Abstract
The filamentous cyanobacterium Planktothrix rubescens frequently occurs in deep and stratified lakes in the temperate region of the northern hemisphere and is a known producer of the hepatotoxic secondary metabolite microcystin. These cyclic heptapepids are synthesized non-ribosomally via large enzyme complexes encoded by the microcystin (mcy) synthetase gene cluster. The occurrence of cyanobacterial strains lacking microcystin but containing the mcy gene cluster has been reported repeatedly; it was shown that this inactivation is due to mutations such as gene deletion events and the insertion of transposable elements. In the present study, twelve lakes in Austria, Germany, and Switzerland were sampled from July 2005 to October 2007, and the proportion of inactive mcy genotypes was quantified in relation to the total population of the red-pigmented filamentous cyanobacterium Planktothrix by means of quantitative PCR. In total, four different mutations were quantified, namely two insertions affecting mcyD, one insertion affecting mcyA, and a deletion within mcyH and mcyA. The mutations occurred over a wide range of the population density (40 – 570,000 filaments L−1) and their abundance was found to be positively correlated with population density. However, on average, all nontoxic mutants were found in a low proportion only (min 0%, mean 6.5% ± 1.1 (SE), max 52% of the total population). The genotype containing the mcyHA deletion had a significantly higher proportion (min 0%, mean 3.7% ± 1, max 52%) when compared with all the genotypes containing insertions within the mcy gene cluster (min 0%, mean 2.8% ± 0.7, max 24%). The results demonstrate that the occurrence of inactive mcy genotypes is linearly related to the population density and selective sweeps of nontoxic mutants did not occur during the transition from prebloom to bloom conditions.
Keywords: Toxicity, microcystin, real-time PCR, gene loss, Planktothrix, transposable elements, microevolution
Introduction
Mass occurrences of bloom-forming toxic cyanobacteria in freshwater are observed worldwide. The filamentous cyanobacterium Planktothrix spp. occurs in the pelagial of lakes and reservoirs and is one of the producers of the toxic heptapeptide microcystin that poses a serious health threat to humans and livestock [4]. These hepatotoxins are synthesized by nonribosomal peptide synthetases (NRPS) via the thio-template mechanism encoded by 9-10 genes constituting the microcystin synthetase (mcy) gene cluster [6, 33, 38]. In the following strains of various cyanobacteria have been repeatedly reported to contain the mcy genes but lack detectable microcystin [13, 16, 21, 24, 39]. For Planktothrix it has been observed that the mcy gene cluster has been frequently inactivated by various mutations, such as insertions of a transposable element or deletions affecting one or two genes of the mcy gene cluster [6]. Recently, we were able to show that those mutations, although they may have arisen independently, are of a relatively recent origin and those nontoxic mutants cannot be discriminated from the strains still containing the intact mcy gene cluster by using additional variable genetic markers [7].
Although the poisoning of aquatic grazers via microcystin has been reported [18, 30], it is unlikely that the microcystins evolved in response to grazing by herbivorous crustaceans. Analyses comparing the phylogenetic trees obtained from housekeeping genes (16S rDNA, rpoC1) and mcyA, mcyD, mcyE revealed a congruent branching suggesting that the evolution of microcystin preceded the first appearance of metazoans, probably by one billion years [28]. However, since toxic effects to other aquatic biota cannot be excluded it might be argued that a specific proportion of microcystin producers in a population is necessary to enable the survival of the individual (nontoxic) cell. Indeed if these nontoxic mutants can be “cheaters,” as in the style of Myxococcus sp. strains that have given up various signalling responses [40], one might expect an over representation of nontoxic mutants under bloom conditions relative to their initial frequency at the beginning of the population development. Alternatively, light limiting conditions as found under dense bloom conditions have been postulated to result in the gradual increase of nontoxic strains at the expense of toxic strains of the genus Microcystis [14]. It has been shown that those inactive mcy genotypes occur under natural conditions for several years [6]. However, it is not clear whether those nontoxic mutants are able to dominate the population under bloom conditions.
It was the aim of the present study to investigate the ecological success of the nontoxic mutants in populations of Planktothrix that vary in population density. The populations that were studied occurred in twelve lakes in the Alps of Austria, Switzerland, and Germany. Due to the stable physical stratification in deep lakes the red pigmented P. rubescens dominates at greater water depths with higher amounts of green light when compared with the light conditions at the water surface [44]. P. rubescens isolated from Lake Zürich was neutrally buoyant in cultures given 6.5 μmol m−2 s−1 in a 12 h light cycle and typically P. rubescens occurs at depths of 9-12 m with low light intensity [44]. Some of the lakes have been repeatedly reported to show Planktothrix blooms (Hallwilersee, Wörthersee, Zürichsee, [12]. Mass developments of the red-pigmented cyanobacteria assigned to P. rubescens have also been reported from North America [8, 25] and the occurrence of cyanobacteria assigned to P. rubescens has also been reported from New Zealand [27]. We hypothesized that under conditions of prebloom development and sparse population densities nonmicrocystin-producing mutants should not be able to occur. Vice versa under conditions allowing blooms to occur nonmicrocystin-producing mutants may be able to flourish and gain selective advantage due to a higher growth rate compared to microcystin producers. The quantitative real-time PCR technique was used to quantify various inactive mcy genotypes and to estimate their share of the total population as described previously [19].
Methods
Study area and sampling
Twelve lakes located in the Alps in Austria, Switzerland, and Germany were sampled from summer 2005 to summer 2007. All the lakes are generally deep and physically stratified (Table 1). The trophic state varied from oligotrophic (Attersee, Offensee, Schwarzensee and Wolfgangsee), and oligo-mesotrophic (Irrsee, Mondsee, Ammersee and Fuschlsee), to mesotrophic (Wörthersee, Zürichsee and Afritzersee) and meso-eutrophic (Hallwilersee). The water column was integrated by collecting one litre of water every 2 metres from the surface to a depth of 20 m. Chlorophyll a and total phosphorus were analysed according to standard methods [11, 47]. For analysing the phytoplankton community, aliquots (100 ml) of the samples were preserved with Lugol’s solution and 2% formaldehyde, each. Additionally, net samples were obtained by pulling a phytoplankton net (30 μm in mesh size) three times from a depth of 20 m to the surface and aliquots were preserved with a final concentration of 2% formaldehyde. For subsequent DNA isolation, aliquots (2-4 L of integrated samples and 20-100 ml of net samples) were filtered onto glass-fibre filters (BMC, Ederol, Vienna, Austria) under vacuum pressure and stored frozen (−20°C).
Table 1.
List of the study lakes in the Alps and their morphometric characters. Concentrations of chlorophyll a, total phosphorous and secchi depths were averaged (mean ± SE) over the sampling period and used to classify the trophic state [42].
| Lake | Country | Latitude | Longitude | Surface area [km2] |
Maximum depth [m] |
Number of samples |
Sampling period |
Trophya | Chl a (mm3 L−1) |
TP (mm3 L−1) |
Secchi (m) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Attersee | AT | 47°54′ N | 13°33′ E | 46.2 | 170.6 | 7 | July 05 - July 07 | O | 0.9 ± 0.2 | 5.0 ± 1.1 | 9.7 ± 1.7 |
| Wolfgangsee | AT | 47°45′ N | 13°25′ E | 12.8 | 114 | 7 | July 05 - July 07 | O | 1.2 ± 0.1 | 4.9 ± 0.5 | 7.3 ± 1.4 |
| Schwarzensee | AT | 47°45′ N | 13°30′ E | 0.48 | 54 | 7 | July 05 - July 07 | O | 0.9 ± 0.1 | 5.6 ± 0.8 | 6.0 ± 0.6 |
| Offensee | AT | 47°45′ N | 13°50′ E | 0.55 | 38 | 7 | July 05 - July 07 | O | 2.2 ± 0.3 | 5.8 ± 0.6 | 8.6 ± 0.7 |
| Ammersee | DE | 47°59′ N | 11°07′ E | 46.6 | 81.1 | 6 | July 05 - June 07 | O-M | 1.5 ± 0.3 | 9.5 ± 1.3 | 4.0 ± 0.6 |
| Fuschlsee | AT | 47°48′ N | 13°16′ E | 2.65 | 67.3 | 7 | Aug. 05 - Aug. 07 | O-M | 2.3 ± 0.3 | 8.1 ± 0.8 | 5.0 ± 0.4 |
| Irrsee | AT | 47°55′ N | 13°18′ E | 3.6 | 32 | 7 | July 05 -July 07 | O-M | 1.7 ± 0.2 | 8.3 ± 0.3 | 4.5 ± 0.7 |
| Mondsee | AT | 47°49′ N | 13°22′ E | 13.8 | 68.3 | 7 | Aug. 05 - Sep. 07 | O-M | 2.7 ± 0.4 | 10 ± 0.5 | 4.5 ± 0.5 |
| Wörthersee | AT | 46°37′ N | 14°07′ E | 19.4 | 85.2 | 6 | July 05 - May 07 | M | 3.9 ± 0.6 | 13 ± 0.7 | 4.9 ± 1.3 |
| Afritzersee | AT | 46°45′ N | 13°46′ E | 0.5 | 22.5 | 6 | Aug. 05 - May 07 | M | 4.5 ± 0.8 | 19.4 ± 2.7 | 4.9 ± 0.3 |
| Zürichsee | CH | 47°15′ N | 08°38′ E | 65.5 | 136 | 7 | Aug. 05 - Oct. 07 | M | 7.1 ± 1.4 | 20.1 ± 3.8 | 4.4 ± 0.4 |
| Hallwilersee | CH | 47°17′ N | 08°12′ E | 10.2 | 47 | 5 | Nov. 05 - Aug. 07 | M-E | 8.5 ± 0.6 | 17.3 ± 1.4 | 2.6 ± 0.3 |
O = oligotrophic, O-M= oligo-mesotrophic, M = mesotrophic, M-E= meso-eutrophic
Cultivation of strains and cell harvesting
The microcystin-producing Planktothrix rubescens strain PCC7821 (L. Gjersj en, Norway) and the inactive microcystin genotypes of Planktothrix rubescens strains No110 and No40 (Mondsee, Austria), No139 (Grabensee, Austria) and No62 (Irrsee, Austria) were grown at 20°C in BG11 medium [29] under constant light conditions (5-15 μmol m−2 s−1, Osram Type L30W/77Fluora) and harvested in the exponential growth phase using vacuum filtration onto glass fibre filters (BMC, Ederol, Vienna, AT). Filters were stored frozen (−20°C) until DNA extraction. Aliquots of the cultures were preserved by adding formaldehyde (final concentration 2%). DNA was extracted and used to relate a series of DNA dilutions (in biovolume equivalents) to the cycle of threshold (Ct value) as measured by real-time PCR.
en, Norway) and the inactive microcystin genotypes of Planktothrix rubescens strains No110 and No40 (Mondsee, Austria), No139 (Grabensee, Austria) and No62 (Irrsee, Austria) were grown at 20°C in BG11 medium [29] under constant light conditions (5-15 μmol m−2 s−1, Osram Type L30W/77Fluora) and harvested in the exponential growth phase using vacuum filtration onto glass fibre filters (BMC, Ederol, Vienna, AT). Filters were stored frozen (−20°C) until DNA extraction. Aliquots of the cultures were preserved by adding formaldehyde (final concentration 2%). DNA was extracted and used to relate a series of DNA dilutions (in biovolume equivalents) to the cycle of threshold (Ct value) as measured by real-time PCR.
Cell counting
Filaments were assigned to the genus Planktothrix according to the morphological criteria characteristic for Planktothrix spp. [1]. Lugol-fixed samples were counted in sedimentation chambers using the inverted microscope technique [41]. At least three transects per chamber were counted at 100× magnification to obtain a count of at least 400 specimens in total [47]. To estimate the biovolume of P. rubescens, 70 filaments from Offensee were measured in diameter using image analysis (Lucia G, on Intrigue Pro Version 4.51). The biovolume was then calculated by assuming the geometric shape of a cylinder and multiplying the mean area of a cylinder by the filament length. The detection limit of Planktothrix in the inverted microscope was one filament per sedimentation chamber (25 ml volume). In order to control for counting errors due to a lack of sedimentation of P. rubescens filaments [43], aliquots preserved in 2% formaldehyde were filtered onto polycarbonate filters (0.45μm, Millipore, Vienna, AT), stained with DAPI, and enumerated using epifluorescence microscopy (Zeiss Axioskop 40). At least two transects per filter at 200× magnification were counted. A significant linear relationship was obtained between the filaments estimated by the sedimentation chamber and by the epifluorescence method: y = −0.033 + 0.9722x (R2 = 0.85, n = 18, min-max = 0 - 4.6 mm3 L−1), where y is the biovolume (mm3 L−1) as determined by the sedimentation method and x is the biovolume (mm3 L−1) as determined by epifluorescence microscopy.
Quantification of genotypes
Quantitative DNA extraction from filters was performed using the standard chloroform-phenol procedure as described [15]. The TaqMan assay (TNA) was used to quantify (i) the total population of Planktothrix via 16S rDNA and (ii) the mcyBA1 genotype (the first adenylation domain of the mcyB gene) that was indicative of all genotypes containing the mcy gene cluster and (iii) four mutations (three insertions of a transposable element and one deletion) resulting in the inactivation of microcystin biosynthesis [6]. In addition, the insertion sequence (IS) element that was found to be inserted into the mcy gene cluster [6] was amplified. The sequence of the IS element was submitted to the IS finder database and denoted ISPlrub1 [35]. All primers and probes were designed during this study from sequences published previously [6].
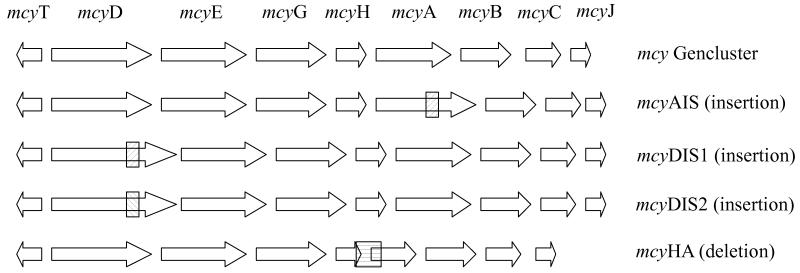
For the design of the TNA amplifying the 16S rDNA of Planktothrix spp. sequences of cyanobacteria from sections I-V that were collected in the ARB software [20] were aligned, and the primers and probes were designed to amplify Planktothrix only. For the amplified region (82 bp), no sequence variation was observed among the 92 entries of the Genbank database (7 April 2008) assigned to Planktothrix spp. according to Suda et al. [37]. To quantify microcystin genotypes, a conserved region within the mcyB gene, based on 27 strains that were sequenced for the first adenylation domain as described (AJ890255 - AJ890282 [17]), was selected. To design the TNAs amplifying specifically the insertions within the mcy gene cluster, primers and probes were located both in the insertion and the flanking mcy gene region (Fig. 1), i.e. for the 1.4 kbp insertions found in the mcyD gene (mcyDIS1, strain No110 and mcyDIS2, strain No139) and in the mcyA gene (mcyAIS, strain No40). Analogously, the primers and probes to detect the deletion were designed to bind upstream and downstream of the deleted mcy region, i.e. for the 1.8 kbp deletion located within mcyH and mcyA (mcyHA, strain No62). For the TNA amplifying the IS element, both primers and probes were located within the IS element.
Figure 1.
Microcystin synthethase (mcy) gene cluster of Planktothrix and the location of mutations resulting in the inactivation of microcystin biosynthesis. mcyAIS, mcyDIS1, mcyDIS2, insertions caused by a transposable (IS) element into genes mcyA and mcyD of the mcy gene cluster, which are transcribed in both directions; mcyHA, deletion of parts of the mcyH and mcyA genes [6].
The primers and probes were designed using the Primer Express 2.0 software (ABI, Table 2). The probes were labelled with a fluorescent reporter dye that was covalently attached to the 5′ end (FAM, 6-carboxyfluorescein) and a fluorescent quencher dye attached to the 3′ end (TAMRA, 6-carboxytetramethylrhodamine). Concentrations of the primers and probes were optimized according to the instruction manual (ABI TaqMan Universal PCR MasterMix). The specificity and robustness of each TNA was tested by adding DNA originating from other organisms as a background. DNA extracted from the Microcystis strains HUB524 and HUB53 was added to the DNA of strain No62 (mcyHA deletion, DNA concentration equivalent to 460 cells template−1) and mixed at ratios of 1:100 and 1:1 (calculated in cell equivalents). To test the specificity of the TNA, which was targeted to 16S rDNA and the IS element, DNA extracts from several field samples that were found to be negative by another TNA amplifying the intergenic spacer region of the phycocyanin operon (PC-IGS) of Planktothrix spp. [33] were tested. Those samples originated from Austria (Längsee, Höllerersee, and Wolfgangsee in August 2006), and contained the DNA of various other cyanobacteria (Anabaena spp., Lyngbya spp., Synechococcus spp.). No background experiments were performed for the TNAs amplifying the insertions in mcyD and mcyA, as the design of these TNAs was considered highly specific.
Table 2.
Oligonucleotide primers and TaqMan probes used for the quantification of the total Planktothrix population, the mcyBA1 genotype, four inactive mcy genotypes and the transposable (IS) element reported to insert into the mcy gene cluster [6]
| TNA | Gene locus | Strain | Forward primer / Reverse primer (5′–3′) | Taqman Probe (5′–3′) | Concentration (fmol μl−1)I |
Annealing T(°C) |
Amplicon (bp) |
|---|---|---|---|---|---|---|---|
| 16S rDNA | 16S rDNA | PCC7821 | ATCCAAGTCTGCTGTTAAAGA / CTCTGCCCCTACTACACTCTAG |
AAAGGCAGTGGAAACTGGAAG | 300/300/250 | 55 | 82 |
| mcyBA1 | mcyBA1 | PCC7821 | ATTGCCGTTATCTCAAGCGAG / TGCTGAAAAAACTGCTGCATTAA |
TTTTTGTGGAGGTGAAGCTCTTTCCTCTGA | 900/900/100 | 60 | 76 |
| mcyDIS1 | 3′end IS- Element, mcyD |
No110 | TTCTTTACTCTTTTCCACCCGACTT / ACAAATTGCTGTTTTTGCGCT |
CGGGAATAGCCCCCCCAAACC | 200/200/100 | 60 | 93 |
| mcyDIS2 | 3′end IS- Element, mcyD |
No139 | TTGAGAATTATGACCCAAAAGTAGGC / ACAAATTGCTGTTTTTGCGCT |
CGGGAATAGCCCCCCCAAACC | 200/200/100 | 60 | 110 |
| mcyAIS | 3′end IS- Element, mcyA |
No40 | TTCTTTACTCTTTTCCACCCGACTT / GAATGAGAGGTAACGGCATTACG |
TGACCAGGGCTGGTTTAGCCAATAGTACA | 200/200/100 | 60 | 168 |
| mcyHA |
mcyHA deletion |
No62 | TCTTCTGGACGGTTTTCTAG / CTTTCCGGGTTTGATGT |
TACAGAATGGGAAAAAATTACTCAAGAGAA | 200/200/250 | 55 | 71 |
| TIB-TM | IS-Element | PCC7821 | ATAGGAGGTATTATCTAAGCAGCAT / GAGGGAAGAAGGTGGTTAGGA |
TCCCTATGGAGTAAGACTTACATTCCCGAT | 200/200/250 | 60 | 176 |
concentrations of the forward primer/reverse primer/TaqMan probe
PCR reactions were initiated by a 10 min hold at 95°C to activate the hot start polymerase, followed by 50 cycles of a two-step PCR, consisting of a denaturation step at 95°C (15 sec) and subsequent annealing and elongation steps at 55°C and 60°C respectively (1 min each). Each measurement was performed in triplicate using the Eppendorf Mastercycler ep realplex system (Eppendorf, Vienna). The 25 μl reaction mix consisted of 12.5 μl TaqMan Universal PCR Master Mix (ABI), 5 μl of DNA template, and variable concentrations of primers and probes (Table 2). To establish the calibration curves, a dilution series of predetermined DNA concentrations from the extracts of strains was prepared and the DNA content in the template (expressed in equivalents of biovolume) was related to the Ct value (defined as the threshold cycle to reach a manually set fluorescence of 100).
Determination of the lower detection limit
Dilution series of purified PCR products obtained from 16SrDNA, mcyBA1, insertions of strains No110 and No139 ranging from 0.1 fmol to 0.1×10−8 fmol were measured by real time PCR as described [34]. The range in dilution corresponded to 3×108 - 0.3 copies template−1. For each of the target regions specific Ct values corresponding to 1 copy template−1 were determined and used to define a lower limit of detection.
Analysis of the TNA results
Fluorescence signals below the limit of detection were set to zero. In general, calibration curves were not extrapolated beyond the highest dilution, which was defined arbitrarily as the limit of quantification corresponding to 18 cells template−1 for mcyDIS2 (strain No139) and 4 cells template−1 for all the other TNAs. The TNA results between zero and the quantification limit were adjusted to the corresponding quantification threshold. Proportions of all the mutations and the IS element were calculated from the cell numbers (biovolume) detected via 16S rDNA-TNA in the same DNA extract. All linear regression curves were fitted using the least square approximation and the associated statistical tests of Sigma Plot 2000 (V 6.10). The data were log10 transformed in order to achieve normal distribution and constant variances. The linear regressions between the total population density (as estimated from 16S rDNA) and the abundance of the mcyBA1 genotype and the mcy insertion/deletion mutants (Fig. 5) were compared in slope and intercept using a general factorial model of Analysis of Variance (ANOVA). The data were modelled as y = μ + βx + ε, where y is the measured abundance of the mcyB genotype or a specific mcv mutant, μ is the overall mean level, β is the effect of the mcyB or mcv mutant genotype, x is the effect of the cells of the total population (as determined from 16S rDNA) as a covariate, and ε is the random deviation, N(0, σ2) [36]. A SPSS statistical package (V 15.0 for Windows) was used for the ANOVA.
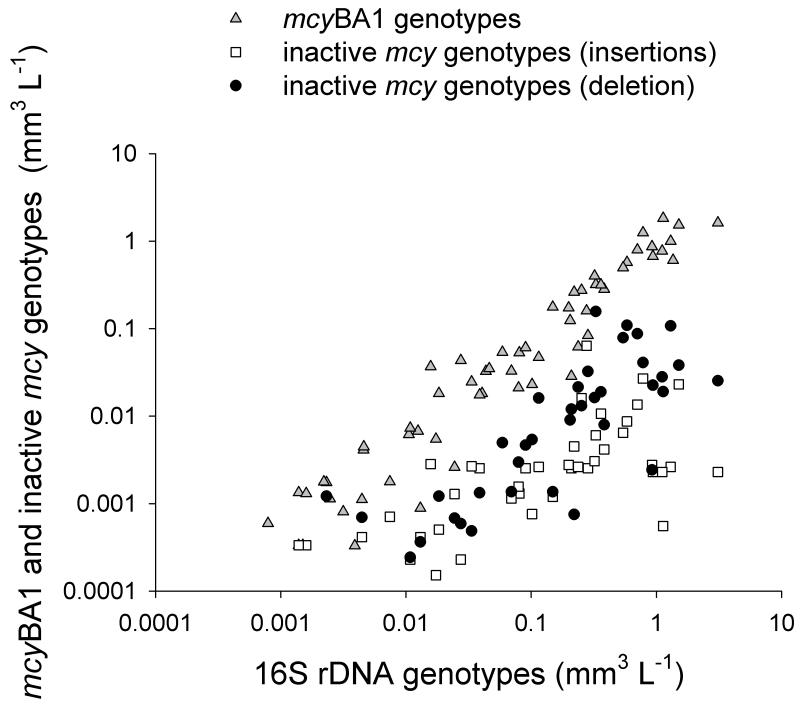
Figure 5.
Relationship between the total population density (in biovolume mm3 L−1) estimated from 16S rDNA and the biovolume estimated from the mcyBA1 genotype (gray triangles), the sum of inactive mcy genotypes containing insertions (white squares) and the inactive genotype containing the deletion (black circles). For details on the calculation of the respective regression curves see text.
Analysis of microcystins and related peptides in single filaments
In order to find out whether nontoxic mutants differ in the production of other related peptides when compared with their microcystin-producing congeners, (109 - 129 single Planktothrix filaments from each lake (a total of 1,230 filaments) were isolated between June 2005 and January 2006. The filaments were isolated under the microscope and analyzed by means of sensitive matrix assisted laser desorption ionisation-time of flight mass spectrometry (MALDI-TOF MS) as described [16]. The automated measurements were performed by Anagnostec GmbH on a Voyager DE Pro workstation (Applied Biosystems) [16, 46]. The following peptides were identified by their molecular mass in accordance with previous studies: Microcystin-LR (molecular weight 995 [M+H]+), desmethyl-microcystin-RR (1024 [M+H]+), desmethyl-microcystin-LR (981 [M+H]+), microcystin-HtyR (1045 [M+H]+), anabaenopeptin B (837 [M+H]+), anabaenopeptin A (844 [M+H]+) and anabaenopeptin F (851 [M+H]+), oscillamide Y (858 [M+H]+), oscillapeptin 1088 (1088 [M+H]+), Cl-aeruginoside 126A (749 [M+H]+), aeruginoside 126A (715 [M+H]+), aeruginosin A (617 [M+H]+), aeruginoside 126B (691 [M+H]+) and aeruginosin 583 (583 [M+H]+) [9, 31, 45, 46]). The absence of the peptides anabaenopeptin B, anabaenopeptin A, and anabaenopeptin F was used to identify those filaments (5%) that dropped out from the analysis due to unknown factors.
Results
Efficiency and specificity of the TaqMan assays
All of the calibration curves that were established by relating the measured Ct values of DNA extracts to the predetermined DNA concentrations in the template (expressed as biovolume equivalents), showed highly significant linear relations and similar amplification efficiencies (Table 3). The DNA mixture of the Planktothrix strain No62 (460 cells template−1) and Microcystis strains HUB524/HUB53 in a ratio 1: 100 revealed a Ct value of 28.55 ± 0.05 (SD), while a hundredfold increase of Microcystis resulted in a Ct value of 28.14 ± 0.19. The same DNA concentration of strain No62 in the absence of Microcystis DNA had a Ct value of 28.37 ± 0.12 and it was concluded that the TNA targeted to the mcyHA deletion was specific. In none of the DNA extracts from the field samples that were found to be free of Planktothrix in the microscope and by the TNA targeted to PC-IGS, the TNAs for 16S rDNA and the IS element revealed amplification that exceeded the fluorescence threshold during 50 cycles. Consequently, both TNAs were considered specific and robust enough to prevent false positive results.
Table 3.
Linear calibration curves of TNA used to quantify the total Planktothrix population, the mcyBA1 genotype, four inactive mcy genotypes and the transposable (IS) element reported to insert into the mcy gene cluster [6].
| TNA | Gene locus | Strain | Calibration curve1 | E (%)2 | R2 | N |
|---|---|---|---|---|---|---|
| 16S | 16S rDNA | PCC7821 | y = 36.9 − 3.836x | 82.3 | 0.998 | 11 |
| mcyBA1 | mcyBA1 | PCC7821 | y = 36.204 − 3.366x | 98.2 | 0.998 | 10 |
| mcyDIS1 | 3′end IS-Element, mcyD | No110 | y = 36.189 − 3.891x | 80.7 | 0.997 | 12 |
| mcyDIS2 | 3′end IS-Element, mcyD | No139 | y = 35.68 − 3.331x | 99.6 | 0.988 | 12 |
| mcyAIS | 3′end IS-Element, mcyA | No40 | y = 36. 73 − 3.378x | 97.7 | 0.993 | 9 |
| mcyHA | mcyHA deletion | No62 | y = 39.927 − 3.677x | 87.1 | 0.985 | 9 |
| TIB-TM | IS-Element | PCC7821 | y = 36.398 − 3.83x | 82.4 | 0.987 | 11 |
y = Ct value (PCR cycle number at the fluorescence threshold of 100), x = amount of template DNA (expressed as log10 of cell number equivalents)
amplification efficiencies (E) were calculated as follows E = (10 −1/slope − 1) × 100
In total, 79 samples were analysed. Except of ten samples, Planktothrix filaments were detected in all the samples during the counting in the microscope. All the samples except of three (Wolfgangsee, 2006 and two samples from Attersee, 2007) gave positive signals for 16S rDNA, indicating the presence of Planktothrix. Comparison of the biovolume estimated using the TNA for 16S rDNA and the biovolume as determined from the microscope revealed a linear relationship following the equation y = −0.46 + 0.86x (R2 = 0.85, n = 68), where y is the log10 biovolume (mm3 L−1) as determined by the TNA and x is the log10 biovolume as determined by the counting in the microscope (Supplemental material, Fig. S1).
Phytoplankton composition in the study lakes
In general, the phytoplankton was composed of species of cyanobacteria, bacillariophyceae, chlorophyceae, cryptophyceae, chrysophyceae, dinophyceae. Cyanobacteria (Planktothrix spp., Anabaena spp., Lyngbya spp.), and bacillariophyceae (Fragilaria spp., Asterionella formosa, and various centric diatoms) were the most abundant groups followed by chrysophyceae (Dinobryon spp.) and cryptophyceae (Cryptomonas spp.). The Planktothrix populations studied revealed a wide range of population sizes (40 – 570,000 filaments L−1 corresponding to 3.8×103 – 5.×107 cells L−1). According to this variation, the lakes were classified as lakes containing a sparse population of Planktothrix sp. only (Attersee, Wolfgangsee and Schwarzensee), an intermediate population density (Offensee, Ammersee, Fuschlsee, Mondsee, Irrsee) and a high population density (Afritzersee, Zürichsee, Wörthersee and Hallwilersee) (Fig. 2). On a relative scale, Planktothrix constituted <10% of phytoplankton biovolume in lakes with a sparse population only (Attersee, Wolfgangsee and Schwarzensee) and 50-100% in lakes showing the highest population density (Afritzersee, Hallwilersee, Wörthersee, Zürichsee). In the other lakes, the biovolume proportions were found to be more variable but Planktothrix never constituted >50% of the phytoplankton.
Figure 2.
Mean (± SE) filament densities from July 2005 to October 2007 in the twelve study lakes as estimated from the inverted microscope technique.
Quantification of mutants containing an inactivated microcystin synthetase gene cluster In four of the twelve investigated lakes, all the mutants were detected (Afritzersee, Mondsee, Wörthersee, Zürichsee). In 10 out of the 12 investigated lakes, at least one inactive genotype was detected in the integrated lake water samples. In some populations, only specific mutations could be detected, i.e. populations from Offensee and Irrsee contained the mcyDIS2 insertion only (Fig. 3). However, the analysis of phytoplankton net samples (resulting in a >1,000 fold concentration of colonial cyanobacteria) revealed the occurrence of other mutations such as the mcyHA deletion in Irrsee (data not shown). In contrast none of the mutants were found in the integrated water samples of Wolfgangsee and Schwarzensee. However, phytoplankton net samples again revealed the occurrence of genotypes containing the mcyDIS2 insertion and it was concluded that the lowest population abundance in Schwarzensee and Wolfgangsee prevented the detection of these mutants in the integrated quantitative lake water samples. It is concluded that nontoxic mutants occurred in all the lakes at all the population densities at least in trace amounts.
Figure 3.
Average (± SE) proportion of inactive mcy genotypes in Planktothrix populations in the twelve study lakes as quantified by real-time PCR. The stars indicate the detection of inactive mcy genotypes in the phytoplankton net samples.
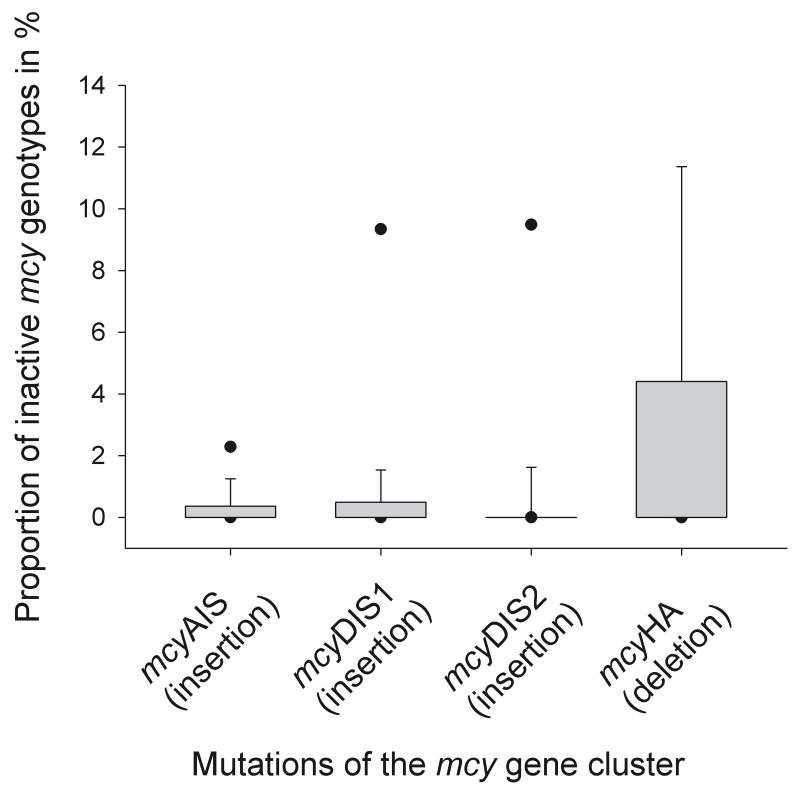
Altogether the mcyHA deletion was detected in 47%, mcyAIS in 37%, mcyDIS1 in 37% and mcyDIS2 in 18% of all the integrated samples. On average, those genotypes containing insertions were found in rather low proportions only (min 0%, mean 2.8% ± 0.7, max 24%). In contrast, the mcyHA deletion was detected more frequently (min 0%, mean 3.7% ± 1, max 52%). Taking all the data together, the difference in proportions between the insertions and the mcyHA deletion was found to be significant (Kruskal-Wallis One Way Analysis of Variance on Ranks, p < 0.001, Fig. 4).
Figure 4.
Percentages of inactive mcy genotypes in populations of Planktothrix (n = 79). Box plots show the median and the 5% - 95% percentiles.
Relationship between the population density and the abundance of the mutants
The IS element potentially inactivating microcystin synthesis was detected in all the lakes (77% of the samples were positive). The abundance of the IS element was linearly related to the population density according to the equation y = −0.69 + 1.1x (R2 = 0.75, n = 60), where y is the DNA concentration expressed in log10 biovolume (mm3 L−1) of the IS element and x is the DNA expressed in log10 biovolume (mm3 L−1) of the total population calculated from the 16S rDNA (Supplemental material, Fig. S2).
Correspondingly, the abundance of the mcyBA1 genotype was linearly correlated with the population density according to the equation: y = −0.09 + 1.14x (R2 = 0.96, n = 76, p < 0.001), where y is the log10 biovolume (mm3 L−1) of the mcyBA1 genotype and x is the log10 biovolume (mm3 L−1) of the total population. In general the abundance of the mutations also was linearly related to the population density. The regression equation for the mcy genotypes containing insertions was y = −2.21 + 0.5x (R2 = 0.55, n = 44, p < 0.001) and for the mcy genotypes containing the mcyHA deletion the regression curve was y = −1.49 + 0.85x (R2 = 0.63, n = 37, p < 0.001), where x is the log10 biovolume (mm3 L−1) of the total population and y is the log10 biovolume (mm3 L−1) of genotypes containing an insertion/deletion (Fig. 5). The regression curves calculated for the mcyBA1 genotype and the mcy mutants containing either the insertions or the mcyHA deletion differed significantly in slope (ANOVA, p ≤ 0.001).
Peptide composition in single filaments
In total, microcystin-containing filaments had a share of 72%, while 28% of the filaments were found without microcystin. Beside microcystins, the most abundant peptides detected were the anabaenopeptins B, A and F (95%), and the aeruginosins (55%). The filaments lacking microcystin had a very similar peptide composition when compared with the peptide composition of the microcystin-producing filaments: anabaenopeptin B (98% in filaments without microcystin vs. 99% in filaments with microcystin), anabaenopeptin A (12% vs. 44%), anabaenopeptin F (70% vs. 90%), oscillamide Y (2% vs. 22%), oscillapeptin 1088 (4% vs. 17%), Cl-aeruginoside 126A (17 vs. 21%), aeruginoside 126A (8% vs. 32%), aeruginosin A (8% vs. 22%), aeruginoside 126B (11% vs. 6%), aeruginosin 583 (18% vs. 12%). None of the peptides occurred with a significantly higher frequency among the microcystin deficient filaments when compared with the microcystin-producing filaments (t-test, p > 0.1).
Discussion
Evolution of the microcystin synthetase gene cluster
In the present study, the abundance of nontoxic microcystin mutants was monitored for two years. Although the proportion of the inactive mcy genotypes was found to vary during the sampling period, no trend of increase or decrease in the proportion through the study period was observed. Instead, we concluded that the abundance of all the nontoxic mutants was primarily related to the population density. These first quantitative estimates confirm previous results, which reported the regular detection of these mutations through the years 2001-2004 [6]. It is generally anticipated that prokaryotic organisms maintain rather small genomes due to a continuous loss of mutated and, therefore, nonfunctional genetic material [22]. However, because the population numbers are huge, only larger gene deletions are considered to provide sufficient selective advantage, so that mutations resulting in gene loss become fixed on a global scale [3]. We recently reported that only larger deletions that lead to the loss of a major part of the mcy gene cluster resulted in the distribution of nontoxic mutants across the European continent [7]. However, this was in the course of rather long time periods, probably spanning millions of years. Rohrlack et al. [31] described the co-occurrence of Planktothrix strains differing in peptide composition over more than 30 years in Lake Steinsfjorden (Norway) suggesting a rather slow evolution of NRPS in Planktothrix. Consequently, successional studies exceeding the usual investigation period of a few years will be needed in order to relate the increase or decrease of specific mcy genotypes and their nontoxic mutants to the evolution of the mcy gene cluster.
Spatial isolation of populations
The results on the average stable proportion of nontoxic mutants are important in order to predict the toxicity of blooms formed by Planktothrix in lakes and reservoirs. It is known that Planktothrix is one of the more efficient invaders and several examples of the sudden appearance of blooms in Europe have been reported [23, 26]. According to the results of this study, inactive mcy genotypes are unlikely to dominate populations by a selective sweep during the transition from prebloom conditions to bloom conditions. Instead it is proposed that these mutants increase only slowly over time and it is not yet possible to predict whether these mutations become fixed on a global scale. One major difficulty to predict their fixation is that populations have been found to diverge in genotype composition due to spatial isolation [17]. In this study, some populations had only one inactive mcy genotype, i.e. in Offensee and Schwarzensee only the mcyDIS2 genotype could be detected. This phenomenon is believed to result from the spatial isolation of populations among different lakes leading to a divergence in genetic population structure for several years. Although this isolation may be linked to a relatively stable co-occurrence of genotypes over decades [31], it is not believed that this effect leads to a complete divergence in the evolution of microcystin synthesis, as genetic exchange has been observed both on a scale of a few kilometres [17] as well as a larger scale covering hundreds of kilometres [2].
Selective pressure on inactive mcy genotypes
From the data presented (Fig. 5) it is concluded that on average the abundance of the inactive mcy genotypes was found linearly related to the population density. For both mcy mutants containing the insertions and the mcyHA deletion the slope of the regression curves was significantly lower when compared with the slope of the regression curve obtained for the mcyBA1 genotype. It is emphasized, however that it would be premature to conclude whether selection may disfavour a single mcy mutant or not. If the nontoxic mutants would be selectively disfavoured due to the lack of microcystin in general then this selective disadvantage is considered to be of relatively minor importance when compared to the overall increase and the distribution of the mcy mutants among most of the study lakes. Alternatively it is possible that the mutants investigated during this study differ in the production of a putative yet undetected peptide that compensates for the lack of the microcystin. In this study the frequency of occurrence as recorded for the most widely distributed anabaenopeptins and aeruginosins did not differ between the two groups. Other more frequent peptides such as cyanopeptolins and microviridins were not considered in this analysis. Consequently, as argued by one reviewer it is possible that those nontoxic mutants are not “cheaters” but rather follow another strategy by producing other peptides replacing the microcystin. In either case we found an astonishing constancy of the share of the nontoxic mutants in relation to a >10,000 fold range in population density. This implies that the selective (dis)advantage to the individual due to the production of a putative peptide should be relatively independent from the density of the total population.
Effects of the deletion or the insertional inactivation of the mcy genes on cellular growth In general, the deletion of parts of the mcy gene cluster was observed more frequently when compared with the insertions (Fig. 4). Studies on gene loss processes generally suggest a mutational bias for deletions that leads to the erosion of the inactivated genetic material. This is considered to counteract the continuous acquisition of genetic material by horizontal gene transfer [22]. It is not yet known whether even smaller deletions as observed in this study have the potential to increase the growth rate due to a reduction in genome size, in contrast to insertions that result in an increased burden of DNA amplification during cell division. On the other hand, as NRPS are large multifunctional enzyme complexes, high metabolic costs solely due to the microcystin synthesis have been proposed. Surprisingly the comparsion of the growth of microcystin deficient mutants and that of the wildtype strain of Microcystis aeruginosa strain PCC7120 did not reveal significant differences [10]. During the study period, the mcyHA deletion was found to have the broadest distribution occurring in lakes that are hundreds of kilometres apart. In the future, comparative growth experiments under controlled conditions in the laboratory between the mutants containing deletions and insertions will need to demonstrate whether the net decrease in gene content can indeed result in higher growth rates.
Distribution of the IS element Plrub1
In this study, the relation between the abundance of the total population and the abundance of the IS element Plrub1 reported to inactivate the mcy gene cluster was linear. It is concluded that this IS element that inactivated microcystin synthesis in Planktothrix is indeed specific for Planktothrix spp. and does not occur in other organisms. It is likely that this IS element is active and contributes to a number of mutations affecting other parts of secondary metabolite synthesis. So far the IS element has been found in all strains assigned to P. rubescens, while green-pigmented strains of P. agardhii differed significantly in the content of the IS element (C. Molitor, R. Kurmayer, unpublished data). It is possible that this IS element constitutes an important factor currently inducing genetic variation among red-pigmented Planktothrix populations in European lakes. It would be an interesting task to find out whether the activity of this IS element can vary and if this may be influenced by specific environmental conditions.
Supplementary Material
Acknowledgements
We would like to thank Guntram Christiansen for his help in molecular biological techniques. We are most grateful to Arno Stöckli (Department Bau, Verkehr und Umwelt, Kanton Aarau, Switzerland), Ferdinand Schanz (University of Zürich), Günther Bruschek and Karl Mayrhofer (BAW Scharfling, Austria) for the provision of water samples. Marcel Erhard did the automated MALDI-TOF MS measurements on peptide composition in single filaments. We appreciated the comments of two anonymous reviewers. This study was financed by the Austrian Science Fund project P18185 “Microevolution of toxin synthesis in cyanobacteria”
References
- 1.Anagnostidis K, Komárek J. Modern approach to the classification system of cyanophytes, 3-Oscillatoriales. Arch Hydrobiol Suppl Algol Stud. 1988;80(50-53):327–472. [Google Scholar]
- 2.Barker GLA, Handley BA, Vacharapiyasophon P, Stevens JR, Hayes PK. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (Cyanobacteria) filaments in the Baltic Sea. Microbiology. 2000;146:2865–2875. doi: 10.1099/00221287-146-11-2865. [DOI] [PubMed] [Google Scholar]
- 3.Berg OG, Kurland CG. Evolution of microbial genomes: sequence acquisition and loss. Mol Biol Evol. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- 4.Chorus I, Bartram J. Toxic cyanobacteria in water. A guide to their public health consequences, monitoring and management. WHO, E & FN Spon; London: 1999. p. 416. [Google Scholar]
- 5.Christiansen G, Fastner J, Erhard M, Börner T, Dittmann E. Microcystin biosynthesis in Planktothrix: genes, evolution, and manipulation. J Bacteriol. 2003;185:564–572. doi: 10.1128/JB.185.2.564-572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen G, Kurmayer R, Liu Q, Börner T. Transposons inactivate biosynthesis of the nonribosomal peptide microcystin in naturally occurring Planktothrix spp. Appl Environ Microbiol. 2006;72:117–123. doi: 10.1128/AEM.72.1.117-123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christiansen G, Molitor C, Philmus B, Kurmayer R. Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol Biol Evol. 2008;25:1695–1704. doi: 10.1093/molbev/msn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmondson WT, Litt AH. Daphnia in Lake Washington. Limnol Oceanogr. 1982;27/2:272–293. [Google Scholar]
- 9.Fastner J, Erhard M, von Döhren H. Determination of oligopeptide diversity within a natural population of Microcystis (Cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol. 2001;67:5069–5076. doi: 10.1128/AEM.67.11.5069-5076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesse K, Dittmann E, Börner T. Consequences of impaired microcystin production for light-dependent growth and pigmentation of Microcystis aeruginosa PCC 7806. FEMS Microbiol Ecol. 2001;37:39–43. [Google Scholar]
- 11.ISO . Water quality - measurement of biochemical parameters - spectrometric determination of the chlorophyll-a concentration. International Organisation for Standardization; Geneve: 1992. p. 12. [Google Scholar]
- 12.Jacquet S, Briand J-F, Leboulanger C, Avois-Jacquet C, Oberhaus L, Tassin B, Vincon-Leite B, Paolini G, Druart J-C, Anneville O, Humbert J-F. The proliferation of the toxic cyanobacterium Planktothrix rubescens following restoration of the largest natural French lake (Lac du Bourget) Harmful Algae. 2005;4:651–672. [Google Scholar]
- 13.Kaebernick M, Rohrlack T, Christoffersen K, Neilan BA. A spontaneous mutant of microcystin biosynthesis: genetic characterization and effect on Daphnia. Environ Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-2920.2001.00241.x. [DOI] [PubMed] [Google Scholar]
- 14.Kardinaal W, Tonk L, Janse I, Hol S, Slot P, Huisman J, Visser P. Competition for light between toxic and nontoxic strains of the harmful cyanobacterium Microcystis. Appl Environ Microbiol. 2007;73:2939–2946. doi: 10.1128/AEM.02892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurmayer R, Christiansen G, Chorus I. The abundance of microcystin-producing genotypes correlates positively with colony size in Microcystis and determines its microcystin net production in Lake Wannsee. Appl Environ Microbiol. 2003;69:787–795. doi: 10.1128/AEM.69.2.787-795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurmayer R, Christiansen G, Fastner J, Börner T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ Microbiol. 2004;6:831–841. doi: 10.1111/j.1462-2920.2004.00626.x. [DOI] [PubMed] [Google Scholar]
- 17.Kurmayer R, Gumpenberger M. Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol Ecol. 2006;15:3849–3861. doi: 10.1111/j.1365-294X.2006.03044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurmayer R, Jüttner F. Strategies for the co-existence of zooplankton with the toxic cyanobacterium Planktothrix rubescens in Lake Zürich. J Plankt Res. 1999;21:659–683. [Google Scholar]
- 19.Kurmayer R, Kutzenberger T. Application of real-time PCR for quantification of microcystin genotypes in a population of the toxic cyanobacterium Microcystis sp. Appl Environ Microbiol. 2003;69:6723–6730. doi: 10.1128/AEM.69.11.6723-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikalsen B, Boison G, Skulberg OM, Fastner J, Davies W, Gabrielsen TM, Rudi K, Jakobsen KS. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J Bacteriol. 2003;185:2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 23.Naselli-Flores L, Barone R, Chorus I, Kurmayer R. Toxic cyanobacterial blooms under a semiarid mediterranean climate: The magnification of a problem. Environ Toxicol. 2007;22:399–404. doi: 10.1002/tox.20268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishizawa T, Asayama M, Fujii K, Harada K, Shirai M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J Biochem. 1999;126:520–529. doi: 10.1093/oxfordjournals.jbchem.a022481. [DOI] [PubMed] [Google Scholar]
- 25.Nürnberg GK, LaZerte BD. An artificially induced Planktothrix rubescens surface bloom in a small kettle lake in Southern Ontario compared to blooms worldwide. Lake Res Manag. 2003;19:307–322. [Google Scholar]
- 26.Padisák J, Scheffler W, Kasprzak P, Koschel R, Krienitz L. Interannual variability in the phytoplankton composition of Lake Stechlin (1994-2000) Arch Hydrobiol/Advanc Limnol. 2003;58:101–133. [Google Scholar]
- 27.Pridmore R, Etheredge M. Planktonic cyanobacteria in New Zealand inland waters: distribution and population dynamics. N Z J Mar Freshw Res. 1987;21:491–502. [Google Scholar]
- 28.Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Börner T, Sivonen K. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci USA. 2004;101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rippka R. Isolation and purification of cyanobacteria. Meth Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 30.Rohrlack T, Dittmann E, Henning M, Börner T, Kohl J-G. Role of microcystins in poisoning and food ingestion inhibition of Daphnia galeata caused by the cyanobacterium Microcystis aeruginosa. Appl Environ Microbiol. 1999;65:737–739. doi: 10.1128/aem.65.2.737-739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohrlack T, Edvardsen B, Skulberg R, Halstvedt CB, Utkilen HC, Ptacnik R, Skulberg OM. Oligopeptide chemotypes of the toxic freshwater cyanobacterium Planktothrix can form subpopulations with dissimilar ecological traits. Limnol Oceanogr. 2008;53:1279–1293. [Google Scholar]
- 32.Rouhiainen L, Vakkilainen T, Siemer BL, Buikema W, Haselkorn R, Sivonen K. Genes coding for hepatotoxic heptapeptides (microcystins) in the cyanobacterium Anabaena strain 90. Appl Environ Microbiol. 2004;70:686–692. doi: 10.1128/AEM.70.2.686-692.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schober E, Kurmayer R. Evaluation of different DNA sampling techniques for the application of the real-time PCR method for the quantification of cyanobacteria in water. Lett Appl Microbiol. 2006;42:412–417. doi: 10.1111/j.1472-765X.2006.01857.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schober E, Werndl M, Laakso K, Korschinek I, Sivonen K, Kurmayer R. Interlaboratory comparison of Taq Nuclease Assays for the quantification of the toxic cyanobacteria Microcystis sp. J Microbiol Meth. 2007;69:122–128. doi: 10.1016/j.mimet.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucl Acid Res. 2006;34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokal R, Rohlf F. Biometry. The principles and practice of statistics in biological research. 3rd edition W.H. Freeman and Company; New York: 1995. p. 886. [Google Scholar]
- 37.Suda S, Watanabe MM, Otsuka S, Mahakahant A, Yongmanitchai W, Nopartnaraporn N, Liu Y, Day JG. Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int J Syst Evol Microbiol. 2002;52:1577–1595. doi: 10.1099/00207713-52-5-1577. [DOI] [PubMed] [Google Scholar]
- 38.Tillett D, Dittmann E, Erhard M, vonDöhren H, Börner T, Neilan BA. Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system. Chemistry and Biology. 2000;7:753–764. doi: 10.1016/s1074-5521(00)00021-1. [DOI] [PubMed] [Google Scholar]
- 39.Tillett D, Parker DL, Neilan BA. Detection of toxigenicity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl Environ Microbiol. 2001;67:2810–2818. doi: 10.1128/AEM.67.6.2810-2818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends Microbiol. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 41.Utermöhl H. Zur Vervollkommnung der quantitativen Phytoplanktonmethodik. Mitt Internat Verein Limnol. 1958;2:1–38. [Google Scholar]
- 42.Vollenweider RA, Kerekes J. Eutrophication of waters. Monitoring, assessment and control. OECD Cooperative programme on monitoring of inland waters (Eutrophication control) Environmental Directorate, OECD; Paris: 1982. p. 154. [Google Scholar]
- 43.Walsby AE, Avery A. Measurement of filamentous cyanobacteria by image analysis. J Microbiol Meth. 1996;26:11–20. [Google Scholar]
- 44.Walsby AE, Ng G, Dunn C, Davis PA. Comparison of the depth where Planktothrix rubescens stratfies and the depth where the daily insolation supports its neutral buoyancy. New Phytol. 2004;162:133–145. [Google Scholar]
- 45.Welker M, Christiansen G, von Döhren H. Diversity of coexisting Planktothrix (cyanobacteria) chemotypes deduced by mass spectral analysis of microystins and other oligopeptides. Arch Microbiol. 2004;182:288–298. doi: 10.1007/s00203-004-0711-3. [DOI] [PubMed] [Google Scholar]
- 46.Welker M, Erhard M. Consistency between chemotyping of single filaments of Planktothrix rubescens (Cyanobacteria) by MALDI-TOF and the peptide patterns of strains determined by HPLC-MS. J Mass Spectrom. 2007;42:1062–1068. doi: 10.1002/jms.1237. [DOI] [PubMed] [Google Scholar]
- 47.Wetzel RG, Likens GE. Limnological analyses. 3rd edition Springer-Verlag; New York: 2000. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.