Summary
The type II secretion system is a multi-protein complex that spans the cell envelope of gram-negative bacteria and promotes the secretion of proteins, including several virulence factors. This system is homologous to the type IV pilus biogenesis machinery and contains five proteins, EpsG-K, termed the pseudopilins that are structurally homologous to the type IV pilins. The major pseudopilin EpsG has been proposed to form a pilus-like structure in an energy-dependent process that requires the ATPase, EpsE. A key remaining question is how the membrane-bound EpsG interacts with the cytoplasmic ATPase, and if this is a direct or indirect interaction. Previous studies have established an interaction between the bitopic inner membrane protein EpsL and EpsE; therefore, in this study we used in vivo cross-linking to test the hypothesis that EpsG interacts with EpsL. Our findings suggest that EpsL may function as a scaffold to link EpsG and EpsE and thereby transduce the energy generated by ATP hydrolysis to support secretion. The recent discovery of structural homology between EpsL and a protein in the type IV pilus system implies that this interaction may be conserved and represent an important functional interaction for both the type II secretion and type IV pilus systems.
Keywords: type IV pilus, prepilin peptidase, PilD
Introduction
The type II secretion (T2S) system is found widely distributed among gram-negative bacteria and is considered a primary virulence system (Cianciotto, 2005, Sandkvist, 2001b). In Vibrio cholerae, this system is encoded by the extracellular protein secretion (eps) operon and is used to secrete several proteins, including the main virulence factor cholera toxin, across the cell envelope (Sandkvist et al., 1997). The complex is composed of at least 12 different proteins, each individually essential for secretion to occur, which form a structure that spans the entire cell envelope (Filloux, 2004). The T2S apparatus is highly homologous to the type IV pilus biogenesis machinery which produces cell surface filaments that are involved in multiple processes, including attachment and colonization, biofilm formation, DNA uptake, twitching motility, and virulence (Craig & Li, 2008).
Specifically, the assembled T2S system consists of the cytoplasmic ATPase, EpsE, an inner membrane platform composed of EpsC, F, L, and M, and an outer membrane secretin pore formed by EpsD (Johnson et al., 2006). The T2S machinery also contains five additional gene products termed the pseudopilins, EpsG, H, I, J and K, that are structurally homologous to the type IV pilins (Alphonse et al., 2010, Kohler et al., 2004, Korotkov et al., 2009, Korotkov & Hol, 2008, Yanez et al., 2008). Additionally, both the T2S pseudopilins and the type IV pilins contain a conserved N-terminus which is recognized and processed by the prepilin peptidase PilD, also referred to as VcpD (Marsh & Taylor, 1998, Strom et al., 1993, Fullner & Mekalanos, 1999). Processing of the pilin subunits is a prerequisite for their incorporation into functional pili (Fullner & Mekalanos, 1999, Marsh & Taylor, 1998, Nunn et al., 1990).
Due to the similarities between the pseudopilins and the type IV pilins, EpsG-K have been suggested to form a pilus-like structure, or pseudopilus, where EpsG is the major and EpsH-K are minor pseudopilins (Campos et al., 2010, Johnson et al., 2006). Over-expression of the EpsG homologs XcpT and PulG in Pseudomonas aeruginosa and Klebsiella oxytoca, respectively, confirmed that the major pseudopilin is indeed capable of assembling into a pilus-like structure that can be detected on the surface of the bacteria (Durand et al., 2003, Vignon et al., 2003). Detection of surface pseudopili required the ATPase and inner membrane platform proteins, and was only observed under over-expression conditions when all other components of the T2S system were held constant. However, during native expression of the complex surface appendages were not observed. Instead, the pseudopilus is hypothesized to span the periplasm and promote secretion by functioning as a piston that can push secreted proteins through the outer membrane pore, or alternatively, by acting as a retractable plug (Filloux, 2004, Hobbs & Mattick, 1993, Sandkvist, 2001a).
The type IV pilins and T2S pseudopilins require the function of an ATPase to polymerize (Camberg & Sandkvist, 2005, Durand et al., 2005, Pelicic, 2008); however, a key question is how the cytoplasmic ATPase of these systems is able to interact with the pilin or pseudopilin subunits that are located within the inner membrane and exposed to the periplasm. A study using P. aeruginosa identified a point mutation in the EpsG homolog, XcpT, which inhibited secretion and could be suppressed by a second point mutation in the EpsE homolog, XcpR (Kagami et al., 1998). Based on this result it has been proposed that EpsG and EpsE are capable of interacting directly, however, molecular mapping of EpsE using subcellular fractionation, domain swapping, and x-ray crystallography have strongly suggested that the region of the suppressor mutation in the EpsE homolog is involved in an interaction with the bitopic inner membrane protein XcpY, the EpsL homolog (Abendroth et al., 2005, Sandkvist et al., 1995). Therefore, EpsL is a plausible candidate to bridge EpsG and EpsE and transduce the energy provided by ATP hydrolysis in order to promote secretion.
In this study we used in vivo cross-linking and co-immunoprecipitation to establish an interaction between the major pseudopilin subunit, EpsG, and EpsL. This interaction occurs in the absence of all other structural components of the apparatus but does require the prepilin peptidase, PilD. Furthermore, when the point mutation originally isolated in XcpT was made in EpsG it severely reduced the amount of cross-linked EpsG-EpsL complex. Based on our findings we propose that EpsL is the linker that allows for association of the ATPase with EpsG, and may function as a mechanical lever to transfer the energy required to support either assembly or disassembly of the pseudopilus.
Results
EpsG and EpsL interact in V. cholerae
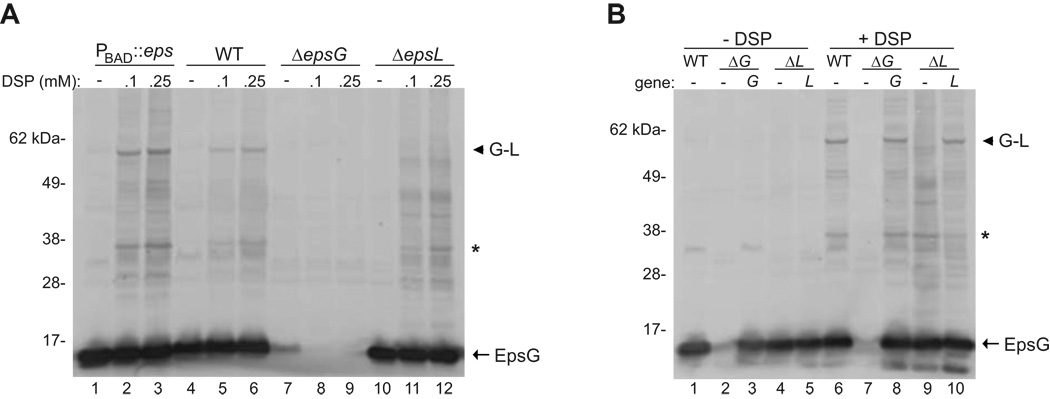
Based on our previous work detailing the interaction between EpsE and the cytoplasmic domain of the membrane protein EpsL (Abendroth et al., 2005, Sandkvist et al., 2000, Sandkvist et al., 1995), we sought to determine if EpsL interacts with EpsG and thus provides a link between the pseudopilus and the ATPase. Previous reports on EpsG homologs have focused on the interactions amongst the five pseudopilins (Douzi et al., 2009, Hu et al., 2002, Kuo et al., 2005), however, a definitive interaction between the major pseudopilin and other components of the T2S apparatus has yet to be revealed. This is likely due to the dynamic nature of the secretion machinery in which transient or weak interactions may be difficult to capture; therefore, we chose to use the membrane permeable cross-linker dithiobis (succinimidyl propionate), DSP, to preserve potential interactions that involve EpsG. Specifically, whole cells were treated with several concentrations of DSP, lysed, and subjected to SDS-PAGE and immunoblotting with anti-EpsG antiserum. Treatment with DSP produced higher molecular weight species in a concentration dependent manner in a wild type strain that were specific for EpsG, as these complexes were absent in a ΔepsG mutant strain (Figure 1A, compare lanes 4–6 with 7–9). The two most prominent cross-linked species were approximately 35 and 60 kDa. The 60 kDa complex was of the appropriate molecular weight to represent a heterodimer of EpsG and EpsL, whose molecular masses are 15 kDa and 45 kDa, respectively. Furthermore, this species was shown to be absent in a ΔepsL mutant strain (Figure 1A). The complex at 35 kDa is presumed to be a dimer of EpsG as it has been well documented that EpsG and its homologs can form dimers even in the absence of other T2S components (Kohler et al., 2004, Korotkov et al., 2009).
Figure 1. In vivo cross-linking of EpsG and EpsL.
(A) Whole cells of V. cholerae TRH7000 wild-type, ΔepsG, ΔepsL, and PBAD::eps (native eps promoter replaced by arabinose inducible promoter and grown in the presence of 0.01% arabinose) were cross-linked using the indicated concentrations of DSP as described in the experimental procedures, and immunoblotted for EpsG. (B) Whole cells of PBAD::eps wild-type containing pMMB67 vector, PBAD::ΔepsG with empty vector or vector encoded epsG, and PBAD::ΔepsL with empty vector or vector encoded epsL were cross-linked with 0.1 mM DSP and immunoblotted for EpsG. The position of EpsG is indicated. An arrow head designates the EpsG-EpsL complex, and an asterisk represents the putative EpsG dimer. The band directly below the putative EpsG dimer is presumed to be cross-reactive as it is also observed in the absence of DSP. The molecular weight markers are shown in kilo Daltons and lane numbers are indicated.
In order to increase the detection of the specific EpsG complexes we also examined the cross-linking profile in a strain where the native eps promoter has been replaced with the PBAD arabinose-inducible promoter, referred to as PBAD::eps (Sikora et al., 2007). This strain allows for specific upregulation of the entire eps operon, resulting in increased levels of all T2S components and thereby maintaining the stochiometry of the secretion complex. Growth of the PBAD::eps strain in the presence of 0.01% arabinose enhanced the detection of cross-linked EpsG complexes while preserving the cross-linking profile that was observed in wild type cells (Figure 1A lanes 1–3); thus, we chose to use the PBAD promoter background to further elucidate the EpsG-EpsL interaction. First, we confirmed that deletions of epsG and epsL in the PBAD::eps background inhibited secretion and complementation with plasmid-encoded epsG and epsL restored secretion in the deletion mutants to near wild-type levels (Figure 2). As expected, we found that deletion of either the epsG or epsL genes in the PBAD::eps strain prevented detection of the 60 kDa cross-linked species with anti-EpsG antibodies (Figure 1B lanes 7 and 9). Furthermore, complementation of the deletion strains with epsG or epsL encoded upon a plasmid restored the detection of the 60 kDa complex (Figure 1B lanes 8 and 10).
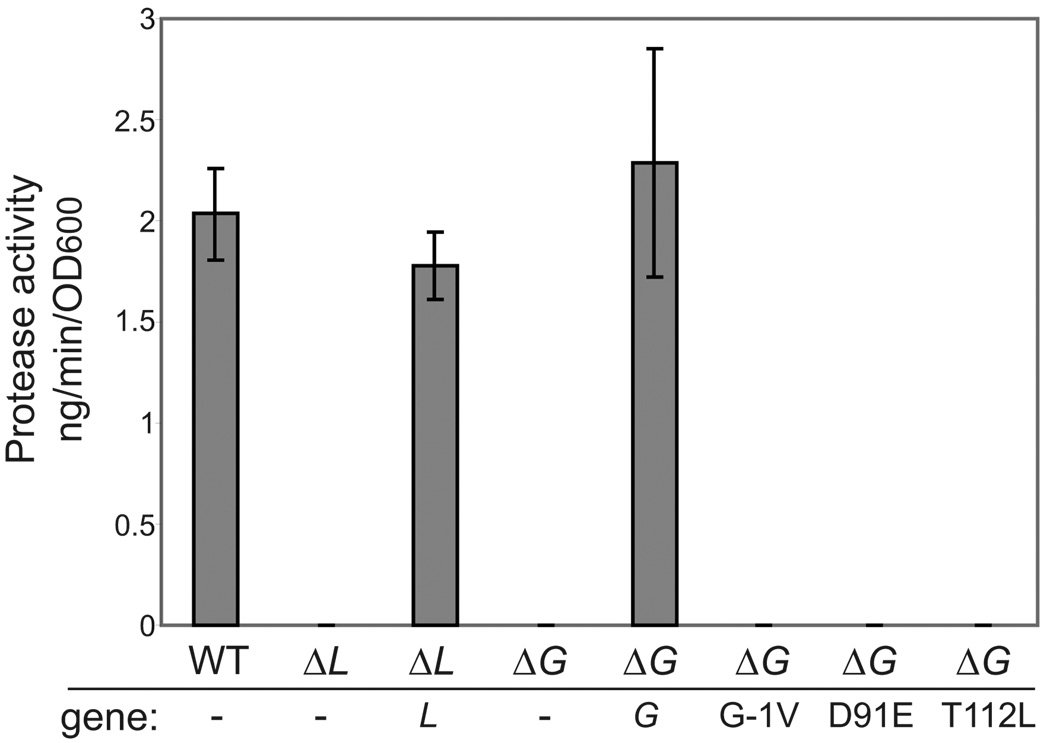
Figure 2. Mutations in EpsG prevent secretion.
Supernatants of over-night cultures of V. cholerae TRH7000 PBAD::eps wild-type, PBAD::ΔepsL containing empty vector or vector encoding wild-type epsL, PBAD::ΔepsG with empty vector, and vector encoded wild-type epsG or mutants were grown over-night at 37°C. Supernatants were separated from the cells and analyzed for the presence of an extracellular protease using the cleavable fluorogenic substrate N-tert-butoxy-canbonyl-Gln-Ala-Arg-7-amido-4-methyl-coumarin as described in the materials and methods. Each sample was assayed in triplicate and the standard error is indicated.
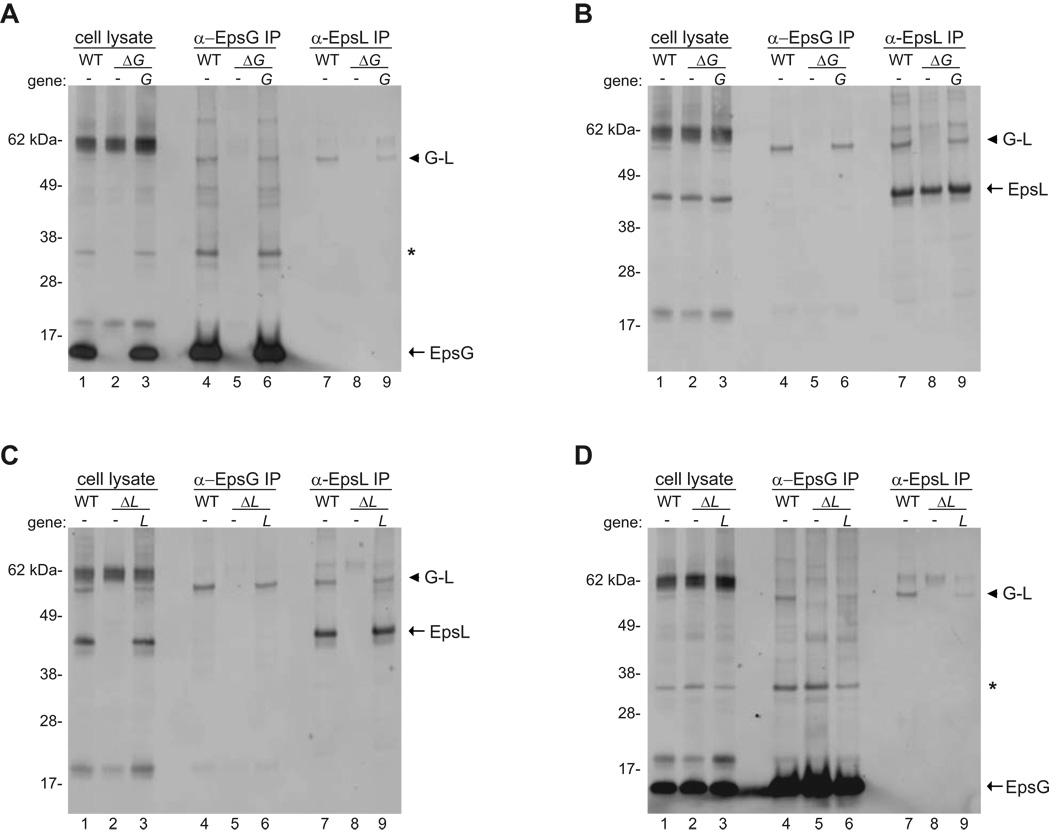
To further verify that the 60 kDa species corresponds to an EpsG-EpsL complex we subjected cross-linked samples to co-immunoprecipitation. Whole cells of PBAD::eps, PBAD::ΔepsG, PBAD::ΔepsL, and their respective complemented strains were cross-linked with 0.25 mM DSP, and cell lysates were prepared and incubated with antibodies specific for either EpsG or EpsL bound to protein G sepharose beads. Following extensive washing the precipitated material was analyzed by SDS-PAGE, and immunoblotted with biotinylated anti-EpsG or anti-EpsL antibodies. The results from the co-immunoprecipitation supported the whole cell cross-linking data by demonstrating that the 60 kDa band was only precipitated from the PBAD::eps and complemented strains. The 60 kDa complex was not pulled down from the epsG and epsL deletion strains when the cell extracts were precipitated with either EpsG or EpsL anti-sera (Figure 3). Precipitating cell extracts with anti-EpsG antibodies followed by immunoblotting for EpsG enriched for monomeric EpsG as well as cross-linked EpsG species, including the 60 kDa complex in wild type and complemented strains (Figure 3A lanes 4 and 6; Figure 3D lanes 4 and 6). Likewise, precipitating samples with anti-EpsL antibodies followed by immunoblotting for EpsL enriched for the EpsL monomer and the 60 kDa protein species (Figure 3B lanes 7 and 9; Figure 3C, lanes 7 and 9). Most intriguingly, when samples were pulled down with anti-EpsG antibodies and immunoblotted for EpsL the only band that was detected was the 60 kDa species (Figure 3B lanes 4 and 6; Figure 3C lanes 4 and 6). Similarly, when samples were immunoprecipitated with anti-EpsL antibodies and immunoblotted for EpsG (Figure 3A lanes 7 and 9; Figure 3D lanes 7 and 9), the only band that was detected was the 60 kDa species. These results confirm that the 60 kDa band represents a cross-linked complex between EpsG and EpsL.
Figure 3. Co-immunoprecipitation of EpsG and EpsL.
Triton X-100 cell extracts were prepared from PBAD::eps wild-type, PBAD::ΔepsG and complemented strain (Panels A and B), and PBAD::ΔepsL and complemented strain (Panels C and D) after cross-linking with 0.25 mM DSP. Cleared cell extracts were immunoprecipitated with either anti-EpsG or anti-EpsL antibodies and subjected to SDS-PAGE and immunoblotting with biotinylated anti-EpsG (Panels A and D) or biotinylated anti-EpsL (Panels B and C) antibodies. In all panels the monomer for EpsG or EpsL is indicated with an arrow and the 60 kDa EpsG-EpsL complex is denoted by an arrow head. In panels immunoblotted for EpsG, the putative EpsG dimer is labeled with an asterisk. The molecular weight markers are shown in kilo Daltons. Lane numbers are indicated.
EpsG processing is required for the interaction with EpsL
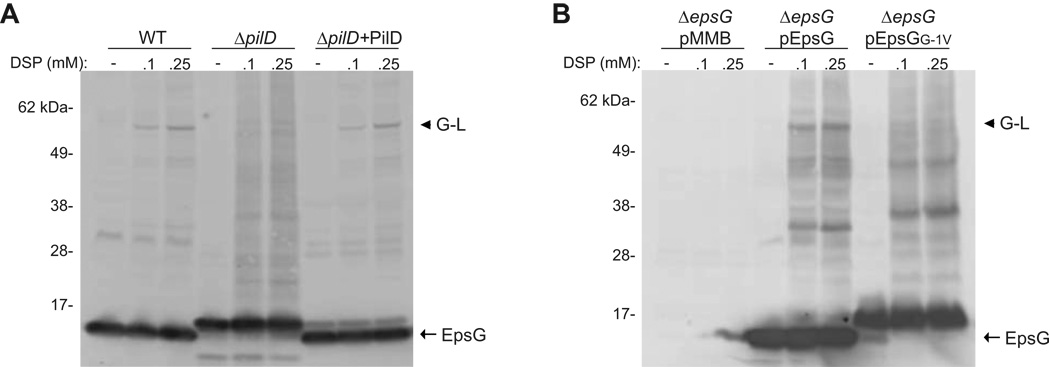
We next wanted to determine if processing of EpsG by the prepilin peptidase, PilD, is required for the interaction with EpsL. The precursors of the T2S pseudopilins and type IV pilins are known to be processed by PilD, a unique peptidase that is responsible for both cleavage and methylation of the newly formed N-terminus (Nunn & Lory, 1993, Pugsley, 1993, Strom et al., 1993). In the absence of pilD secretion through the T2S apparatus is inhibited, indicating that processing of the pseudopilins is necessary for these proteins to be functional (Fullner & Mekalanos, 1999, Marsh & Taylor, 1998). In order to address if PilD is required for EpsG and EpsL to associate we performed in vivo cross-linking using whole cells of wild-type and a pilD deficient strain of V. cholerae, followed by SDS-PAGE and immunoblotting for EpsG. The ΔpilD mutant strain did not exhibit the 60 kDa cross-linked complex observed in the wild-type strain although a very faint band slightly larger than the EpsG-EpsL complex was detected that may be indicative of unprocessed EpsG associating with EpsL (Figure 4A). However, the intensity of this band was greatly reduced in comparison to the band comprising EpsL and the mature, processed form of EpsG suggesting that the affiliation between unprocessed EpsG and EpsL was much less efficient. Furthermore, upon complementation of the ΔpilD strain the position and intensity of the EpsG-EpsL cross-linked species was restored to the levels observed in the wild-type cells.
Figure 4. Processing of EpsG is necessary for EpsG-EpsL cross-linking.
(A) Whole cells of V. cholerae C6706 wild-type, ΔpilD, and ΔpilD complemented with pACYC184 expressing pilD were cross-linked with the indicated concentrations of DSP, and immunoblotted with antibodies specific for EpsG. (B) Whole cells of PBAD::ΔepsG expressing wild-type epsG, or epsG with mutation to residue G-1 were cross-linked and analyzed as described above. The processed monomer of EpsG is represented by an arrow, and the EpsG-EpsL complex is labeled with an arrow head. Position of molecular weight markers is shown.
To further verify that processing of EpsG is required for cross-linking with EpsL, we also examined an EpsG mutant that cannot be processed by PilD. Previous studies on type IV pilins and the EpsG homolog, PulG, from K. oxytoca demonstrated that substitution of the glycine residue (referred to as G-1) at the cleavage site G-F prevented cleavage by PilD, and inhibited pilus formation and secretion, respectively (Pugsley, 1993, Strom & Lory, 1991). We substituted EpsG residue G-1 for valine and expressed the mutant gene from a plasmid in the PBAD::ΔepsG strain. Similar to the finding in K. oxytoca, EpsGG-1V was not processed (Figure 4B) and unable to support secretion of protease through the T2S apparatus (Figure 2). We next performed in vivo cross-linking with PBAD::ΔepsG expressing wild type EpsG or EpsGG-1V, and subjected samples to SDS-PAGE and immunoblotting for EpsG. As was observed for the pilD deficient strain, the EpsGG-1V mutant did not exhibit the 60 kDa cross-linked band indicative of an EpsG-EpsL interaction (Figure 4B). From these data we concluded that processing of EpsG by the prepilin peptidase is necessary for the association with EpsL.
EpsG and EpsL interact in the absence of all other T2S components
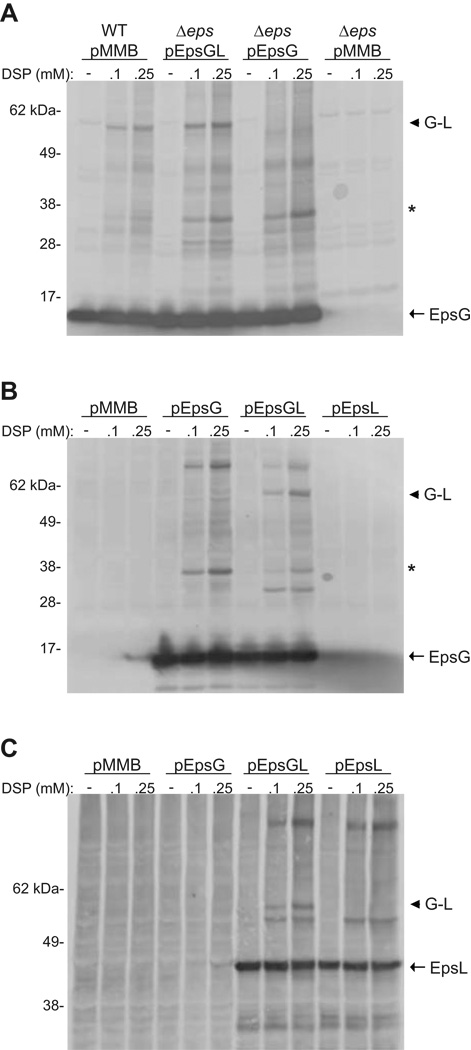
Since our data signified that processing by the prepilin peptidase is a prerequisite for EpsG to interact with EpsL, we wanted to examine whether any other components of the T2S machinery are essential for the interaction to occur. We first determined the in vivo cross-linking pattern of EpsG in strains deficient for each individual gene encoded by the eps operon and found that EpsG and EpsL still interacted, indicating that no specific component was required for their association (data not shown). To further explore the requirements necessary for EpsG and EpsL to assemble we expressed either epsG alone (pEpsG), or in conjunction with epsL (pEpsGL), in a V. cholerae strain where the entire eps operon has been deleted (Δeps) but still encodes the prepilin peptidase, PilD (Sikora et al., 2007). Whole cells were incubated with DSP, subjected to SDS-PAGE, immunoblotted for EpsG, and the cross-linking profiles were compared to that of wild-type cells. As shown in Figure 5A, EpsG in the presence of only EpsL was sufficient for the 60 kDa complex to be restored and the band migrated in accordance to the EpsG-EpsL protein species observed in the wild-type strain. Expression of epsG alone did not result in the 60 kDa cross-linked band; however, when EpsG was produced alone or with EpsL we did observe other EpsG protein species, including the intense band around 35 kDa which is presumed to be an EpsG dimer. Despite the presence of these additional EpsG specific bands, the 60 kDa band was only observed in the presence of EpsL.
Figure 5. EpsG and EpsL interact in the absence of other T2S components.
(A) Whole cells of V. cholerae TRH7000 wild-type, and TRH7000 with the entire eps operon removed (Δeps) expressing plasmid-encoded epsG, or epsG and epsL (pEpsGL) were cross-linked with the indicated concentrations of DSP and immunoblotted for EpsG. (B and C) Whole cells of E. coli MC1061 with pACYC184-pilD and pMMB67 encoding epsG, epsL, or epsG and epsL concomitantly were cross-linked with the indicated concentrations of DSP and probed with antibodies specific for EpsG (B) or EpsL (C). The monomer for EpsG (Panels A and B) and EpsL (Panel C) is indicated by an arrow. An arrow head designates the EpsG-EpsL complex, and an asterisk represents the putative EpsG dimer in Panels A and B. Positions of molecular weight markers are indicated.
Although we were capable of establishing an EpsG-EpsL interaction in the absence of other Eps components in V. cholerae, we wanted to additionally determine if any other proteins specific to V. cholerae are required for EpsG and EpsL to form a complex by performing the cross-linking experiments in E. coli. Because processing of EpsG by PilD was required for the EpsG-EpsL association, we tested E. coli expressing pilD from a plasmid as well as an additional vector containing either epsG (pEpsG), epsL (pEpsL), or both epsG and epsL (pEpsGL). Whole cells were cross-linked, and analyzed by SDS-PAGE and immunoblotting with either EpsG or EpsL antisera. As was observed in V. cholerae, the 60 kDa protein species was only detected when both EpsG and EpsL were present, suggesting that no other protein unique to V. cholerae is required for their interaction (Figure 5B). In E. coli we also saw other EpsG specific protein species that were present in the absence of EpsL, including the proposed dimer at 35 kDa. Interestingly, the 35 kDa band and another EpsG complex located at approximately 80kDa were reduced when epsL was expressed with epsG. This may imply that EpsG preferentially associates with EpsL and that the addition of EpsL titrated EpsG away from other protein complexes.
In V. cholerae we were unable to determine the cross-linking profile for EpsL due to cross-reactive bands recognized by our polyclonal EpsL antibodies, unless the cross-linking procedure was followed by immunoprecipitation and immunoblotting. However, in E. coli we were able to address whether an EpsG and EpsL interaction could be detected by immunoblotting with anti-EpsL antibodies. When comparing cells expressing EpsL either alone or in the presence of EpsG the only additional species that was detected when EpsG and EpsL were co-produced migrated to approximately 60 kDa, implying a specific EpsG-EpsL association (Figure 5C). Our findings that expression of epsG and epsL concomitantly in either V. cholerae Δeps or E. coli results in an EpsG-EpsL cross-linked complex suggests that they do not require any additional members of the T2S machinery in order to associate. Furthermore, these data establish that the cross-linked EpsG species observed at 60 kDa in wild type V. cholerae is indicative of an EpsG-EpsL complex.
Mutation of EpsG residue T112 alters the interaction with EpsL
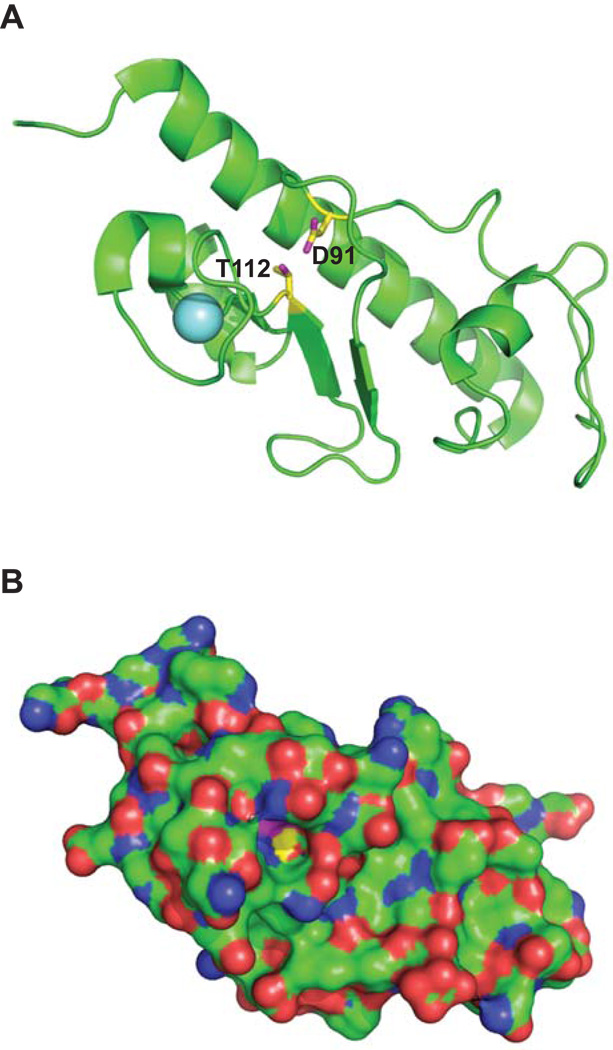
To further explain the original finding by Kagami et al. that showed that the threonine to leucine mutation isolated in XcpT, the P. aeruginosa homolog of EpsG, was suppressed by a second mutation in the EpsE homolog, XcpR, we created the equivalent residue change, EpsGT112L, as was reported for XcpT (Kagami et al., 1998). The recently solved structure for EpsG indicated that the side chain of residue T112 is within hydrogen bond distance to residue D91 and mutation of either residue should have a similar effect on EpsG (Figure 6A) (Korotkov et al., 2009); therefore, we also substituted D91 for glutamic acid. The mutant genes were expressed from a plasmid in the PBAD::ΔepsG strain and we addressed whether the mutant proteins would interfere with secretion through the T2S apparatus by testing for the presence of a secreted protease in the culture supernatant. As shown in Figure 2, neither EpsGT112L nor EpsGD91E were able to support secretion of the protease, indicating that mutation of either residue resulted in a non-functional protein. The residue changes reduced the detection of EpsG protein seen by immunoblotting, implying that the mutant proteins may be less stable (data not shown).
Figure 6. Location and interaction between residues Asp91 and Thr112 in EpsG.
V. cholerae EpsG (Korotkov et al., 2009; PDB code 3FU1) depicted with the side chains of D91 and T112 colored with yellow carbons and magenta oxygens. The light blue sphere is a bound calcium ion. (A) View approximately perpendicular to the EpsG helix axis with the backbone cartoon in green and the side chains of D91 and T112 as sticks. (B) Surface representation of the same view with the side chains of D91 and T112 with the same color code as in Panel A, the other atoms are colored green for carbons, blue for nitrogens and red for oxygens. A deep cavity is apparent where only a few atoms of D91 and T112 are visible. Figure prepared with PyMOL (DeLano, 2002).
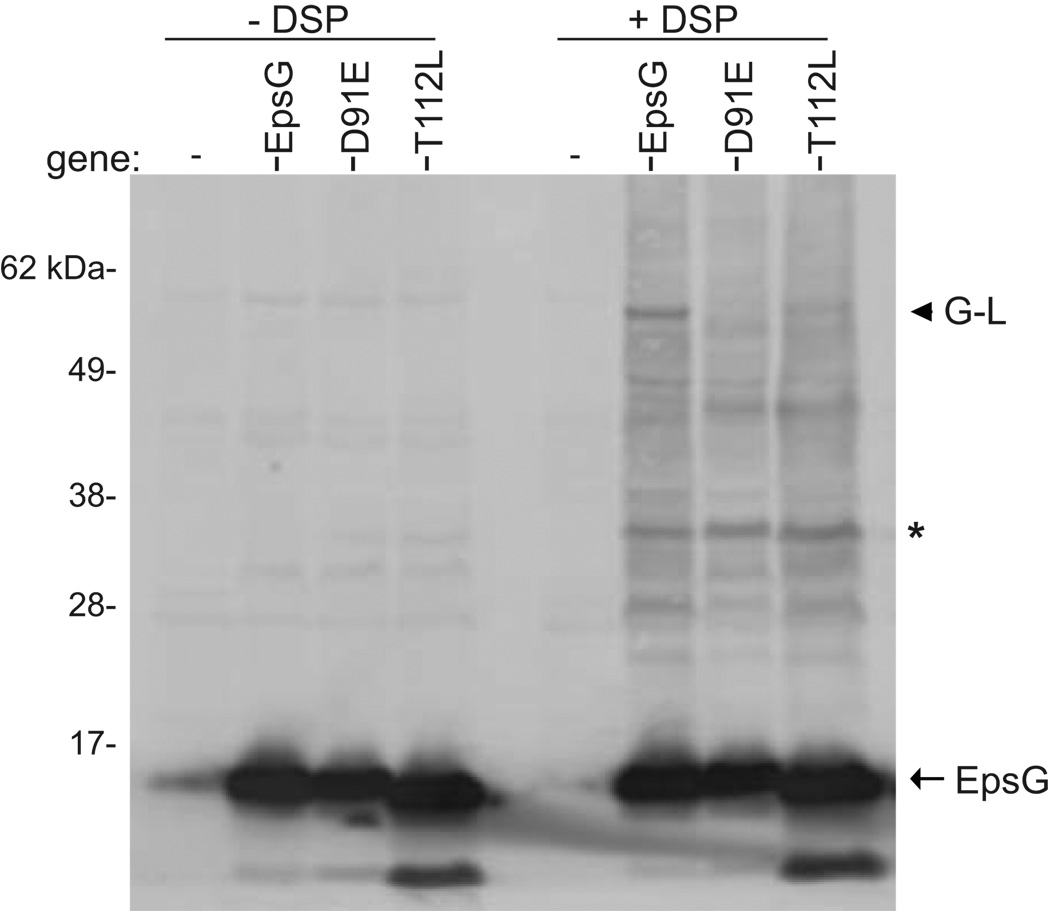
In order to examine if replacement of residues T112 or D91 affected the ability of EpsG to cross-link with EpsL we needed to produce the mutant proteins at the same level as wild type EpsG in PBAD::ΔepsG. We first determined the amount of IPTG required to produce the equivalent amount of all EpsG variants. Whole cells were then incubated with or without 0.1 mM DSP, and subjected to SDS-PAGE and immunoblotting with anti-EpsG antiserum. Neither mutant protein was able to cross-link with EpsL at the level observed in the complemented strain with both EpsGT112L and EpsGD91E exhibiting significantly reduced amounts of the EpsG-EpsL 60 kDa band (Figure 7). While wild type EpsG was found in the 60 kDa complex with EpsL the non-functional mutant proteins accumulated in a 45 kDa complex, similar to the cross-linking profile observed in an epsL deficient strain. Taken in conjunction with the protease secretion data for the point mutations, our cross-linking result implies that the changes in EpsG residues T112 or D91 prevent or alter the EpsG-EpsL interaction. However, since the side chains of T112 and D91 are barely surface exposed and occur at the bottom of a deep crevice (Figure 6B) they may not mediate a direct contact with EpsL. Therefore, replacement of these residues likely has an indirect effect on the EpsG-EpsL interaction.
Figure 7. Replacement of residues D91 or T112 alters the cross-linking of EpsG with EpsL.
Whole cells of PBAD::ΔepsG expressing wild-type epsG, or epsG with mutation to either residue D91 or T112 were incubated in the absence or presence of 0.1 mM DSP and subjected to SDS-PAGE and immunoblotting with anti-EpsG antibodies. The EpsG monomer is designated by an arrow, the EpsG-EpsL complex is indicated by an arrow head, and a predicted EpsG dimer is labeled with an asterisk. Molecular weight markers are indicated.
Discussion
In order to better understand how the T2S apparatus assembles and promotes secretion through the gram-negative cell envelope, we have continued to examine the specific protein interactions that occur within this secretion complex. The T2S machinery is likely dynamic, therefore, in this study we elected to use in vivo cross-linking in whole cells in order to capture interactions within their native environment. Using this methodology, we have established a protein interaction between the major pseudopilin, EpsG, and the inner membrane component, EpsL. Additionally, we have determined the T2S components necessary for the EpsG-EpsL interaction, thus allowing us to begin to dissect the sequential order of events that leads to the incorporation of EpsG into the T2S complex, or pseudopilus. Previous studies have shown that homologs of EpsG are cotranslationally inserted into the inner membrane using the Signal Recognition Particle and SEC pathway, and do not require any components of the T2S inner membrane platform in order to be targeted to the inner membrane (Arts et al., 2007, Francetic et al., 2007). Following insertion into the inner membrane, our results indicate that EpsG must be processed by the prepilin peptidase before it can interact with EpsL. The processing results in the removal of 7 cytoplasmically exposed N-terminal residues of EpsG (Fullner & Mekalanos, 1999, Marsh & Taylor, 1998). The processing and methylation events, both performed by PilD, have been speculated to cause a conformational change that alters the way that EpsG is presented within the inner membrane, thus enabling EpsG to interact with other components of the T2S apparatus (Nunn & Lory, 1993, Strom et al., 1993). This hypothesis is supported by our finding that processing is a prerequisite for EpsG to cross-link with EpsL, suggesting that it is the mature form of EpsG that is recognized by EpsL. Furthermore, we show that no other T2S component is required for the interaction between EpsG and EpsL. This suggests that EpsL may be the first protein that EpsG interacts with as it is incorporated into the T2S machinery, and that EpsL may be responsible for recruiting EpsG to this complex.
One implication as to the importance of the interaction between EpsG and EpsL is that EpsL may function to link the cytoplasmic ATPase, EpsE, with EpsG. The interaction between EpsE and the cytoplasmic domain of EpsL is necessary and sufficient to recruit EpsE to the inner membrane (Abendroth et al., 2005, Sandkvist et al., 1995). In addition, this interaction allows EpsE to adopt an active oligomeric state and in vitro studies have demonstrated that EpsL stimulates the ATPase activity of EpsE (Camberg et al., 2007). Recent findings from our laboratory and others have shown that members of the type II/IV secretion ATPase family to which EpsE belongs undergo dynamic changes as they bind and hydrolyze ATP (Patrick et al., submitted, Misic et al., 2010, Planet et al., 2001, Robien et al., 2003, Satyshur et al., 2007, Savvides et al., 2003, Yamagata & Tainer, 2007). Due to the presence of a flexible linker, the entire N-terminal domain (NTD) of EpsE undergoes a large movement relative to the nucleotide-binding C-terminal domain. The ensuing conformational changes do not only have substantial effects on the arrangement of subunits within the EpsE hexamer, but may also affect EpsL, which is interacting with a sub-domain of the NTD (Abendroth et al., 2005). The conversion of chemical energy to mechanical work, translated through EpsL, may in turn promote assembly of the pseudopilus. In support of this suggestion, Py et al. have shown that the interaction between the cytoplasmic domain of the EpsL and EpsE homologs, OutL and OutE, in Dickeya dadantii (previously classified as Erwinia chrysanthemi) results in a conformational change in the periplasmic domain of OutL (Py et al., 2001).
We found that the T112L and D91E substitutions in EpsG severely reduced the amount of cross-linked EpsG-EpsL complex and negatively affected the secretion of protease via the T2S machinery. Sequence comparison of EpsG with other T2S homologs showed that T112 is highly conserved and is found as either a threonine or serine, while D91 is conserved as an aspartic acid in all homologs examined (Korotkov et al., 2009). The structure of EpsG revealed that the side chain hydroxyl of T112 forms a solvent exposed hydrogen bond with one of the carboxylate oxygens from D91 at the bottom of a deep crevice (Figure 6) (Korotkov et al., 2009). The second carboxylate atom of D91 is engaged in a hydrogen bond with the main chain NH group of Asn95. The mutation T112L disrupts the hydrogen bond between T112 and D91. Due to close packing of residues at this site the substitution D91E could also affect this hydrogen bond as well as the one between D91 and Asn95 and thus alter the conformation of loop 90–96. The resultant conformational change may explain why introducing an additional atom through the conservative replacement D91E is sufficient to have a negative effect on EpsG. In summary, the T112L and D91E mutations may cause a perturbation of the native fold and thus indirectly affect the EpsG-EpsL interaction. In preliminary attempts to identify the site(s) of interaction between EpsG and EpsL we have replaced every individual lysine residue in EpsG (data not shown). Only in one of the mutants (EpsGK70A) was the EpsG-EpsL cross-linking reduced, but not abolished (data not shown). Therefore, future studies will be needed to determine the precise site of interaction between EpsG and EpsL, which could occur either between the exposed periplasmic globular domains or within the membrane spanning portion of the proteins.
Due to the structural similarities between the pseudopilins, several intriguing questions remain regarding the minor pseudopilins, EpsH-K, which are proposed to form a cap on the tip of the pseudopilus (Douzi et al., 2009, Korotkov & Hol, 2008). It has yet to be determined if the minor pseudopilins also interact with EpsL; however, if the role of EpsL is to recruit EpsG to the T2S complex then it may also serve as a recruitment factor for one or more of the structurally related minor pseudopilins. Support for this suggestion comes from a yeast two hybrid study of T2S proteins from D. dadantii that detected an interaction between the EpsJ and EpsL homologs, OutJ and OutL (Douet et al., 2004). It is possible that the minor pseudopilins also require an interaction with EpsL and energy provided by EpsE in order to become incorporated into the pseudopilus. Energy transduced by EpsL may be required to extract the N-terminal hydrophobic alpha-helix of both the major and minor pseudopilins from the inner membrane as the pseudopilus is formed in the periplasmic compartment.
A related mechanism to link the pilin subunits to the ATPase is likely to exist for the type IV pilus biogenesis machinery. Recently, the structure of P. aeruginosa PilN was found to be homologous to that of the periplasmic domain of EpsL (Sampaleanu et al., 2009). PilN was suggested to interact with the cytoplasmic protein PilM, which has a predicted actin-like fold similar to the cytoplasmic domain of EpsL, in order to form a complex that is functionally equivalent to EpsL (Ayers et al., 2009, Abendroth et al., 2004). The findings that PilM and PilN, as well as homologs for EpsF and EpsM are required to form the type IV pilus suggests that the inner membrane complex of the type IV pilus machinery may be analogous to the one formed by the T2S apparatus (Ayers et al., 2009, Carbonnelle et al., 2006). Misic et al. have speculated that the type IV pilins may either directly interact with the ATPase or that their interaction may be mediated by the inner membrane complex composed of PilN, and the EpsF and EpsM homologs (Misic et al., 2010). Based on our findings, we propose that the PilM/PilN complex interacts with the pilin subunits and links them to the ATPase to support pilus assembly. At this point it is not understood why the EpsL equivalent is split into two proteins in the type IV pilus biogenesis system in P. aeruginosa, but it may be related to the requirement for several different ATPases to support pilus assembly and disassembly that lead to twitching motility (Pelicic, 2008). However, comparable to the T2S system, non-retractile type IV pili such as the toxin co-regulated pilus (TCP) in V. cholerae may only require the function of one ATPase. The cytoplasmic ATPase TcpT of the TCP system has been shown to interact with TcpR, a bitopic inner membrane protein that has suggested homology to EpsL (Tripathi & Taylor, 2007). Similar to the relationship between EpsE and EpsL, the interaction between TcpT and TcpR is responsible for bringing the ATPase to the inner membrane (Tripathi & Taylor, 2007). Intriguingly, the first 100 amino acids of TcpT are involved in the interaction with TcpR which is in agreement to the region of EpsE that interacts with EpsL, thus, it is likely that TcpR may interact directly with the pilin to support assembly.
In this study we have furthered our understanding of the T2S system by establishing a protein interaction between the major pseudopilin EpsG and the inner membrane component EpsL. Interestingly, every condition examined that disrupted or altered the interaction between EpsG and EpsL also prevented secretion, validating the importance of their association within the T2S complex. Future work will be needed to completely understand the implications of this interaction; however, we propose that EpsL functions to link EpsG to the ATPase, providing a means by which energy is transduced between the two compartments in order for the pseudopilus to assemble.
Experimental procedures
Bacterial strains and growth conditions
All strains and plasmids used in this study are listed in Table 1. Strains were grown in Luria-Bertani (LB) broth at 37°C. V. cholerae TRH7000 and its derivatives were cultured in the presence of 100 µg/ml thymine. V. cholerae PBAD::eps strains were grown in the presence of 0.01% arabinose. Antibiotics (Sigma-Aldrich) were used at the following concentrations: ampicillin at 100 µg/ml, carbenicillin at 200 µg/ml, chlormanphenicol at 2 µg/ml, and kanamycin at 50 µg/ml. Expression of eps genes was induced with isopropanyl-β-D-thiogalactopyranoside (IPTG) at the specified concentrations.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Features | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| TRH7000 | El Tor strain, wild-type for type II secretion | Hirst et al., 1984 |
| ΔepsG | in frame replacement of epsG with CmR | Lybarger et al., 2009 |
| ΔepsL | in frame replacement of epsL with aph-3 (KmR) | Lybarger et al., 2009 |
| PBAD::eps | TRH7000 PBAD::eps | Sikora et al., 2007 |
| PBAD::ΔepsG | TRH7000 PBAD::eps with in frame replacement of epsG with CmR | This study |
| PBAD::ΔepsL | TRH7000 PBAD::eps with in frame replacement of epsL with aph-3 (KmR) | This study |
| Δeps | TRH7000 in frame replacement of epsC-N with CmR | Sikora et al., 2007 |
| C6706str2 | El Tor o1, Inaba, SmR | Thelin & Taylor, 1996 |
| ΔpilD (JM313) | C6706str with in frame deletion of pilD (CmR) | Marsh & Taylor, 1998 |
| E. coli | ||
| MC1061 | F- lac- K-12 laboratory strain | Casadaban & Cohen, 1980 |
| MM294/pRK2013 | helper strain for conjugations | Meselson & Yuan, 1968 |
| Plasmids | ||
| pMMB67 | low copy number IPTG inducible vector (ApR) | Furste et al., 1986 |
| pMMB872 | pMMB67 with BstEII sites in lacIQ and repC removed | This study |
| pMMB917 | epsG in pMMB872 | This study |
| pEpsG | epsG in pMMB67 | Lybarger et al., 2009 |
| pEpsGD91E | substitution of residue D91 to glutamic acid in pMMB917 | This study |
| pEpsGT112L | substitution of residue T112 to leucine in pEpsG | This study |
| pEpsGG-1V | substitution of residue G-1 to valine in pEpsG | This study |
| pEpsL (pMS44) | epsL in pMMB67 | Sandkvist et al., 1995 |
| pEpsGL | epsGepsL in pMMB67 | This study |
| pACYC184 | low copy number vector (CmR, TcR) | Chang & Cohen, 1978 |
| pPilD (pJM294) | pilD in pACYC184 | Marsh & Taylor, 1998 |
Construction of deletion strains
PBAD::ΔepsG was constructed by replacing chromosomal epsG with the cat gene conferring chloramphenicol resistance in PBAD::eps using the conditions previously described for creating TRH7000 ΔepsG (Lybarger et al., 2009). Simlarly, PBAD::ΔepsL was constructed in PBAD::eps by replacing chromosomal epsL with aph-3 conferring kanamycin resistance using the conditions previously described for creating TRH7000 ΔepsL (Sikora et al., 2007).
Plasmid construction and conjugation
pEpsGL was constructed by excising epsG cloned in PCR-script (Stratagene) using the restriction sites PstI and SalI. The excised epsG fragment was cloned into similarly digested pMS44 using T4 DNA Ligase (New England BioLabs) in order to concomitantly express full length EpsG upstream of full length EpsL.
T112L mutation of epsG was introduced with QuickChange II site-directed mutagenesis kit (Stratagene) using previously constructed pEpsG as a template (Lybarger et al., 2009). Primers used for the site change were 5′-ggcacgattgatgtgttcctgttaggtgcggacggtcaag-3′ and 5′-cttgaccgtccgcacctaacaggaacacatcaatcgtgcc-3′. G-1V mutation of epsG was made similarly using the primers 5′-tgcgtaaacaaacggtctttaccctgctcgaagtaatg-3′ and 5′-cattacttcgagcagggtaaagaccgtttgtttacgca-3′.
pMMB872 was created for downstream subcloning by sequential removal of the BstEII sites from pMMB67 using the mutagenesis technique described previously (Allemandou et al., 2003). The primer pairs used to remove the site from lacIQ were 5′-cagcgcgatttgctggtggcccaatgcgaccaga-3′ and 5′-gggctggacggtaactgagttcgcggcgggca-3′ (pMMBO109-1), and 5′-P-tgctccacgcccagtcgcgtaccgtcttcatgg-3′ and 5′-cgagacaggccctgcggggctgcacacgcgcc-3′ (pMMBO109-2). The primer pairs used to remove the site from repC were 5′-cttgcccgccgcgaactcagttaccgtccagcccag-3′ and pMMBO109-1 and 5′P-cgcgaccagctccggcaacgcctcgcgcac-3′ and pMMBO109-2. pMMB917 was generated by inserting epsG as a EcoRI-SalI fragment into pMMB872.
D91E was introduced with the primers 5′-aagcgtctgcctaaagaaccttggggtaacgac-3′ and 5′-gtcgttaccccaaggttctttaggcagacgctt-3′ by the same Stratagene technology described for T112L using truncated epsG encoding residues G25 through Q127 and the stop codon inserted in the vector pET28b (Novagen, Madison WI) as a template. A BstEII – KpnI fragment of the mutated gene was then excised from pET28b and used to replace the similar fragment in pMMB917. Sequences were verified at the University of Michigan DNA Sequencing Core.
All plasmid constructs were introduced into their bacterial strains by using the conjugative helper strain MM294/pRK2013 (Meselson & Yuan, 1968). Transconjugants were selected for antibiotic resistance encoded by the vector. All parental strains also received empty vectors except for C6706 ΔpilD which would not grow in the presence of vector only.
In vivo cross-linking
Overnight cultures were diluted in fresh media. The following concentrations of IPTG were used to induce the plasmid constructs equivalent to the PBAD::eps chromosomal EpsG or EpsL levels: PBAD::ΔepsG with pEpsG, pEpsGG-1V, pEpsGD91E, or pEpsGT112L at 15, 40, 40, and 20 µM, respectively, and PBAD::ΔepsL with pEpsL at 10 µM. E. coli MC1061 containing pEpsG, pEpsL, or pEpsGL were induced with 10 µM IPTG. TRH7000 Δeps containing pEpsG or pEpsGL were induced with 10 and 100 µM IPTG, respectively.
Diluted cultures were grown to an optical density at 600 nm between 1.5 and 2.0. Cells were harvested by centrifugation at 3,500 × g for 5 minutes and concentrated 8 fold by suspension in 500 µl of phosphate-buffered saline. Samples were cross-linked using dithiobis (succinimdyl propionate) (Pierce) for 1 hour at room temperature. The reactions were quenched for 10 minutes with 25 µl of 1 M Tris pH 8.0. Samples were centrifuged 8,000 × g for 5 minutes and suspended in 500 µl 50 mM Tris pH 8.0. To lyse, cells were incubated with 10 µl of 10 mg/ml lysozyme and 10 µl of 1 mg/ml DNase for 10 minutes followed by sonication at 30% amplitude for 10 seconds using one second pulses using the Vibra Cell Ultrasonic Processor (Sonics and Materials, Inc.). Samples were boiled for 5 minutes in SDS loading buffer and analyzed on NuPAGE 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen) at the following concentrations: TRH7000 wild type and MC1061 were loaded at 10 µl of OD600=2.0, all PBAD::eps strains, Δeps, and C6706 wild type and the ΔpilD complemented strain were loaded at 10 µl of OD600=1.5, TRH7000 ΔepsG and ΔepsL, and C6706 ΔpilD were loaded at 10 µl of OD600=1.0. Gels were transferred to nitrocellulose membranes in NuPAGE transfer buffer and blocked in phosphate buffered-saline containing 3% BSA. Membranes were incubated with polyclonal antisera against EpsG (1:150,000 in Tris-buffered saline with 0.1% Tween-20) or EpsL (1:40,000 in Tris buffered saline with 0.1% Tween-20), followed by horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (BioRad) diluted 1:15,000 in Tris-buffered saline containing 0.1% Tween-20. Blots were developed with ECL Plus Western blotting detection reagent (GE Healthcare) and protein was visualized using a Typhoon Trio variable mode imager system and Image Quant software.
Triton X-100 extraction and co-immunoprecipitation
Cultures were grown, cross-linked, and quenched as described above. Following quenching samples were subjected to Triton X-100 extraction as previously described (Johnson et al., 2007). 100 µl of OD600=2.0 of the Triton X-100 extracted material was immunoprecipitated with anti-EpsL or anti-EpsG antiserum bound to protein G-sepharose as previously detailed (Sandkvist et al., 1999). Antiserum labeled protein G sepharose beads bound to the cell extract were washed twice in high salt Triton X-100 buffer (Tris-buffered saline containing 500 mM NaCl, 0.01% Triton X-100, and 1M EDTA). Samples were then washed once in low salt buffer Triton X-100 buffer (Tris-buffered saline containing 150 mM NaCl, 0.01% Triton X-100, and 1M EDTA), followed by a final wash in Tris-buffered saline. 25 µl of Laemmli buffer was added to the washed beads and boiled for 10 minutes prior to centrifugation. Immunoprecipitated samples were divided equally and analyzed on NuPAGE 4–12% Bis-Tris polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked in Tris-buffered saline containing 5% milk, incubated with either biotinylated EpsG (1:5,000) or biotinylated EpsL antibodies (1:10,000) in Tris-buffered saline with 0.1% Tween-20, followed by incubation with horseradish peroxidase-conjugated streptavidin at 1:25,000 in Tris-buffered saline with 0.1% Tween-20. Blots were developed with ECL Plus Western blotting detection reagent (GE Healthcare) and protein was visualized using a Typhoon Trio variable mode imager system and Image Quant software.
Protease secretion assay
Secretion of extracellular protease was determined as previously described (Sikora et al., 2007). The amount of fluorescence was compared to a standard curve of cleaved substrate in order to determine the nanograms of protease secreted.
Acknowledgements
We thank Ron Taylor for the vcpD mutant strain JM313 and plasmid pJM294 and Konstantin Korotkov for stimulating discussions. This project was supported by the awards RO1AI049294 and RO1AI081705 (to M.S.) and RO1AI34501 (to W.G.J.H) from the National Institute of Allergy and Infectious Diseases as well as funds from MSU Center for Microbial Pathogenesis (to M.B.). M.D.G. was supported in part by National Institutes of Health training grant AI007528.
References
- Abendroth J, Bagdasarian M, Sandkvist M, Hol WG. The structure of the cytoplasmic domain of EpsL, an inner membrane component of the type II secretion system of Vibrio cholerae: an unusual member of the actin-like ATPase superfamily. Journal of molecular biology. 2004;344:619–633. doi: 10.1016/j.jmb.2004.09.062. [DOI] [PubMed] [Google Scholar]
- Abendroth J, Murphy P, Sandkvist M, Bagdasarian M, Hol WG. The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. Journal of molecular biology. 2005;348:845–855. doi: 10.1016/j.jmb.2005.02.061. [DOI] [PubMed] [Google Scholar]
- Allemandou F, Nussberger J, Brunner HR, Brakch N. Rapid Site-Directed Mutagenesis Using Two-PCR-Generated DNA Fragments Reproducing the Plasmid Template. J Biomed Biotechnol. 2003;2003:202–207. doi: 10.1155/S1110724303209141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alphonse S, Durand E, Douzi B, Waegele B, Darbon H, Filloux A, Voulhoux R, Bernard C. Structure of the Pseudomonas aeruginosa XcpT pseudopilin, a major component of the type II secretion system. J Struct Biol. 2010;169:75–80. doi: 10.1016/j.jsb.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M. Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J Bacteriol. 2007;189:2069–2076. doi: 10.1128/JB.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers M, Sampaleanu LM, Tammam S, Koo J, Harvey H, Howell PL, Burrows LL. PilM/N/O/P proteins form an inner membrane complex that affects the stability of the Pseudomonas aeruginosa type IV pilus secretin. Journal of molecular biology. 2009;394:128–142. doi: 10.1016/j.jmb.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Camberg JL, Johnson TL, Patrick M, Abendroth J, Hol WG, Sandkvist M. Synergistic stimulation of EpsE ATP hydrolysis by EpsL and acidic phospholipids. The EMBO journal. 2007;26:19–27. doi: 10.1038/sj.emboj.7601481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Sandkvist M. Molecular analysis of the Vibrio cholerae type II secretion ATPase EpsE. J Bacteriol. 2005;187:249–256. doi: 10.1128/JB.187.1.249-256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos M, Nilges M, Cisneros DA, Francetic O. Detailed structural and assembly model of the type II secretion pilus from sparse data. Proceedings of the National Academy of Sciences of the United States of America. 2010 doi: 10.1073/pnas.1001703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle E, Helaine S, Nassif X, Pelicic V. A systematic genetic analysis in Neisseria meningitidis defines the Pil proteins required for assembly, functionality, stabilization and export of type IV pili. Molecular microbiology. 2006;61:1510–1522. doi: 10.1111/j.1365-2958.2006.05341.x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ, Cohen SN. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. Journal of molecular biology. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianciotto NP. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005;13:581–588. doi: 10.1016/j.tim.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Craig L, Li J. Type IV pili: paradoxes in form and function. Curr Opin Struct Biol. 2008;18:267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific LLC; 2002. [Google Scholar]
- Douet V, Loiseau L, Barras F, Py B. Systematic analysis, by the yeast two-hybrid, of protein interaction between components of the type II secretory machinery of Erwinia chrysanthemi. Research in microbiology. 2004;155:71–75. doi: 10.1016/j.resmic.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Douzi B, Durand E, Bernard C, Alphonse S, Cambillau C, Filloux A, Tegoni M, Voulhoux R. The XcpV/GspI pseudopilin has a central role in the assembly of a quaternary complex within the T2SS pseudopilus. The Journal of biological chemistry. 2009;284:34580–34589. doi: 10.1074/jbc.M109.042366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Bernadac A, Ball G, Lazdunski A, Sturgis JN, Filloux A. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J Bacteriol. 2003;185:2749–2758. doi: 10.1128/JB.185.9.2749-2758.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E, Michel G, Voulhoux R, Kurner J, Bernadac A, Filloux A. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. The Journal of biological chemistry. 2005;280:31378–31389. doi: 10.1074/jbc.M505812200. [DOI] [PubMed] [Google Scholar]
- Filloux A. The underlying mechanisms of type II protein secretion. Biochimica et biophysica acta. 2004;1694:163–179. doi: 10.1016/j.bbamcr.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP. Signal Recognition Particle-Dependent Inner Membrane Targeting of the PulG Pseudopilin Component of a Type II Secretion System. J Bacteriol. 2007;189:1783–1793. doi: 10.1128/JB.01230-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullner KJ, Mekalanos JJ. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infection and immunity. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furste JP, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Hirst TR, Sanchez J, Kaper JB, Hardy SJ, Holmgren J. Mechanism of toxin secretion by Vibrio cholerae investigated in strains harboring plasmids that encode heat-labile enterotoxins of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:7752–7756. doi: 10.1073/pnas.81.24.7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs M, Mattick JS. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Molecular microbiology. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- Hu NT, Leu WM, Lee MS, Chen A, Chen SC, Song YL, Chen LY. XpsG, the major pseudopilin in Xanthomonas campestris pv. campestris, forms a pilus-like structure between cytoplasmic and outer membranes. The Biochemical journal. 2002;365:205–211. doi: 10.1042/BJ20020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Abendroth J, Hol WG, Sandkvist M. Type II secretion: from structure to function. FEMS microbiology letters. 2006;255:175–186. doi: 10.1111/j.1574-6968.2006.00102.x. [DOI] [PubMed] [Google Scholar]
- Johnson TL, Scott ME, Sandkvist M. Mapping critical interactive sites within the periplasmic domain of the Vibrio cholerae type II secretion protein EpsM. J Bacteriol. 2007;189:9082–9089. doi: 10.1128/JB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami Y, Ratliff M, Surber M, Martinez A, Nunn DN. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Molecular microbiology. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- Kohler R, Schafer K, Muller S, Vignon G, Diederichs K, Philippsen A, Ringler P, Pugsley AP, Engel A, Welte W. Structure and assembly of the pseudopilin PulG. Molecular microbiology. 2004;54:647–664. doi: 10.1111/j.1365-2958.2004.04307.x. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Gray MD, Kreger A, Turley S, Sandkvist M, Hol WG. Calcium is essential for the major pseudopilin in the type 2 secretion system. The Journal of biological chemistry. 2009;284:25466–25470. doi: 10.1074/jbc.C109.037655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkov KV, Hol WG. Structure of the GspK-GspI-GspJ complex from the enterotoxigenic Escherichia coli type 2 secretion system. Nat Struct Mol Biol. 2008;15:462–468. doi: 10.1038/nsmb.1426. [DOI] [PubMed] [Google Scholar]
- Kuo WW, Kuo HW, Cheng CC, Lai HL, Chen LY. Roles of the minor pseudopilins, XpsH, XpsI and XpsJ, in the formation of XpsG-containing pseudopilus in Xanthomonas campestris pv. campestris. Journal of biomedical science. 2005;12:587–599. doi: 10.1007/s11373-005-7372-3. [DOI] [PubMed] [Google Scholar]
- Lybarger SR, Johnson TL, Gray MD, Sikora AE, Sandkvist M. Docking and assembly of the type II secretion complex of Vibrio cholerae. J Bacteriol. 2009;191:3149–3161. doi: 10.1128/JB.01701-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JW, Taylor RK. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Molecular microbiology. 1998;29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Misic AM, Satyshur KA, Forest KT. P. aeruginosa PilT Structures with and without Nucleotide Reveal a Dynamic Type IV Pilus Retraction Motor. Journal of molecular biology. 2010 doi: 10.1016/j.jmb.2010.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn DN, Lory S. Cleavage, methylation, and localization of the Pseudomonas aeruginosa export proteins XcpT, -U, -V, and -W. J Bacteriol. 1993;175:4375–4382. doi: 10.1128/jb.175.14.4375-4382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelicic V. Type IV pili: e pluribus unum? Molecular microbiology. 2008;68:827–837. doi: 10.1111/j.1365-2958.2008.06197.x. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Molecular microbiology. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- Py B, Loiseau L, Barras F. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO reports. 2001;2:244–248. doi: 10.1093/embo-reports/kve042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. Journal of molecular biology. 2003;333:657–674. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- Sampaleanu LM, Bonanno JB, Ayers M, Koo J, Tammam S, Burley SK, Almo SC, Burrows LL, Howell PL. Periplasmic domains of Pseudomonas aeruginosa PilN and PilO form a stable heterodimeric complex. Journal of molecular biology. 2009;394:143–159. doi: 10.1016/j.jmb.2009.09.037. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. Biology of type II secretion. Molecular microbiology. 2001a;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- Sandkvist M. Type II secretion and pathogenesis. Infection and immunity. 2001b;69:3523–3535. doi: 10.1128/IAI.69.6.3523-3535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M, Bagdasarian M, Howard SP, DiRita VJ. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. The EMBO journal. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M, Hough LP, Bagdasarian MM, Bagdasarian M. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol. 1999;181:3129–3135. doi: 10.1128/jb.181.10.3129-3135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M, Keith JM, Bagdasarian M, Howard SP. Two regions of EpsL involved in species-specific protein-protein interactions with EpsE and EpsM of the general secretion pathway in Vibrio cholerae. J Bacteriol. 2000;182:742–748. doi: 10.1128/jb.182.3.742-748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandkvist M, Michel LO, Hough LP, Morales VM, Bagdasarian M, Koomey M, DiRita VJ, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyshur KA, Worzalla GA, Meyer LS, Heiniger EK, Aukema KG, Misic AM, Forest KT. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. The EMBO journal. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora AE, Lybarger SR, Sandkvist M. Compromised outer membrane integrity in Vibrio cholerae Type II secretion mutants. J Bacteriol. 2007;189:8484–8495. doi: 10.1128/JB.00583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom MS, Nunn DN, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelin KH, Taylor RK. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infection and immunity. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SA, Taylor RK. Membrane association and multimerization of TcpT, the cognate ATPase ortholog of the Vibrio cholerae toxin-coregulated-pilus biogenesis apparatus. J Bacteriol. 2007;189:4401–4409. doi: 10.1128/JB.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignon G, Kohler R, Larquet E, Giroux S, Prevost MC, Roux P, Pugsley AP. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol. 2003;185:3416–3428. doi: 10.1128/JB.185.11.3416-3428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata A, Tainer JA. Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. The EMBO journal. 2007;26:878–890. doi: 10.1038/sj.emboj.7601544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez ME, Korotkov KV, Abendroth J, Hol WG. Structure of the minor pseudopilin EpsH from the Type 2 secretion system of Vibrio cholerae. Journal of molecular biology. 2008;377:91–103. doi: 10.1016/j.jmb.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]