasmic sperm injection". Singapore Med. J. 2008;49(3):221–227. [PubMed] [Google Scholar]
- 15.Balaban B, Yakin K, Urman B. "Randomized comparison of two different blastocyst grading systems". Fertil. Steril. 2006;85(3):559–563. doi: 10.1016/j.fertnstert.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Dokras A, Sargent I L, Barlow DH. "Human blastocyst grading: an indicator of developmental potential?". Hum. Reprod. 1993;8(12):2119–2127. doi: 10.1093/oxfordjournals.humrep.a137993. [DOI] [PubMed] [Google Scholar]
- 17.Gardner D K, Surrey E, Minjarez D, Leitz A, Stevens J, Schoolcraft W B. "Single blastocyst transfer: a prospective randomized trial". Fertil. Steril. 2004;81(3):551–555. doi: 10.1016/j.fertnstert.2003.07.023. [DOI] [PubMed] [Google Scholar]
- 18.Ning F, Delhomme D, LeCun Y, Piano F, Bottou L, Barbano P E. "Toward automatic phenotyping of developing embryos from videos"; IEEE Trans. Image Proc; 2005. pp. 1360–1371. [DOI] [PubMed] [Google Scholar]
- 19.Hamahashi S, Onami S, Kitano H. "Detection of nuclei in 4D nomarski DIC microscope images of early caenorhabditis elegans embryos using local image entropy and object tracking". BMC Bioinformatics. 2005 May;6 doi: 10.1186/1471-2105-6-125. [Online]. Available from: http://www.biomedicalcentral.com/1471-2105/6/125/abstract/ [Accessed: 18th May 2010]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng W, Cosman P, Berry CC, Feng Z, Schafer W R. “Automatic tracking, feature extraction and classification of C. elegans phenotypes". IEEE Trans. Biomed. Eng. 2004;51(10):1811–1820. doi: 10.1109/TBME.2004.831532. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Peng H. “Automatic recognition and annotation of gene expression patterns of fly embryos". Bioinformatics. 2007;23(5):589–596. doi: 10.1093/bioinformatics/btl680. [DOI] [PubMed] [Google Scholar]
- 22.Peng H. "Bioimage informatics: a new area of engineering biology". Bioinformatics. 2008 Jul;24(17):1827–1836. doi: 10.1093/bioinformatics/btn346. [Online]. Available from: http://bioinformatics.oxfordjournal.org/cgi/reprint/24/17/1827. [Accessed: 19th May 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perdersen U D, Olsen O F, Olsen N H. "A multiphase variational level set approach for modelling human embryos". Proceedings of the 2nd IEEE Workshop on variational, Geometric and Level Set Methods.2003. [Google Scholar]
- 24.Zhao H K, Chan T, Merriman B, Osher S. "A variational level set approach to multiphase motion". J. Comput. Phys. 1996;127:179–195. [Google Scholar]
- 25.Hoffman R, Gross L. "The modulation contrast microscope". Nature. 1975;254:586–588. doi: 10.1038/254586a0. [DOI] [PubMed] [Google Scholar]
- 26.Hnida C, Engenheiro E, Ziebe S. "Computer-contrelled, multilevel, morphometric analysis of blastomere size as biomarker of fragmentation and multinuclearity in huma embryos". Hum. Reprod. 2004;19(2):288–293. doi: 10.1093/humrep/deh070. [DOI] [PubMed] [Google Scholar]
- 27."FertMorph", [Online] Available: http://www.ihmedical.com/products.aspx?id=fmorph-s . 2010. May 13, [Accessed: May, 13 2010].
- 28.Argeholm IE, Hnida C, Cruger DG, Berg C, Bruun-Petersen G, Kolvraa S, Ziebe S. "Nuclei size in relation to nuclear status and aneuploidy rate for chromosomes in donated four cells embryos". J. Assist. Reprod. Genet. 2008;25:95–102. doi: 10.1007/s10815-008-9199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giusti A, Corani G, Gambardella L, Magli C, Gianaroli L. "Blasotmere segmentation and 3D morphology measurements of early embryos from hoffman modulation contrast image stacks". 2010 IEEE International Symposium on Biomedical Imaging; 13-17 April 2010; Rotterdam,The Netherlands. [Google Scholar]
- 30.Beuachat A, Thevenaz P, Unser M, Ebner T, Senn A, Urner F, Germond M, Sorzano COS. "Quantitative morphometrical characterization of human pronuclear zygotes". Hum. Reprod. 2008;23(9):1983–1992. doi: 10.1093/humrep/den206. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson A, Overgaard N C, Heyden A. "Automatic segmentation of zona pellucida in hmc images of human embryos". Proceedings of the 17th International Conference on Pattern Recognition (ICPR'04); 2004. pp. 23–26. [Google Scholar]
- 32.Morales D A, Bengoetxea E, Larranaga P. "Automatic segmentation of zona pellucida in human embryo images applying an active contour model". Proceedings of the 12th Annual Conference on Medical Image Understanding and Analysis; 2008. pp. 209–213. [Google Scholar]
- 33.Klaus-Ruediger P. Fifty Second Annual Meeting Microscopy Society America. London, UK: Informa Healthcare; 2004. "Digital differential hysteresis image processing displays what microscope acquires but the eye can't see"; pp. 416–417. [Google Scholar]
- 34.Gardner D K, Stevens J, Sheehan C B, Schoolcraft WB. Human preimplantation embryo selection. London, UK: Informa Healthcare; 2007. "Analysis of blastocyst morphology"; pp. 79–87. [Google Scholar]
- 35.Kock A D, Smuts M P, Madden J D, Rodriguez A J, Chantilis S J, Meintjes M. "Digital image analysis of blastocysts. Morphometrics correlations with pregnancy outcome". Fertil. Steril. 2006;86:51–52. [Google Scholar]
- 36.Kawamoto K, Houlihan C A, Balas E A, Lobach D F. "Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success". BMJ. 2005 Mar; doi: 10.1136/bmj.38398.500764.8F. [Online]. Availble from: www.bmj. com/cgi/content/abstract/bmj.38398.500764.8Fv1. [Accessed: 18th May 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manna C, Patrizi G, Rahman A, Sallam H. "Experimental results on the recognition of embryos in human assisted reproduction". Reprod. BioMed. 2004;8(4):460–469. doi: 10.1016/s1472-6483(10)60931-5. [Online]. Availble from: www.rmbojournal.com. [Accessed: 18th May 2010] [DOI] [PubMed] [Google Scholar]
- 38.Patrizi G, Manna C, Moscatelli C, Nieddu L. "Pattern recognition methods in human-assisted reproduction". Int. Trans. in Opin. Res. 2004;11(4):365–379. [Google Scholar]
- 39.Jurisica I, Mylopoulos J, Glasgow J, Shapiro H, Casper R F. "Case-based reasoning in ivf: prediction and knowledge mining". Artif. Intel. Med. 1998 Jan;12(1):1–24. doi: 10.1016/s0933-3657(97)00037-7. [DOI] [PubMed] [Google Scholar]
- 40.Slade S. "Case-based reasoning: a research paradigm". AI. Mag. 1991;12(1):45–55. [Google Scholar]
- 41.Wettschereck D, Dietterich T G. "An experimental comparison of the nearest neighbor and nearest hyperrectangle algorithms". Mach. Learn. 1995 Apr;19(1):5–27. [Google Scholar]
- 42.Morales D A, Bengoetxea E, Larranaga P, Garcia M, Franco Y, Fresnada M, Merino M. “Baysian classification for the selection of in vitro human embryos using morphological and clinical data". Comput. Method Program Biomed. 2008 May;90(2):104–116. doi: 10.1016/j.cmpb.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 43.Cestnik B, Kononenko I, Bratko I. Progress in Machine Lea. Wilmslow, U.K: Sigma Press; 1987. “ASSISTANT-86: a knowledge elicitation tool for sophisticated users"; pp. 31–45. [Google Scholar]
- 44.Langley P, Sage S. "Induction of selective Bayesian classifier". Proceedings of the 10th Conference on Uncertainty in Artificial Intelligence; Seattle, WA. 1994. pp. 399–406. [Google Scholar]
- 45.Kononenko I. "Semi-naive Bayesian classifiers". Proceedings of the 6th European Working Session on Learning; 1991. pp. 206–219. [Google Scholar]
- 46.Friedman N, Geiger D, Goldsmidt M. “Bayesian network classifiers". Mach. Learn. 1997; 29:131–163. [Google Scholar]
- 47.Sahami M. "Learning limited dependence Bayesian classifier". Proceedings of the 2nd International Conference on Knowledge Discovery and Data Mining; 1996. pp. 335–338. [Google Scholar]
- 48.Freund Y, Schapire RE. "A short introduction to boosting". J. Jpn. Soc. Artif. Intel. 1999;4. no. 5:771–780. [Google Scholar]
- 49.Lum VLF, Leow WK, Chen Y, Howe TS, Png MA. "Combinig classifiers for bone fracture detection in x-rays images". IEEE International Conference on Image Processing; 2005. pp. 1149–1152. [Google Scholar]
- 50.Osowski S, Siroic R, Siwek K. "Genetic algorithm for integration of ensemble of classifiers in arrhithymia recognition"; IEEE Instrumentation and Measurement Technology Conference; 2009. pp. 1496–1500. [Google Scholar]
- 51.Kittler J, Hatef M, Duin RPW, Matas J. “On combining classifiers". IEEE Trans PAMI. 1998;20(3):226–239. [Google Scholar]
- 52.Richter K S, Harris D C, Daneshmand S T, Shapiro B S. “Quantitave grading of a human blastocyst: optimal inner cell mass size and shape". Fertl. Steril. 2001;76(6):1157–1167. doi: 10.1016/s0015-0282(01)02870-9. [DOI] [PubMed] [Google Scholar]
- 53.Neuber E, Mahutte N G, Arici A, Sakkas D. "Sequential embryo assessment outperforms investigator-driven morphological assessment at selecting a good quality blastocyst". Fertil. Steril. 1985;85(3):794–796. doi: 10.1016/j.fertnstert.2005.08.064. [DOI] [PubMed] [Google Scholar]