Abstract
Unlike most organ systems, which have evolved to maintain homeostasis, the brain has been selected to sense and adapt to environmental stimuli by constantly altering interactions in a gene network that functions within a larger neural network. This unique feature of the central nervous system provides a remarkable plasticity of behavior, but also makes experimental investigations challenging. Each experimental intervention ramifies through both gene and neural networks, resulting in unpredicted and sometimes confusing phenotypic adaptations. Experimental dissection of mechanisms underlying behavioral plasticity ultimately must accomplish an integration across many levels of biological organization, including genetic pathways acting within individual neurons, neural network interactions which feed back to gene function, and phenotypic observations at the behavioral level. This dissection will be more easily accomplished for model systems such as Drosophila, which, compared with mammals, have relatively simple and manipulable nervous systems and genomes. The evolutionary conservation of behavioral phenotype and the underlying gene function ensures that much of what we learn in such model systems will be relevant to human cognition. In this essay, we have not attempted to review the entire Drosophila memory field. Instead, we have tried to discuss particular findings that provide some level of intellectual synthesis across three levels of biological organization: behavior, neural circuitry and biochemical pathways. We have attempted to use this integrative approach to evaluate distinct mechanistic hypotheses, and to propose critical experiments that will advance this field.
Introduction
Like embryonic development, complex behaviors are shaped by selective pressure and are remarkably conserved across the animal phyla. Such a genetic perspective provides the conceptual motivation to use ‘simple’ model systems initially to discover specific genes involved in a given behavioral phenomenon and then to manipulate each gene’s expression or function to unravel the relevant molecular and neuronal mechanism(s). This reductionist goal of understanding behavioral traits, however, couldn’t be more difficult. The brain is a unique organ, which has evolved to be plastic. Other organs are homeostatic machines, designed to maintain body functions within relatively narrow limits as the organism is exposed to varying environments. The brain, in contrast, has evolved to perceive the outside world and to change its structure and function in response to specific experiences. Brain plasticity invokes ensembles of neuronal circuits and cellular mechanisms to register a new experience and to change an animal’s behavioral response(s) adaptively. The unit functions of individual gene products ramify through intricate gene networks and complex neural networks that continually feed back to each other in response to environmental stimuli or experimental manipulation [1]. The nature of the organ has therefore provided us with an amazing capacity for behavioral plasticity but at the same time has hampered our attempts to understand our own incredible behavioral complexity.
Herein lies the value of Drosophila as a model system for the study of learning and memory as behavioral manifestations of brain plasticity. The genetic tools developed for Drosophila provide a powerful means to establish causal links from genes to neural networks to behavior. Genetic analyses in flies has enabled unbiased screens for gene mutations that affect behavior. Initial ‘hits’ have led to subsequent molecular-genetic and reverse-genetic experiments to identify the gene sequence, establish its spatiotemporal pattern of expression and suggest underlying biochemical pathways and neurocircuitry. More sophisticated genetic tools have then permitted functional manipulations of sets of phenotypically related genes to distinguish their respective roles in the behavioral biology (dissection of learning/memory), systems biology (dissection of neural circuitry) and cell biology (dissection of biochemical pathways) of brain plasticity.
We do not intend to review all the genes thought to be involved in Drosophila learning and memory (for reviews, see [2–4]). Rather, we want to discuss particular sets of genes and tools that have provided intellectual synthesis across various levels of biological organization. We consider the field to be at a critical juncture in the endeavor. Integrative data now exist to support conceptual synthesis of findings from behavioral, anatomical, and biochemical dissection of memory. From the available data, two disparate hypotheses have emerged about how gene and neural networks subserve memory in flies. Our intent is to inform the reader of these hypotheses, and ultimately to suggest critical experiments that will distinguish between them.
Genetic Dissection of Memory
Drosophila has been shown to be capable of an array of learning tasks. Among these, the most robust is a Pavlovian assay in which the animals learn to associate a conditioned stimulus (CS, odors) with an unconditioned stimulus (US, footshock). Consequently, the field has focused a great deal of research effort on understanding this particular brand of behavioral plasticity. In this review, we limit our discussions to this task and to a related appetitive Pavlovian task in which a sugar reward is substituted for the shock punishment [5,6]. Many of the extant learning/memory mutants originally identified with the olfactory task also perform poorly in other behavioral tasks. Hence, we expect that some of the underlying biochemical mechanisms are likely to apply in the neural circuits sub-serving those other learned behaviors as well.
Olfactory learning in Drosophila shows many of the behavioral properties generally described for Pavlovian learning in other animals, including acquisition, extinction, CS/US saliency, order dependence, temporal specificity, conditioned excitation, conditioned inhibition and CS/US pre-exposure effects ([7–10]; M. Del Vecchio and T.T., unpublished data; for review, see [11]). The same is true for memory formation. Early experiments demonstrated an anesthesia-sensitive phase of memory formation (ASM) that was followed by an anesthesia-resistant phase (ARM) lasting up to one day after a single training session [5,12–16]. Massed training (10 sessions administered one immediately after the other) produces even stronger memory retention, lasting for about three days, but such memory is not sensitive to the protein synthesis inhibitor cycloheximide. In contrast, spaced training (10 sessions with a 15 minute rest interval between each) yields a protein-synthesis-dependent memory lasting at least one week [16]. In short, repetition produces better memory, and spaced repetition is best. Together, these experimental manipulations of normal fruit flies suggested that an early, labile memory is ‘consolidated’ over time into a long-lasting, stable memory — as in other animals, including humans.
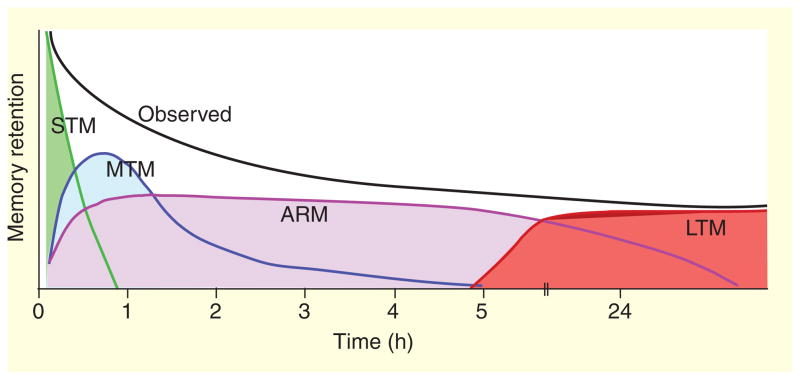
Study of mutant and transgenic flies has genetically dissected olfactory memory formation further into four distinct phases: short-term memory (STM), middle-term memory (MTM), anesthesia-resistant memory (ARM) and long-term memory (LTM) [16]. This model of multiple memory phases (Figure 1) originated as an inference from the literature [15]. In Aplysia, STM of sensitization appeared to correlate with training-induced elevation of cyclic AMP (cAMP) in neurons sub-serving the behavioral response (reviewed in [17]) and in flies the dunce and rutabaga mutants identified cAMP components in STM [8,18–24]. Thus, STM in flies was assumed to correspond to high learning levels immediately after training and to decay away within 60 min, while ARM was shown to appear slowly, reaching asymptotic levels within two hours after training [13,14,16]. These two memory phases, however, did not appear to explain the observed memory retention after one training session. MTM was postulated to exist, based on the assumption that STM, MTM and ARM act additively to produce the observed memory retention curve.
Figure 1. Dissection of memory phases.
At the behavioral level, the observed decay of memory appears relatively seamless (black). Experimental disruptions in numerous animal species including humans, however, reveal temporally, mechanistically and anatomically distinct phases underlying memory retention. In Drosophila, at least four mechanistically distinct phases have been described. These are short-term memory (STM; green), middle-term memory (MTM; blue) anesthesia-resistant memory (ARM; purple) and long-term memory (LTM; red).
Key evidence for the existence of MTM came from experiments on amnesiac mutants [15,25]. The first clue came simply from comparing the retention curves of normal flies and amnesiac mutants [8]. amnesiac mutant flies show near-normal memory retention immediately after a single training session and again around seven hours later. In between these time points, memory retention in the mutants is appreciably lower than normal. The second clue came from ‘reversal retention’ experiments. Earlier experiments had revealed that normal flies were capable of ‘reversal learning’ [8]. In this paradigm, flies first learned that octanol was paired with shock and methylcyclohexanol was not. In a subsequent training experience, the animals then had to relearn the opposite relationship, i.e., that methylcyclohexanol now was associated with shock and octanol was not. This second training session can thus be used as a ‘disruptor’ of the first odor–shock association. When this reversal is applied at different time points after training, a reversal learning sensitive window is revealed. This reversal-sensitive phase occurs after STM and before ARM, implying that MTM is preferentially disrupted. Importantly, the reversal retention curves of normal flies and amnesiac mutants were indistinguishable, suggesting that the amnesiac mutation and reversal learning each specifically disrupt MTM.
Independent evidence from experiments on animals with a temperature-sensitive mutation in DC0 (which encodes a catalytic subunit of cAMP-dependent protein kinase, PKA) also suggests the existence of a genetically distinct MTM [26]. An earlier study reported that constitutive mutants of DC0 had learning defects [27], as do the temperature-sensitive DC0ts mutants. A shift from permissive to restrictive temperature shortly before behavioral experiments, however, further disrupted memory retention in DC0ts mutants, and the temperature-shift-specific effect was indistinguishable from the amnesiac memory curve. Together, these data suggest that amnesiac (which has some similarity to pituitary adenylyl cyclase activating peptide, PACAP) and DC0 participate in the formation of MTM in normal flies. Early memory can therefore be genetically dissected into functionally distinct STM and MTM phases.
In chicks and rats, pharmacological treatments distinguished STM, MTM and LTM phases, with LTM appearing to be both anesthesia resistant and protein synthesis dependent (see [28] for review). Similar treatments in flies began to suggest that ARM and LTM are mechanistically separate entities. Inhibition of protein synthesis has no effect on ARM under conditions that block LTM [16]. Massed repetition of training produces more ARM but does not induce LTM, while the latter is uniquely induced by the same number of repetitions delivered in a spaced fashion [16]. Genetic dissection of memory formation in Drosophila clearly establishes that ARM and LTM are distinct. Disruptions of the transcription factors dCREB2, Adf1, or Notch block LTM without affecting ARM [29–32]. Conversely, radish mutants are deficient in ARM, but LTM remains intact [16,29]. These findings indicate that long-lasting memory can be genetically dissected into functionally distinct ARM and LTM phases, which co-exist in normal flies.
Genetic Dissection of Neuronal Circuitry
As in higher organisms, the central nervous system of insects is composed of highly specialized sets of neuronal structures. One of the most obvious structures in the insect brain is the mushroom body (MB). This complex neuronal structure was the first brain region shown to play a role in insect olfactory learning [33–35] (reviewed in [4,36,37]), and much of the field has since focused on MB structure and function, as well as on a single biochemical pathway believed to act within MBs (see section on dissection of biochemical pathways). There now exists a wealth of convergent data that demonstrate a critical role for cAMP signaling within the MB in at least early memory of olfactory associations [4,38–41].
The historical focus on the MB was in part due to the ease with which this structure is visualized, making its study simpler than other less organized neuropilar structures [42]. For this reason, tools to drive tissue-specific expression of transgenes have been well developed for the study of MB biochemistry and function. We will place these MB studies into the context of growing evidence for a more complex neural circuitry and gene network that underlie olfactory memory formation in this system.
The Anatomy of Mushroom Bodies
The intricate and beautiful architecture of the MB was first studied in detail in large insects [43]. In adult Drosophila, MBs consist of approximately 2,500 neurons per brain hemisphere. The intrinsic Kenyon cells are organized in a very distinct architecture. Their cell bodies are located in the dorsal posterior of the brain surrounding their main dendritic projections to a neuropil field called the calyx. Kenyon cell axons form a bundle called the peduncle, which projects anteriorly and ventrally (reviewed in [44]; Figure 2A). This axon bundle then bifurcates to form two major branches, one of which projects horizontally and the other vertically. The horizontal branch is further subdivided into three lobes — the β, β′ and γ lobes. The vertical branch consists of α and α′ lobes. MB structure is composed of at least three Kenyon cell types: one that projects its axons only to the α and β lobes, a second type that projects its axons to α′ and β′ lobes and a third that projects to the γ-lobe only [45]. This subdivision of axon projections is not merely an arbitrary descriptive partition, but also reflects developmental [45] and perhaps functional distinctions [39,46,47].
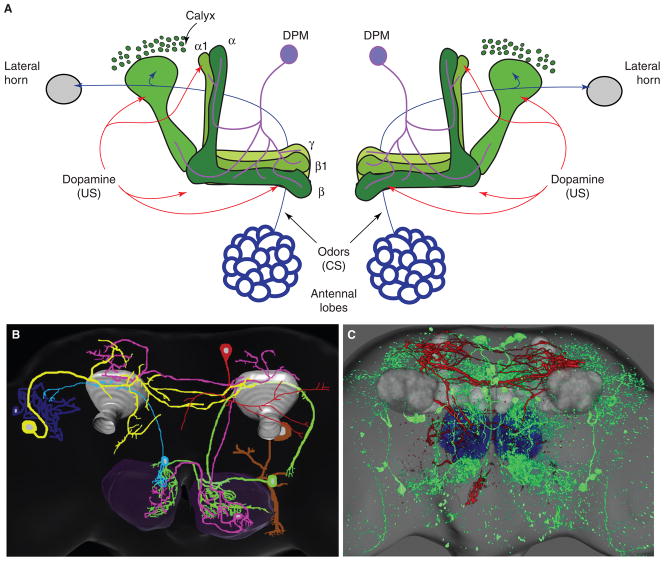
Figure 2. Dissection of neural circuits.
(A) Anterior view of the neural circuitry involved in olfactory associative memory. The mushroom body (MB) is believed to be a site where CS (odors) and US (electric shock) are associated. The primary olfactory processing center is the antennal lobe (AL; blue). The CS (odor) is conveyed out of the antennal lobe to the lateral horn (LH; gray) and MB (green) by several different projection neuron tracts, the mACT, oACT and iACT (blue arrows). The US is believed to reach the MB via dopaminergic inputs to calyx and lobes (red arrows). Neuromodulation by dorsal paired medial neurons (DPM; purple) is required after training, for memory consolidation. The calyx contains the dendritic field of the MB. MB axon terminals are contained in the lobes. MBs consist of three types of Kenyon cells: α, β neurons, whose axonal projections comprise the α and β lobes (dark green), α′ β′ neurons, whose projections enter the α′ β′ lobes (green), and γ neurons whose projections form the γ lobes (light green). (B) The radish circuit. Schematic showing several neuron types labeled by the radish-C133 Gal4 driver [59]. Functional inhibition of the radish-reporter neurons disrupts memory, indicating that neurons outside of MB play a role. DAL (yellow) and DPM2 (red) neurons each send fibers into the dorsal protocerebrum. APSP (pink) and APN (green) neurons send fibers from the AL (dark gray) to the MB calyx (grey) as well as to uncharacterized parts of the dorsal protocerebrum. (C) Posterior/dorsal view of additional non-MB neurons that may participate in memory formation. mampas and murashka were identified in a behavioral screen for mutants with defective one-day memory after spaced training [78]. mampus Gal4-driven GFP labels a few neurons (red) whose fibers project from AL (blue) to MB calyx (grey) as well as to uncharacterized portions of the dorsal protocerebrum. This ‘mampus neuron’ appears similar to the APSP neuron identified with the C133 Gal4 driver (see (B)). murashka Gal4-driven GFP (green) labels a series of neurons, some of which project fibers into the calyx.
MBs Are a Site of CS–US Convergence
Consistent with their role in olfactory associative learning, MB Kenyon cells appear to receive input from several different sensory modalities, including olfaction, electric shock and taste (for example, sugar) [6,48,49]. In Drosophila, the most obvious neuronal inputs to MBs are from the primary olfactory system. The organization of the insect olfactory system is remarkably similar to that of mammals [50,51]. As in mammals, the olfactory receptor neurons each primarily express one receptor type [52,53]. These sensory neurons project axons to the antennal lobes, which appear homologous in structure and function to the olfactory bulb in vertebrates. Like the olfactory bulb, antennal lobes consist of discrete structures called glomeruli, each of which generally receives inputs from neurons expressing a single receptor type. Glomeruli are approximately spherical structures that are composed of many synapses between (i) sensory neurons, (ii) local interneurons, which are mostly GABAergic, and (iii) acetylcholinergic projection neurons that send axons outside of the antennal lobes. Like mitral cells in the vertebrate olfactory bulb, dendrites of projection neurons are generally restricted to a single glomerulus.
In insects, there are several types of projection neuron that carry information out of the antennal lobes. Medial and outer antenno-cerebral tracts (mACT and oACT, respectively) project to an ill-defined region of the brain called the lateral horn. Another set of projection neurons carry information along the inner ACT (iACT) directly to the MB, as well as to the lateral horn [43,54,55]. In Drosophila, all of these major projection neurons appear to be cholinergic [56,57]. Besides the cholinergic projection neurons from the olfactory system, there also are GABAergic neurons that synapse onto the Kenyon cell dendrites [58]. Several additional neuron types that are uncharacterized in terms of neurotransmitter type or function also appear to project from the antennal lobes to additional regions of the dorsal protocerebrum, including MBs [59].
In addition to cholinergic inputs from the antennal lobes, MBs also appear to receive dopaminergic and octopaminergic inputs both to the calyx and to the lobes (dendrites and axons). The dopaminergic neurons appear to convey the electric shock US [6]. In contrast, the octopaminergic neurons appear to mediate the sugar US for appetitive association of odors [6].
The early descriptive studies of MB anatomy indicated that MB Kenyon cells could be a site of convergence of US and CS stimuli for olfactory learning. This observation was consistent with the finding that dunce and rutabaga, which were the first memory mutants to be molecularly identified (see section of dissection of biochemical pathways), each encode proteins that are preferentially, but not exclusively, expressed in MBs [12,20,24,60–63]. Since then, several additional genes with roles in olfactory memory have been identified with preferential expression in the MB Kenyon cells [27,60,61,64–67]. In fact, a screen for genes whose expression levels are somewhat elevated in MBs led to identification of a few whose functions are required for olfactory memory [66–68].
Subsequent interventionist manipulations of MB function confirmed a role for this neural structure in olfactory associative learning. Ablation of MB structure during development, for example, results in animals that are able to sense and respond to the stimuli, but are unable to form behavioral associations [34]. More recently, the Gal4 expression system has permitted manipulation of individual gene function with some specificity in MB. This bi-partite expression system relies on a panel of ‘driver lines’, each of which expresses the yeast Gal4 transcriptional activator in specific neuronal sub-types. These Gal4 ‘driver lines’ then are crossed with flies containing a Gal4-responsive UAS-promoter-driven transgene [69]. There are now numerous cases where manipulation of gene function in MBs has been shown to be capable of perturbing performance in the Pavlovian olfactory learning assay [10,38,39,70,71].
rutabaga Function Is Required in MB for Early Memory
One of the clearest cases where relevant gene function has been mapped to MBs is that of rutabaga. The STM defect of rutabaga mutants can be rescued completely by driving expression in MBs of a rutabaga+ cDNA transgene [39,72]. This initial finding recently has been extended to the demonstration of a physiologically relevant role for rutabaga protein in adult MB neurons by combined spatial and temporal control of rutabaga expression [41,71]. This spatiotemporal control was accomplished by use of two independent approaches: the so-called TARGET system, which relies on a temperature-sensitive Gal80 repressor to control the temporal activation of a spatially restricted Gal4 driver [40], and the GeneSwitch method, which relies on a Gal4 driver that responds to progesterone rather than temperature [41]. This modified Gal4 driver activates UAS-driven transgenes when the drug RU486 is administered.
These studies also hint at a functional subdivision of MB lobes because they suggest that the γ lobe in particular subserves STM [39]. This conclusion is based on a detailed analysis of rutabaga rescue using eight different MB-expressing Gal4 drivers that yield overlapping expression in different parts of the MB. Six of these MB lines yield some degree of rutabaga rescue and also show some degree of expression in γ lobes. In contrast, two MB drivers that do not rescue STM of rutabaga mutants lack γ lobe expression. There are two caveats of interpretation, however, which limit the strength of this conclusion. First, the two Gal4 lines for which γ lobe expression and rutabaga rescue are absent each show rather weak expression in the other lobes. Thus the failure to rescue with these two lines might result from insufficient levels of rutabaga+ expression. Second, the two Gal4 lines whose expression appears restricted to γ lobes actually show only a partial rescue of the rutabaga memory defect. In contrast, full rescue is seen only with the Gal4 lines, whose expression includes all the lobes of the MB. This raises the possibility that the other MB lobes play an additional role.
These limitations aside, this elegant paper succeeds in presenting a distinct hypothesis: that γ lobes subserve STM. A complementary hypothesis is presented by a study of α-lobe absent (ala) mutants, which appear to have normal γ lobes but abnormal α/α′ and β/β′ lobes. These mutants have normal STM, but defective LTM [46,47]. The authors of this study thus hypothesize that LTM is stored in α/α′ lobes. Though intriguing, we consider this conclusion to be premature for four reasons.
First, the anatomical defects in ala mutants are extremely variable, ranging from nearly normal MB structure, to a complete absence of β/β′ or α/α′ lobes. In this mutant strain, 36% of flies lack β/β′ lobes, while only 4.5% lack α/α′ lobes. To correlate memory deficits with anatomical defects, the authors imaged brains after identifying flies that responded to the CS correctly (i.e., by avoidance) or incorrectly. Performance indices were then calculated separately for individuals with different classes of MB defect. Only the flies classified as deficient in the α/α′ lobes scored poorly for LTM. Because such a small percentage of animals fell into the α/α′ lobes missing class, however, the total number that were tested for LTM was only 53 flies. In this population-based assay, a single performance index normally relies on testing two groups of 100 flies. In most studies in the literature, a minimum sample size of n = 6 experiments (roughly 1,200 animals per data point) are performed. Thus the statistical rigor of this finding is uncertain.
Second, the distinctions between phenotypic classes are oversimplified. Animals that lack α/α′ lobes, for instance, also exhibit midline fusions of the β/β′ lobes, suggesting that development of these structures is interdependent. This is not surprising given the fact that α and β lobes are made up of different branches originating from the same group of neurons. This is also the case for α′ and β′ lobes, which derive from a bifurcation of α′/β′ neurons. The developmental defect underlying the α/α′ lobe absent class of animals thus could result either from a loss of α/α′ axon branches, or from lack of bifurcation of α/α′ from β/β′ during development. Either way, classification of the underlying defect as α/α′ lobe specific is not accurate.
Third, even if the interpretation that α/α′ lobes play a role in LTM is correct, this relationship might be indirect. Developmental defects in α/α′ lobes, for instance, not only would disrupt storage within MB, but also would disrupt output from these lobes to other anatomical regions of the brain.
Finally, a general word of caution is required when using developmental mutants to assess circuit requirements for memory formation. In many cases, observation of structural defects at the gross anatomical level (for example, the presence or absence of a MB lobe) may not provide the level of resolution necessary to understand the anatomical basis of a functional defect. In the case of the ala mutant, for instance, there may be additional wiring defects throughout the brain that are not visible at the gross structural level. This possibility makes forging functional links between circuitry and behavior tenuous at best.
In the case of LTM, which requires gene expression for its formation, identifying the anatomical locus of activity-induced transcription would shed light on the site of plasticity. More generally, linking anatomical circuits with memory phases will be aided by investigation of the spatial requirements for expression of relevant genes (as was done for rutabaga and STM). For LTM, this has only been investigated for the Notch gene [31,32] and the data are consistent with a requirement for Notch function in MBs because expression of a Notch-inhibiting RNAi transgene in MBs disrupts LTM [32]. It is worth mentioning, however, that only one Gal4 driver was used in this study and it is not entirely specific for MB Kenyon cells. The use of additional MB Gal4 drivers was not possible because they all killed the flies or caused other phenotypes when used to express the Notch RNAi transgene (Steven de Belle, personal communication). In addition, the requirement of Notch function in MBs supports the hypothesis that MBs are part of the LTM circuit, but does not directly address the question of where the relevant gene transcription occurs.
Extra MB circuitry
Focus on MBs, while fruitful, may have resulted in a neglect of other relevant neural circuitry — a classic example of searching for lost keys ‘under the street lamp’. Behavioral screens for memory mutants, however, are anatomically unbiased, thereby providing the opportunity to find keys in the dark. Consequently, this approach has broadened our understanding of both the neural circuitry and the genetic networks involved. Two notable examples bear witness to this perspective.
Expression studies of the amnesiac memory mutant [73–75] provided one of the first hints that the circuitry underlying olfactory memory might include neurons outside of the MBs. amnesiac expression identified dorsal paired medial (DPM) neurons, which are extrinsic to the MBs but which nevertheless send projections to the MB lobes [76]. These large neurons coat the lobes and peduncle with synapses that have been suggested to co-release amnesiac peptide and acetylcholine [77], although this has not been demonstrated. Transgenic expression of amnesiac, a putative neuropeptide (see section on dissection of biochemical pathways), in these neurons rescues the amnesiac memory phenotype, establishing a link between DPM cell function and amnesiac-dependent memory.
As was the case for amnesiac, the radish gene has also broadened our view of the anatomical underpinnings of olfactory memory. A Gal4 driver (C133) inserted in the radish gene (but see section on dissection of biochemical pathways) reveals a very complex pattern of expression in a number of different neuron types, but does not label Kenyon cells [59]. Detailed analysis of individual neurons comprising the C133 pattern has documented numerous novel cell types. Though MB Kenyon cells are not among these, several neurons appear to project to MBs, antennal lobes, and lateral horn (Figure 2B). Although expression patterns of Gal4 drivers do not always correspond to expression of the nearby gene, C133-driven expression of radish+ is sufficient to rescue the radish memory defect. Thus at least a subset of these neurons is involved in radish-dependent ARM.
Both amnesiac and radish were identified in a behavioral screen for memory mutants and have begun to extend the circuitry of olfactory memory beyond MBs. A large-scale forward mutagenesis has recently identified nearly 60 new mutants with defects in one-day memory after spaced training [78]. Significantly, many of these mutations were generated using Gal4 driver transposons. Reporter expression patterns have revealed a wealth of new information. A number of these memory mutant Gal4 drivers label MB Kenyon cells, which is not surprising given past anatomical screens for memory mutants [68] and the prominent role that MBs play in memory processing. More interestingly, however, several of these Gal4 drivers do not yield expression in MBs at all, but instead label various neurons dispersed throughout the central brain. Some of these newly identified neurons send fibers into MBs or other potentially important brain regions [78] (Figure 2C). For instance, some appear similar to neurons labeled by the radish C133 Gal4 driver (A.S. Chiang, J.D. and T.T., unpublished observations). Others appear to interconnect MBs with potentially relevant neuropil structures such as the lateral horn, which may participate in olfactory processing [54,55,79], or the central complex, which is believed to play a critical role in motor output programs [80,81]. Functional manipulation of these new memory genes, and of the neurons in which they are expressed, promises to expand our knowledge of the olfactory memory circuit.
Neural Function
Gene discovery, and reverse genetic manipulations of gene function have established a role in memory for MB Kenyon cells, as well as for some neurons extrinsic to MBs. The behavioral dissection of memory formation in many studies, however, reveals functionally distinct temporal phases of memory processing. Do they all reside in MBs, or do different memory phases involve additional circuitry? Chronic disruptions of genes (mutants) or cells (lesions) cannot address this question because the temporal requirements for neuronal functions are missed. While transcription-based methods for gene induction such as TARGET [40] and GeneSwitch [41] provide some degree of temporal and spatial control of gene modulation, the kinetics of these methods are generally orders of magnitude slower than neural activity. Thus, these strategies are not ideal to dissect the temporal requirements of individual memory phases. A more rapid and reversible approach is to use temperature-sensitive mutants [78].
Unlike transcription-based approaches, the effects of temperature-sensitive (TS) mutants usually derive from amino acid substitutions that result in temperature-dependent conformational changes in protein structure and function, with functional disruptions ensuing rapidly. The Shibire-TS approach takes advantage of this mechanism to connect neural activity with memory-induced behavioral responses. This approach, which is currently unique to Drosophila, uses a dominant-negative TS mutation in Shibire, the fly dynamin homolog. Dynamin is required for vesicle endocytosis, which is rate limiting for recycling of neurotransmitter vesicles [82]. The Shibire-TS system works by first overexpressing the mutant Shibire-TS transgene in a spatially restricted pattern using various Gal4 drivers [83]. At permissive temperature, dynamin function and neurotransmission are relatively normal. When raised to the restrictive temperature, however, defective dynamin function blocks vesicle-recycling-dependent neurotransmission within a minute. This disruption of neural activity is entirely reversible, again resuming within minutes of being returned to the permissive temperature [70,82,83]). When this transgene is expressed in photoreceptors, for example, flies are rapidly rendered blind at restrictive temperature, but rapidly recover vision upon return to permissive temperature [83]. Similarly, expression of Shibire-TS throughout much of the central nervous system yields reversible control of locomotion [70].
A potential caveat to this approach is that dynamin also is required for other cell functions that use vesicle endocytosis, such as receptor trafficking [82]. Nonetheless, the effect on neurotransmission is clear, and the approach has been used with some success to investigate the temporal requirements for MB neurons [10,70,71], DPM cells [76,77], radish-C133 expressing neurons [59] and dopaminergic neurons, which are believed to convey the electric shock US [6].
Expression of Shibire-TS in MBs was used to dissect requirements for MB neurotransmission during acquisition, storage or retrieval of olfactory memory. These experiments demonstrated that blocking dynamin function during acquisition, when the odor and shock are associated, has no effect on memory formation, so long as dynamin function is restored prior to memory retrieval. In contrast, memory performance was completely disrupted when MB output was blocked during retrieval. This same result was obtained for memory measured 5, 30, 60, or 180 min after training in the aversive paradigm [10,70,71], and also has been observed with the appetitive assay [6].
The observation that synaptic transmission in MBs is not required during acquisition suggests a model [70,71,84] in which the initial CS-US association requires inputs to MB Kenyon cells, but does not require synaptic transmitter release from MB neurons themselves. In contrast, memory retrieval requires MB output, presumably to drive the behavioral responses of the animal. This model also predicts that, in contrast to MB Kenyon cells, neurotransmission in the cells that convey the US inputs would be required only during acquisition, and not during retrieval, because the US stimulus is only presented to the animals during training. In the case where electric shock is used as the US, dopaminergic inputs to MBs appear to satisfy this prediction [6]. In contrast, dopamine does not appear to mediate the US in the case of sugar reward learning. Instead, this appetitive US appears to require octopaminergic inputs to MBs. Together, these functional manipulations of the circuit suggest that the acquisition (initial association) occurs at an anatomical site that is on or upstream of MB. The available data do not address the question of whether this plasticity might be post-synaptic (i.e. in calyx), pre-synaptic (i.e. in lobes) or both [84].
These findings also do not exclude the possibility that additional CS–US associations occurring upstream of MB might also contribute. Recently, a form of plasticity was in fact observed in a set of projection neurons. Yu et al. [85] used the fluorescent indicator synapto-pHluorin to image neurotransmitter release and observed an increase in synaptic release from projection neurons at a specific glomerulus after odor was paired with shock. In contrast, application of either stimulus alone did not induce this effect, consistent with the idea that the underlying mechanism may be associative. Based on these findings, the authors of this study propose that plasticity in projection neurons may play a role in learning. While this idea remains an intriguing possibility, the behavioral relevance of the observed plasticity still needs to be established. Given the role of dopamine in mediating the US, it would be important to determine whether dopaminergic fibers innervate the antennal lobes. More direct behavioral evidence of an associative role for antennal lobe neurons comes from localized cooling of antennal lobes after an appetitive conditioning assay in the honeybee [35]. In this case, the octopaminergic VUM neuron has been shown to convey the US stimulus both to antennal lobes and to the MBs (reviewed in [86]). For the Pavlovian assay in Drosophila, the available data are consistent with the idea that initial CS–US associations occur in MB Kenyon cells, with the interesting possibility that additional associations occur upstream (e.g. in antennal lobes).
It is important to note, however, that all of the above studies focus on early memory (within the first 3 hours), leaving the circuitry underlying ARM and LTM an open question. A recent study attempted to use the Shibire-TS approach to dissect these forms of consolidated memory [47]. These authors used Gal4 drivers to express Shibire-TS in various subsets of MB neurons and then attempted to determine which lobes are involved in ARM or LTM. The authors observed defective levels of ARM when they disrupted dynamin function in MBs. Based on this finding, they argued that ARM is stored in MBs. We see two serious problems with the interpretation of this experiment. First, levels of ARM were not tested at permissive temperature. This is a critical control experiment because of the potential for leaky effects of overexpressing the Shibire-TS transgene, even at permissive temperature. Accordingly the difference between performance at permissive and restrictive temperature is more relevant than the absolute performance levels [82]. In the absence of this control experiment, it is not possible to assess whether the defect in ARM was due to an acute inhibition of dynamin function, a chronic developmental effect, or genetic background differences between the strains used.
An even more serious problem with this study, however, is that in all cases, the authors measured memory performance under conditions where MB transmission was chronically inhibited even during retrieval. Several studies already have documented that MB output is required during retrieval at this time point [10,70,71]. Hence the reduced levels of residual ARM observed are most easily explained not as a defect in ARM per se, but as an inhibition of memory retrieval. Together, the above two logical flaws not only undermine the conclusion that ARM is localized in MBs, but obviously invalidate any arguments about lobe specificity for ARM storage.
This same study also investigated the role of MB neurotransmission in retrieval of LTM. The Shibire-TS transgene was again used to block dynamin-mediated transmission in MBs and the animals were given five spaced training sessions at the permissive temperature. Memory retrieval then was measured, and found to be defective 24 hours later at the restrictive temperature when the transgene was expressed in α,α′, β, and β′ lobes. In contrast, expression in γ lobes appeared to have no effect. These data raise the interesting possibility that retrieval of LTM requires MB output from α, α′, β, or β′ lobes. But here again, this conclusion is undermined by the lack of control experiments performed at permissive temperature. Thus in our view, the anatomical circuitry responsible for storage and retrieval of ARM and LTM remains an open question.
Taken together, each of the various manipulations of MB function are consistent with a fairly simple cellular model in which Kenyon cells are the sole coincidence detectors that integrate US and CS stimuli. Experimental manipulation of cells extrinsic to MBs, however, has begun to suggest a more complex circuitry. In the case of amnesiac-expressing DPM cells, dynamin activity is not required for learning, but instead is required for an intermediate form of memory [76]. This is generally consistent with amnesiac being an MTM mutant [2]. It should be noted, however, that amnesiac is a putative neuropeptide (see section on dissection of biochemical pathways). Unlike small-molecule neurotransmitters, which are released via a dynamin-sensitive mechanism requiring rapid vesicle recycling, neuropeptides are released via dense core vesicles, a mechanism which is much less sensitive to dynamin-mediated recycling. The Shibire-TS inhibition of DPM cells is thus unlikely to block release of amnesiac neuropeptide. The fact that dynamin inhibition in these neurons disrupts memory therefore suggests that DPM neurons actually co-release a small molecule neurotransmitter other than amnesiac, which is more sensitive to Shibire-TS. In fact there is some evidence that DPM cells express choline-acetyltransferase, suggesting that they are cholinergic [77].
One of the most interesting findings regarding DPM cell function is the observation that dynamin-dependent output from these neurons is only required after training and before testing [37,77]. These data suggest that DPM cells are not involved in the CS–US association, but instead play a neuromodulatory role in some memory function within MB neurons, on another MB extrinsic neuron, or even on MB Kenyon cell output. DPM cells are thus the first example in this system of a neuron type that is not required for CS–US association, but instead is involved in memory processing after the association is formed.
The DPM cells hint at the possibility that the circuitry for memory storage may extend beyond MB Kenyon cells. Another clue comes from a Gal4 driver that is inserted in the radish gene (but see section on dissection of biochemical pathways). This Gal4 driver is expressed outside MBs and includes numerous neuron types, some of which innervate MBs [59] (Figure 2B). Functional manipulation with Shibire-TS of these ‘radish neurons’ raises the possibility that memory consolidation requires transfer of activity out of MBs into this novel circuit because transient inhibition of activity in these neurons causes increasingly severe memory defects when the disruption is performed at progressively later time points [59]. This hypothesis could be tested more rigorously if the relevant subset of neurons is identified. Regardless, these data suggest a non-MB circuit requirement for a later phase of memory.
Taken together, the findings from functional manipulation of the ‘memory circuit’ with Shibire-TS indicate that this simple form of associative learning requires a substantially complex circuitry that extends beyond MBs. The initial CS–US association likely occurs in MBs and possibly upstream (for example, in antennal lobes). STM and MTM also may involve plasticity in MB Kenyon cells, and there is some evidence for compartmentalization of function within MBs. The evidence for a role of γ lobes in at least a part of the rutabaga-dependent memory is currently the best example of this. By contrast, the circuitry underlying ARM and LTM are largely unknown.
Genetic Dissection of Biochemical Pathways
Mutants can be arranged into a genetic pathway of memory formation (Figure 3), logically implying some sort of temporal sequence to the underlying biochemistry, as is the case for memory phases at the behavioral level (Figure 1). Likewise, genetic components can sometimes be organized into cell signaling pathways. What is not clear, however, is whether each of the biochemical functions associated with the various genes all occur in the same cells (circuits) or are parsed across different circuits (and memory phases). As mentioned above, analysis of gene function is also confounded by any developmental versus physiological effects (or both) that result from the various biochemical dysfunctions [87,88]. With such caveats in mind, an initial attempt can be made to arrange the existing learning/memory mutants into biochemical and behavioral pathways.
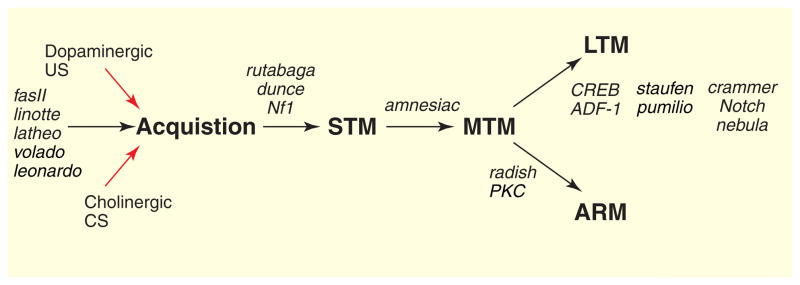
Figure 3. Dissection of biochemical pathways.
One of the hallmarks of memory is that consolidation occurs through a series of mechanistically distinct phases which can be processed sequentially or in parallel. In Drosophila, single gene mutants appear to selectively disrupt different memory phases. These mutants can be arranged into a genetic pathway of memory formation, with different mutants preferentially affecting learning/acquisition (fasII, linotte, latheo, volado and leonardo), STM (rutabaga, dunce, Nf1), MTM (amnesiac), ARM (radish) and LTM (CREB, ADF-1, staufen, pumilio, crammer, Notch, nebula). This genetic dissection implies a temporal sequence to the underlying biochemistry, but individual genes also may act at more than one phase, and in more than one part of the neural circuitry.
Several genes currently are considered to be involved in learning on the basis of the observations that mutant memory retention levels are lower than normal at the earliest measurable retention interval and mutant memory decay rates are similar to those of normal flies across all successive retention intervals [15,25]. With this operational definition, ‘learning genes’ appear to include latheo, linotte, 14-3-3 (leonardo), scabrous (volado), fasII and DC0 (PKA) [27,66,67,89–91]. ‘STM genes’ are those where mutant retention levels immediately after training are lower than normal and mutant memory decay rates are faster than normal during the first 30 min after training [15,25]: these include dunce and rutabaga [8,39–41]. ‘MTM genes’ are those where mutant retention levels immediately after training and seven hours later are nearly normal and mutant memory retention levels in between these two retention intervals are lower than normal. The amnesiac and DC0ts genes represent this class [8,26,76,92]. The radish gene is the only known ‘ARM gene’ [14,16,59] and is characterized by retention levels that are lower than normal at all time intervals and severe reduction of cold-shock-resistant memory. Finally, ‘LTM genes’ are those where mutants show defects in the protein-synthesis-dependent memory uniquely produced by spaced training [16,93]. To date, these include dCREB2, Adf1 (nalyot), Notch, crammer and nebula [29–32,94,95].
This genetic dissection of memory suggests not only sequential steps in the processing of olfactory memory but also parallel steps, because LTM and ARM appear to form independently of each other (see section on genetic dissection of memory) [16]. Recently, an alternative hypothesis has been presented [47] in which ARM and LTM are not processed in parallel, but instead are ‘mutually exclusive’. In this alternative model, ARM forms after massed training but not after spaced training and LTM forms only after spaced training. The authors of this study suggest that ARM actually prevents LTM after massed training, and likewise LTM induction inhibits formation of ARM after spaced training. This hypothesis derives primarily from the observation that in ala mutant animals, spaced training actually yielded lower levels of 5 hour memory (presumably ARM, although they do not demonstrate this) than did a single training session.
This hypothesis seems unlikely for three reasons. First, unlike ala mutants, wild-type animals form more memory after spaced training than after either one training session or repeated massed training [16]. Second, the additional component of memory that is formed after spaced training appears to consist of both ARM and LTM because either inhibition of protein synthesis (with cycloheximide) or inhibition of CREB-mediated transcription (with induced overexpression of a dominant-negative CREB isoform) each reduce performance to levels similar to that seen after massed training [16,29]. In contrast, memory after massed training is unaffected by these interventions. Thus if memory after spaced training consisted only of CREB-dependent LTM, one would need to argue that each of these disruptions cause only a partial inhibition of CREB-dependent gene expression. In this case, the residual memory after spaced training would be CREB-dependent LTM that escaped disruption. A third line of experimentation, however, appears to rule this out: radish mutants are not only deficient in ARM measured after massed training, but exhibit reduced memory after spaced training [16]. This strongly suggests that memory after spaced training includes a radish-dependent ARM component. Taken together, these data largely refute the ‘exclusive memory hypothesis’.
This same study also suggested that ARM is independent of cAMP signaling. This conclusion is drawn from their observation of normal levels of ARM in rutabaga adenylyl cyclase mutants (see below). The authors cite Dudai et al. [13] as confirmation of this observation. Such confirmation is false, however, because this earlier study demonstrated that, although some ARM is induced in rutabaga mutants, its levels are significantly lower than normal. While this discrepancy could derive from the fact that these studies relied on different alleles of rutabaga, the conclusion that the formation of ARM does not depend on cAMP signaling needs clarification.
Gene discovery by forward mutagenesis has now identified mutations in genes whose function might participate in several different signaling pathways. To date, however, only the cAMP second messenger pathway clearly has been ‘hit’ with multiple mutants. The dunce and rutabaga genes (involved in STM) encode a cAMP-specific phosphodiesterase (PDE) [22] and a calcium-sensitive adenylyl cyclase (AC) [63], respectively. Mutations of both the catalytic and regulatory subunits of PKA yield defects in learning or MTM [26,27,96]. dCREB2 (involved in LTM) encodes a cAMP response element binding protein (transcription factor) [97]. Neurofibromatosis 1 (Nf1), which encodes a Ras-specific GTPase-activating protein, also appears to act in the cAMP pathway [98]. In addition to its role as a tumor suppressor gene, NF1 appears to be associated with learning disabilities in humans. In Drosophila, Nf1 mutants have reduced STM [98].
The amnesiac gene (involved in MTM) also may encode a neuropeptide with some homology to pituitary adenylyl cyclase activated peptide (PACAP), which could act as a G-protein-coupled inducer of AC [99]. The connection between amnesiac and cAMP signaling remains tentative, however, because the amnesiac open reading frame actually may encode several different peptides. At this point it is unclear which of these are functionally relevant. It also is worth noting that the nature of the molecular lesion associated with the amnesiac1 mutation remains unknown despite a report by Feany and Quinn [74] to have found a point mutation in the amnesiac1 transcript. Moore et al. [75] failed to detect any point mutation in amnesiac1 and claimed, rather, that the DNA sequence obtained from amnesiac+ flies of Feany and Quinn was in error, thereby producing an incorrect coding sequence of the amnesiac+ gene and an apparent point mutation in amnesiac1. To date, this critical error in Feany and Quinn [74] has not been clarified or corrected by the authors.
CREB’s role in Drosophila LTM also has been questioned recently. Perazzona et al. [100] were unable to reproduce the enhancement of memory originally reported by Yin et al. [101] after induced overexpression of a CREB activator in the C28 transgenic strain. Perazzona et al. [100] describe a point mutation in transgenic sequence of CREB activator. This point mutation would be predicted to form a truncated, nonfunctional protein, thereby providing an apparent molecular rational for the failure to enhance memory with this transgenic strain/construct. It is worth noting that Yin claims that the C28 transgene is indeed able to produce a protein product by internal initiation (J.C. Yin, personal communication). This hypothesis could explain the results of Davis et al. [102], who also reported an enhancement of developmental plasticity of the neuromuscular junction using the very same C28 CREB activator strain. This critical demonstration of functional ‘activity’ from the C28 transgene by an independent group was not cited by Perazonna et al. [100]. As it stands, the exact mechanism by which the C28 transgene is able to enhance both memory and neuromuscular junction plasticity needs clarification. Finally, Perazzona et al. [100] also reported a ‘leaky expression’ of the heat-shock responsive CREB blocker in the 17-2 transgenic strain, an effect that has not been seen by other groups [29,103–105]. This suggests that differences in general experimental protocols (behavior, heat shock, etc.) and/or changes in genetic background may underlie their results.
In spite of the controversial status of some of these studies, progress continues to extend the involvement of cAMP signalling both upstream of adenylyl cyclase and downstream of CREB. dNR1, the Drosophila homolog of the mammalian NMDAR1 receptor, has recently been shown to be involved in olfactory learning and subsequent LTM formation [106]. These observations are similar to studies of vertebrate models that suggest that activity-dependent, CREB-mediated transcriptional responses are initiated by NMDAR activation (reviewed in [107]. The finding that NMDAR activation also underlies learning and memory in flies suggests that glutamatergic transmission might also play a role in signaling onto CREB in flies.
The role of different neurotransmitter systems in olfactory memory in flies appears to be complex. In addition to the observed role of NMDAR and the proposed role of amnesiac neuropeptide(s), the projection neurons that convey olfactory information to the MB appear to be largely cholinergic. An early report suggested that dopamine or serotonin (or both) might also play a role because temperature-sensitive mutants of the Dopa decarboxylase (Ddc) gene showed defective shock avoidance learning at restrictive temperature [23]. This observation proved not to be reproducible, however, even after much effort using both the original olfactory shock-avoidance task and the Pavlovian task [108]. Fortunately, dopamine has again reared its head as the ‘associative’ ligand for olfactory shock-avoidance learning. Schwaerzel et al. [6] used the Shibire-TS approach in combination with a promoter from the tyrosine hydroxylase gene [109] to demonstrate that dopaminergic transmission plays a role, probably to mediate the electric shock US.
Another productive advance in the biochemistry of memory formation lies in the identification of genetic components that act downstream of CREB. Dubnau et al. [78] employed the complementary ‘genomic’ approaches of a large-scale behavioral screen for mutants defective in one-day memory after spaced training (which would identify genes involved developmentally or physiologically in learning, STM, MTM, ARM or LTM) with DNA microarray technology to identify genes in normal flies that are transcriptionally regulated and thus would identify genes involved physiologically during LTM formation. The pumilio translational repressor gene was found with both approaches: it is transcriptionally upregulated during memory formation after spaced training, and two independent transposon-mediated mutations of pumilio yielded mutants with defective one-day memory after spaced training. From developmental studies, pumilio was known to be part of a gene pathway involved in regulating the spatial distribution of mRNA translation. In addition to pumilio, four additional components of this pathway, staufen, orb, moesin and eIF-2G were also transcriptionally regulated during LTM formation, while transposon-mediated lesions were found in or near two additional components, oskar (norka mutants) and eIF-5C (krasavietz mutants) [78].
As a further test of the hypothesis that this cellular pathway is involved in LTM formation in Drosophila, memory formation was evaluated in temperature-sensitive mutants of staufen [78]. One-day memory after spaced training proved normal in staufents mutants trained, stored and tested at permissive temperature. In contrast, one-day memory after spaced training was disrupted in staufents mutants that were trained and tested at permissive temperature but merely stored at restrictive temperature during the 24-hour retention interval between training and testing. These data suggest that the molecular machinery required for mRNA transport and local translation itself is regulated in response to LTM formation.
Three other LTM genes, less well integrated into cAMP signaling, deserve brief mention because of their potential neurobiological importance. The first, nebula, is a highly conserved member of the family of calcipressin-like calcineurin inhibitors. nebula mutants appear to disrupt LTM formation specifically [95]. Intriguingly, this gene also is homologous to Down’s syndrome critical region1 gene (DSCR1), thereby suggesting a direct connection between memory processing and this form of mental retardation. The second gene is crammer (cer) [94], encoding an inhibitor of a subfamily of cysteine proteinases named cathepsins. Interestingly, cer may have a role in glial cells that surround the MBs. Finally, Notch is a classic neurogenic gene mediating signaling pathways involved in several developmental processes. In neurons, it regulates neurite outgrowth in vivo [110,111], in tissue culture [112], and in the developing Drosophila central nervous system [113,114]. Conditional disruption of Notch causes deficits specific for LTM formation [31,32,115]. Recently, Alagille syndrome (mental retardation) and Cadasil Syndrome (dementia) have been linked to deficits in the Notch pathway [116,117]. Notch also is a substrate of presenilin (γ-secretase), thereby also linking it to Alzheimer’s disease (dementia) [118].
Additional complexity of biochemical signaling is suggested for ARM. radish recently has been shown to encode a PLA2 (phospholipase A2) [59]. Cleavage of membrane phospholipids by some vertebrate homologs yields arachidonic acid, which itself can activate atypical protein kinase C (aPKC) [119–121]. Davis [4] reports an unpublished claim by another group that radish does not encode PLA2. While we are unable to evaluate the unpublished data supporting this alternative claim, we nevertheless can suggest additional experiments to support our conclusion that radish encodes a PLA2. First, demonstration that expression of the PLA2 cDNA can rescue the original radish1 allele would bolster the observed rescue of the transposon allele (C133). Second, direct biochemical measurement of PLA2 function in adult brain tissue should determine whether this enzyme is defective in mutant animals.
In addition to PLA2, aPKCζ is implicated in ARM. Overexpression of a constitutively active form of aPKCζ enhances memory in both wild-type flies and also improves the memory of radish mutants, suggesting that PKC acts downstream of radish [103]. This finding also is consistent with identification of radish as a PLA2 (see above).
Critical Issues
Deriving mechanistic insight from the considerable array of genes involved in memory is confounded by the fact that functionally distinct gene networks act within functionally distinct neural networks to subserve behavior. The Drosophila memory field is now at a critical juncture where we are moving past identifying learning and memory genes to this more integrative effort. Now, we as a field must capitalize upon the unique genetic tools available in Drosophila to forge mechanistic links between levels of biological organization. Models of how biochemical signaling pathways participate in learning and memory need to go beyond thinking about synaptic plasticity between two neurons to an understanding of how a network of genes acting in a network of neurons subserves the full richness of memory.
Three major issues continue to dog genetic dissection of memory in this model system. First, most mutants studied to date carry gene disruptions that are present during development and (often) expressed widely. Consequently, we do not know whether the observed effects on memory reflect acute physiological dysfunctions or rather derive from developmental abnormalities [39,87]. In addition, we do not in most cases know the relevant circuits in which each of the genes function — and we do not believe necessarily that ‘relevant function’ will correspond with ‘preferential expression’. Second, with the exception of the cAMP pathway, most of the genes that have been identified in this system have not been conceptually integrated into a functional gene network. Consequently, mechanistic insight is currently limited to disconnected hypotheses suggested by individual genes. Third, while progress has been made in expanding our knowledge of the anatomical circuitry relevant to this phenotype, there are few cases where we have forged functional links between genetic units of function, neural circuit components, and behavioral manifestations of memory consolidation (such as memory phases). As a result of these conceptual gaps in our understanding, we cannot currently distinguish between two disparate notions of how information is processed during memory formation. At one extreme is an anatomically and biochemically simple hypothesis, in which association of shock and odor occurs solely in/upstream of MB Kenyon cells, is consolidated solely in MB Kenyon cells, and relies principally upon cAMP signaling and additional genes whose action directly impinges upon cAMP signaling. In this model, the behavioral manifestation of memory phases derives directly from sequential cellular modifications with different kinetics and distinct pharmacological sensitivities. In this model, all phases of memory rely on modifications occurring within the same set of neurons. We do not believe that the current data rule out an alternative memory transfer hypothesis, in which consolidation involves a dynamic interaction within a larger circuit of which MB Kenyon cells are a component. In this latter hypothesis, different memory phases can rely upon distinct signaling pathways acting in distinct anatomical foci.
So far, the hypothesis that has been favored in the literature [4,122] is that memory at all time points resides upstream of and within MB Kenyon cells. This MB-centric view probably derives from the fact that there is as yet no conclusive evidence to support the alternative more complex hypothesis. The memory systems model needs serious consideration, however, for three reasons. First, in most species and tasks the initial anatomical site of association is not the permanent focus of plasticity. Instead, one of the hallmarks of memory consolidation in other species is that the circuit requirements for memory storage generally shift over time to involve additional foci [123–130]. There are already some hints that this may be the case in Drosophila. For example, there is one report that a structural alteration in or near a part of the central complex is correlated with LTM [131]. This report is strictly correlative and has not been shown to be functionally relevant.
Second, additional gene discovery also has identified several memory genes that may be expressed exclusively outside of MB [59,76,78]. Functional manipulation of these neurons has indicated that they also are part of a broader memory consolidation circuit. In the case of the ‘radish reporter neurons’ there is direct evidence that transient inhibition at progressively later time points leads to increasingly severe memory defects [59]. This observation suggests a temporally graded requirement for a neural activity in a circuit that is outside of MB Kenyon cells.
Finally, there are simply no data which distinguish the MB-centric and memory transfer hypotheses. Until such data exist, we remain agnostic on this question, but we wish to resist the emerging dogma that rejects without experimental evidence the interesting possibility that MBs are only a temporary storage structure and are only a portion of a larger neural circuit for olfactory memory. We end this essay by proposing three critical lines of investigation that will distinguish between the hypotheses.
First, the anatomical focus and temporal requirement of each gene’s action must be determined for each phase of memory. To give one example, acute expression of the rutabaga+ cDNA in MBs has been shown to be sufficient to rescue the rutabaga STM defect [39–41]. But this result does not address the site of rutabaga-independent memory (because rutabaga animals do in fact have residual levels of memory) and it does not address the question of rutabaga’s role in other memory phases.
Second, cAMP-independent pathways of learning have not been genetically or anatomically dissected. By generating double-mutant combinations with rutabaga, it should be possible to determine which of the existing mutants primarily disrupt the residual learning in rutabaga null animals. Here again, investigation of the anatomical site and time of action of these additional biochemical pathways will be critical.
Finally, the anatomical circuitry for LTM must be determined. The memory transfer hypothesis strongly predicts (i) a requirement for CREB-mediated transcription in a non-MB locus, (ii) a time period after training when output from MB would be required to consolidate memory at such a distal locus and (iii) an involvement in LTM retrieval of output from the site where the relevant CREB-mediated transcription occurs.
Our understanding of memory as a biological phenomenon derives necessarily from investigation at several different levels of organization. When findings from each level are considered alone, they are confusing. Only by a ‘vertical integration’ across levels of biological organization can we begin to understand the behavioral phenotype in a mechanistic sense. The experimental tools are now in place to accomplish this for Pavlovian learning in flies.
Acknowledgments
We would like to thank Ann-Shyn Chiang for providing the image in figure 2C, and also thank the NIH and Dart NeuroGenomics Inc. for funding to T.T. and the NIH, Dart NeruoGenomics Inc. and Beckman foundation for funding to J.D.
References
- 1.van Swinderen B, Greenspan RJ. Flexibility in a gene network affecting a simple behavior in Drosophila melanogaster. Genetics. 2005;169:2151–2163. doi: 10.1534/genetics.104.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dubnau J, Tully T. Gene discovery in Drosophila: new insights for learning and memory. Annu Rev Neurosci. 1998;21:407–444. doi: 10.1146/annurev.neuro.21.1.407. [DOI] [PubMed] [Google Scholar]
- 3.Waddell S, Quinn WG. Flies, genes, and learning. Annu Rev Neurosci. 2001;24:1283–1309. doi: 10.1146/annurev.neuro.24.1.1283. [DOI] [PubMed] [Google Scholar]
- 4.Davis R. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 5.Tempel BL, Bonini N, Dawson DR, Quinn WG. Reward learning in normal and mutant Drosophila. Proc Natl Acad Sci USA. 1983;80:1482–1486. doi: 10.1073/pnas.80.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, Heisenberg M. Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci. 2003;23:10495–10502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quinn WG, Harris WA, Benzer S. Conditioned behavior in Drosophila melanogaster. Proc Natl Acad Sci USA. 1974;71:708–712. doi: 10.1073/pnas.71.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tully T, Quinn WG. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J Comp Physiol [A] 1985;157:263–277. doi: 10.1007/BF01350033. [DOI] [PubMed] [Google Scholar]
- 9.Tanimoto H, Heisenberg M, Gerber B. Experimental psychology: event time turns punishment to reward. Nature. 2004;430:983. doi: 10.1038/430983a. [DOI] [PubMed] [Google Scholar]
- 10.Schwaerzel M, Heisenberg M, Zars T. Extinction antagonizes olfactory memory at the subcellular level. Neuron. 2002;35:951–960. doi: 10.1016/s0896-6273(02)00832-2. [DOI] [PubMed] [Google Scholar]
- 11.Dubnau J. Neurogenetic dissection of conditioned behavior: evolution by analogy or homology? J Neurogenet. 2003;17:295–326. doi: 10.1080/01677060390441859. [DOI] [PubMed] [Google Scholar]
- 12.Quinn WG, Dudai Y. Memory phases in Drosophila. Nature. 1976;262:576–577. doi: 10.1038/262576a0. [DOI] [PubMed] [Google Scholar]
- 13.Dudai Y, Corfas G, Hazvi S. What is the possible contribution of Ca2+-stimulated adenylate cyclase to acquisition, consolidation and retention of an associative olfactory memory in Drosophila. J Comp Physiol [A] 1988;162:101–109. doi: 10.1007/BF01342707. [DOI] [PubMed] [Google Scholar]
- 14.Folkers E, Drain P, Quinn WG. Radish, a Drosophila mutant deficient in consolidated memory. Proc Natl Acad Sci USA. 1993;90:8123–8127. doi: 10.1073/pnas.90.17.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tully T, Boynton S, Brandes C, Dura JM, Mihalek R, Preat T, Villella A. Genetic dissection of memory formation in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1990;55:203–211. doi: 10.1101/sqb.1990.055.01.022. [DOI] [PubMed] [Google Scholar]
- 16.Tully T, Preat T, Boynton SC, Del Vecchio M. Genetic dissection of consolidated memory in Drosophila. Cell. 1994;79:35–47. doi: 10.1016/0092-8674(94)90398-0. [DOI] [PubMed] [Google Scholar]
- 17.Kandel ER. The molecular biology of memory storage: a dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 18.Dudai Y, Jan YN, Byers D, Quinn WG, Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc Natl Acad Sci USA. 1976;73:1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis RL, Kiger JA., Jr Dunce mutants of Drosophila melanogaster: mutants defective in the cyclic AMP phosphodiesterase enzyme system. J Cell Biol. 1981;90:101–107. doi: 10.1083/jcb.90.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byers D, Davis RL, Kiger JA., Jr Defect in cyclic AMP phosphodiesterase due to the dunce mutation of learning in Drosophila melanogaster. Nature. 1981;289:79–81. doi: 10.1038/289079a0. [DOI] [PubMed] [Google Scholar]
- 21.Davis RL, Davidson N. Isolation of the Drosophila melanogaster dunce chromosomal region and recombinational mapping of dunce sequences with restriction site polymorphisms as genetic markers. Mol Cell Biol. 1984;4:358–367. doi: 10.1128/mcb.4.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen CN, Denome S, Davis RL. Molecular analysis of cDNA clones and the corresponding genomic coding sequences of the Drosophila dunce+ gene, the structural gene for cAMP phosphodiesterase. Proc Natl Acad Sci USA. 1986;83:9313–9317. doi: 10.1073/pnas.83.24.9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tempel BL, Livingstone MS, Quinn WG. Mutations in the dopa decarboxylase gene affect learning in Drosophila. Proc Natl Acad Sci USA. 1984;81:3577–3581. doi: 10.1073/pnas.81.11.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livingstone MS, Sziber PP, Quinn WG. Loss of calcium/calmodulin responsiveness in adenylate cyclase of rutabaga, a Drosophila learning mutant. Cell. 1984;37:205–215. doi: 10.1016/0092-8674(84)90316-7. [DOI] [PubMed] [Google Scholar]
- 25.Tully T, Bolwig G, Christensen J, Connolly J, DelVecchio M, DeZazzo J, Dubnau J, Jones C, Pinto S, Regulski M, et al. A return to genetic dissection of memory in Drosophila. Cold Spring Harb Symp Quant Biol. 1996;61:207–218. [PubMed] [Google Scholar]
- 26.Li W, Tully T, Kalderon D. Effects of a conditional Drosophila PKA mutant on olfactory learning and memory. Learn Mem. 1996;2:320–333. doi: 10.1101/lm.2.6.320. [DOI] [PubMed] [Google Scholar]
- 27.Skoulakis EM, Kalderon D, Davis RL. Preferential expression in mushroom bodies of the catalytic subunit of protein kinase A and its role in learning and memory. Neuron. 1993;11:197–208. doi: 10.1016/0896-6273(93)90178-t. [DOI] [PubMed] [Google Scholar]
- 28.DeZazzo J, Tully T. Dissection of memory formation: from behavioral pharmacology to molecular genetics. Trends Neurosci. 1995;18:212–218. doi: 10.1016/0166-2236(95)93905-d. [DOI] [PubMed] [Google Scholar]
- 29.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 30.DeZazzo J, Sandstrom D, de Belle S, Velinzon K, Smith P, Grady L, DelVecchio M, Ramaswami M, Tully T. nalyot, a mutation of the Drosophila myb-related Adf1 transcription factor, disrupts synapse formation and olfactory memory. Neuron. 2000;27:145–158. doi: 10.1016/s0896-6273(00)00016-7. [DOI] [PubMed] [Google Scholar]
- 31.Ge X, Hannan F, Xie Z, Feng C, Tully T, Zhou H, Zhong Y. Notch signaling in Drosophila long-term memory formation. Proc Natl Acad Sci USA. 2004;101:10172–10176. doi: 10.1073/pnas.0403497101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Presente A, Boyles RS, Serway CN, de Belle JS, Andres AJ. Notch is required for long-term memory in Drosophila. Proc Natl Acad Sci USA. 2004;101:1764–1768. doi: 10.1073/pnas.0308259100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 34.de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. [DOI] [PubMed] [Google Scholar]
- 35.Erber J, Masuhr TH, Menzel R. Localization of short-term memory in the brain of the bee. Physiol Entomol. 1980;5:343–358. [Google Scholar]
- 36.Davis RL, Han KA. Neuroanatomy: mushrooming mushroom bodies. Curr Biol. 1996;6:146–148. doi: 10.1016/s0960-9822(02)00447-5. [DOI] [PubMed] [Google Scholar]
- 37.Dubnau J, Chiang AS, Tully T. Neural substrates of memory: from synapse to system. J Neurobiol. 2003;54:238–253. doi: 10.1002/neu.10170. [DOI] [PubMed] [Google Scholar]
- 38.Connolly JB, Roberts IJ, Armstrong JD, Kaiser K, Forte M, Tully T, O’Kane CJ. Associative learning disrupted by impaired Gs signaling in Drosophila mushroom bodies. Science. 1996;274:2104–2107. doi: 10.1126/science.274.5295.2104. [DOI] [PubMed] [Google Scholar]
- 39.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 40.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of mMemory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 41.Mao Z, Roman G, Zong L, Davis RL. Pharmacogenetic rescue in time and space of the rutabaga memory impairment by using Gene-Switch. Proc Natl Acad Sci USA. 2004;101:198–203. doi: 10.1073/pnas.0306128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ito K, Suzuki K, Estes P, Ramaswami M, Yamamoto D, Strausfeld NJ. The organization of extrinsic neurons and their implications in the functional roles of the mushroom bodies in Drosophila melanogaster Meigen. Learn Mem. 1998;5:52–77. [PMC free article] [PubMed] [Google Scholar]
- 43.Strausfeld NJ. Atlas of an insect brain. New York: Springer-Verlag; 1976. [Google Scholar]
- 44.Heisenberg M. Mushroom body memoir: from maps to models. Nat Rev Neurosci. 2003;4:266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 45.Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- 46.Pascual A, Preat T. Localization of long-term memory within the Drosophila mushroom body. Science. 2001;294:1115–1117. doi: 10.1126/science.1064200. [DOI] [PubMed] [Google Scholar]
- 47.Isabel G, Pascual A, Preat T. Exclusive consolidated memory phases in Drosophila. Science. 2004;304:1024–1027. doi: 10.1126/science.1094932. [DOI] [PubMed] [Google Scholar]
- 48.Liu L, Wolf R, Ernst R, Heisenberg M. Context generalization in Drosophila visual learning requires the mushroom bodies. Nature. 1999;400:753–756. doi: 10.1038/23456. [DOI] [PubMed] [Google Scholar]
- 49.Jefferis GS, Marin EC, Stocker RF, Luo L. Target neuron prespecification in the olfactory map of Drosophila. Nature. 2001;414:204–208. doi: 10.1038/35102574. [DOI] [PubMed] [Google Scholar]
- 50.Shipley MT, Ennis M. Functional organization of olfactory system. J Neurobiol. 1996;30:123–176. doi: 10.1002/(SICI)1097-4695(199605)30:1<123::AID-NEU11>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 51.Hallem EA, Carlson JR. The odor coding system of Drosophila. Trends Genet. 2004;20:453–459. doi: 10.1016/j.tig.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 52.Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999;96:725–736. doi: 10.1016/s0092-8674(00)80582-6. [DOI] [PubMed] [Google Scholar]
- 53.Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron. 1999;22:327–338. doi: 10.1016/s0896-6273(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 54.Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 2002;109:243–255. doi: 10.1016/s0092-8674(02)00700-6. [DOI] [PubMed] [Google Scholar]
- 55.Wong AM, Wang JW, Axel R. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 2002;109:229–241. doi: 10.1016/s0092-8674(02)00707-9. [DOI] [PubMed] [Google Scholar]
- 56.Gorczyca MG, Phillis RW, Budnik V. The role of tinman, a mesodermal cell fate gene, in axon pathfinding during the development of the transverse nerve in Drosophila. Development. 1994;120:2143–2152. doi: 10.1242/dev.120.8.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuyama K, Meinertzhagen IA, Schuermann FW. Synaptic connections of cholinergic antennal lobe relay neurons in the brain of Drosophila melanogaster. J Comp Neurol. 2003;466:299–315. doi: 10.1002/cne.10867. [DOI] [PubMed] [Google Scholar]
- 58.Yasuyama K, Meinertzhagen IA, Schurmann FW. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. J Comp Neurol. 2002;445:211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]
- 59.Chiang AS, Blum A, Barditch J, Chen YH, Chiu SL, Regulski M, Armstrong JD, Tully T, Dubnau J. radish encodes a phospholipase-A2 and defines a neural circuit involved in anesthesia-resistant memory. Curr Biol. 2004;14:263–272. doi: 10.1016/j.cub.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Han PL, Levin LR, Reed RR, Davis RL. Preferential expression of the Drosophila rutabaga gene in mushroom bodies, neural centers for learning in insects. Neuron. 1992;9:619–627. doi: 10.1016/0896-6273(92)90026-a. [DOI] [PubMed] [Google Scholar]
- 61.Nighorn A, Healy MJ, Davis RL. The cyclic AMP phosphodiesterase encoded by the Drosophila dunce gene is concentrated in the mushroom body neuropil. Neuron. 1991;6:455–467. doi: 10.1016/0896-6273(91)90253-v. [DOI] [PubMed] [Google Scholar]
- 62.Bellen HJ, Gregory BK, Olsson CL, Kiger JA., Jr Two Drosophila learning mutants, dunce and rutabaga, provide evidence of a maternal role for cAMP on embryogenesis. Dev Biol. 1987;121:432–444. doi: 10.1016/0012-1606(87)90180-1. [DOI] [PubMed] [Google Scholar]
- 63.Levin LR, Han PL, Hwang PM, Feinstein PG, Davis RL, Reed RR. The Drosophila learning and memory gene rutabaga encodes a Ca2+/Calmodulin-responsive adenylyl cyclase. Cell. 1992;68:479–489. doi: 10.1016/0092-8674(92)90185-f. [DOI] [PubMed] [Google Scholar]
- 64.Han KA, Millar NS, Davis RL. A novel octopamine receptor with preferential expression in Drosophila mushroom bodies. J Neurosci. 1998;18:3650–3658. doi: 10.1523/JNEUROSCI.18-10-03650.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han KA, Millar NS, Grotewiel MS, Davis RL. DAMB, a novel dopamine receptor expressed specifically in Drosophila mushroom bodies. Neuron. 1996;16:1127–1135. doi: 10.1016/s0896-6273(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 66.Grotewiel MS, Beck CD, Wu KH, Zhu XR, Davis RL. Integrin-mediated short-term memory in Drosophila. Nature. 1998;391:455–460. doi: 10.1038/35079. [DOI] [PubMed] [Google Scholar]
- 67.Skoulakis EM, Davis RL. Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron. 1996;17:931–944. doi: 10.1016/s0896-6273(00)80224-x. [DOI] [PubMed] [Google Scholar]
- 68.Han PL, Meller V, Davis RL. The Drosophila brain revisited by enhancer detection. J Neurobiol. 1996;31:88–102. doi: 10.1002/(SICI)1097-4695(199609)31:1<88::AID-NEU8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 69.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 70.Dubnau J, Grady L, Kitamoto T, Tully T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature. 2001;411:476–480. doi: 10.1038/35078077. [DOI] [PubMed] [Google Scholar]
- 71.McGuire SE, Le PT, Davis RL. The role of Drosophila mushroom body signaling in olfactory memory. Science. 2001;293:1330–1333. doi: 10.1126/science.1062622. [DOI] [PubMed] [Google Scholar]
- 72.Zars T, Wolf R, Davis R, Heisenberg M. Tissue-specific expression of a type I adenylyl cyclase rescues the rutabaga mutant memory defect: in search of the engram. Learn Mem. 2000;7:18–31. doi: 10.1101/lm.7.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aceves-Pina EO, Booker R, Duerr JS, Livingstone MS, Quinn WG, Smith RF, Sziber PP, Tempel BL, Tully TP. Learning and memory in Drosophila, studied with mutants. Cold Spring Harb Symp Quant Biol. 1983;48:831–840. doi: 10.1101/sqb.1983.048.01.086. [DOI] [PubMed] [Google Scholar]
- 74.Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- 75.Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 76.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103:805–813. doi: 10.1016/s0092-8674(00)00183-5. [DOI] [PubMed] [Google Scholar]
- 77.Keene AC, Stratmann M, Keller A, Perrat PN, Vosshall LB, Waddell S. Diverse odor-conditioned memories require uniquely timed dorsal paired medial neuron output. Neuron. 2004;44:521–533. doi: 10.1016/j.neuron.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 78.Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol. 2003;13:286–296. doi: 10.1016/s0960-9822(03)00064-2. [DOI] [PubMed] [Google Scholar]
- 79.Tanaka NK, Awasaki T, Shimada T, Ito K. Integration of chemosensory pathways in the Drosophila second-order olfactory centers. Curr Biol. 2004;14:449–457. doi: 10.1016/j.cub.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 80.Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci. 1993;13:1852–1861. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Strauss R. The central complex and the genetic dissection of locomotor behaviour. Curr Opin Neurobiol. 2002;12:633–638. doi: 10.1016/s0959-4388(02)00385-9. [DOI] [PubMed] [Google Scholar]
- 82.Kitamoto T. Targeted expression of temperature-sensitive dynamin to study neural mechanisms of complex behavior in Drosophila. J Neurogenet. 2002;16:205–228. doi: 10.1080/01677060216295. [DOI] [PubMed] [Google Scholar]
- 83.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. J Neurobiol. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 84.Dubnau J, Tully T. Functional anatomy: from molecule to memory. Curr Biol. 2001;11:R240–R243. doi: 10.1016/s0960-9822(01)00115-4. [DOI] [PubMed] [Google Scholar]
- 85.Yu D, Ponomarev A, Davis RL. Altered representation of the spatial code for odors after olfactory classical conditioning; memory trace formation by synaptic recruitment. Neuron. 2004;42:437–449. doi: 10.1016/s0896-6273(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 86.Stevenson PA, Sporhase-Eichmann U. Localization of octopaminergic neurones in insects. Comp Biochem Physiol A. 1995;110:203–215. doi: 10.1016/0300-9629(94)00152-j. [DOI] [PubMed] [Google Scholar]
- 87.Dauwalder B, Davis RL. Conditional rescue of the dunce learning/memory and female fertility defects with Drosophila or rat transgenes. J Neurosci. 1995;15:3490–3499. doi: 10.1523/JNEUROSCI.15-05-03490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, et al. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 89.Boynton S, Tully T. latheo, a new gene involved in associative learning and memory in Drosophila melanogaster, identified from P element mutagenesis. Genetics. 1992;131:655–672. doi: 10.1093/genetics/131.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dura JM, Preat T, Tully T. Identification of linotte, a new gene affecting learning and memory in Drosophila melanogaster. J Neurogenet. 1993;9:1–14. doi: 10.3109/01677069309167272. [DOI] [PubMed] [Google Scholar]
- 91.Cheng Y, Endo K, Wu K, Rodan AR, Heberlein U, Davis RL. Drosophila fasciclinII is required for the formation of odor memories and for normal sensitivity to alcohol. Cell. 2001;105:757–768. doi: 10.1016/s0092-8674(01)00386-5. [DOI] [PubMed] [Google Scholar]
- 92.DeZazzo J, Xia S, Christensen J, Velinzon K, Tully T. Developmental expression of an amn(+) transgene rescues the mutant memory defect of amnesiac adults. J Neurosci. 1999;19:8740–8746. doi: 10.1523/JNEUROSCI.19-20-08740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mery F, Kawecki TJ. A cost of long-term memory in Drosophila. Science. 2005;308:1148. doi: 10.1126/science.1111331. [DOI] [PubMed] [Google Scholar]
- 94.Comas D, Petit F, Preat T. Drosophila long-term memory formation involves regulation of cathepsin activity. Nature. 2004;430:460–463. doi: 10.1038/nature02726. [DOI] [PubMed] [Google Scholar]
- 95.Chang KT, Shi YJ, Min KT. The Drosophila homolog of Down’s syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc Natl Acad Sci USA. 2003;100:15794–15799. doi: 10.1073/pnas.2536696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goodwin SF, Del Vecchio M, Velinzon K, Hogel C, Russell SR, Tully T, Kaiser K. Defective learning in mutants of the Drosophila gene for a regulatory subunit of cAMP-dependent protein kinase. J Neurosci. 1997;17:8817–8827. doi: 10.1523/JNEUROSCI.17-22-08817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yin JC, Wallach JS, Wilder EL, Klingensmith J, Dang D, Perrimon N, Zhou H, Tully T, Quinn WG. A Drosophila CREB/CREM homolog encodes multiple isoforms, including a cyclic AMP-dependent protein kinase-responsive transcriptional activator and antagonist. Mol Cell Biol. 1995;15:5123–5130. doi: 10.1128/mcb.15.9.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- 99.Zhong Y. Mediation of PACAP-like neuropeptide transmission by coactivation of Ras/Raf and cAMP signal transduction pathways in Drosophila. Nature. 1995;375:588–592. doi: 10.1038/375588a0. [DOI] [PubMed] [Google Scholar]
- 100.Perazzona B, Isabel G, Preat T, Davis RL. The role of cAMP response element-binding protein in Drosophila long-term memory. J Neurosci. 2004;24:8823–8828. doi: 10.1523/JNEUROSCI.4542-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 102.Davis GW, Schuster CM, Goodman CS. Genetic dissection of structural and functional components of synaptic plasticity. III. CREB is necessary for presynaptic functional plasticity. Neuron. 1996;17:669–679. doi: 10.1016/s0896-6273(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 103.Drier EA, Tello MK, Cowan M, Wu P, Blace N, Sacktor TC, Yin JC. Memory enhancement and formation by atypical PKC activity in Drosophila melanogaster. Nat Neurosci. 2002;5:316–324. doi: 10.1038/nn820. [DOI] [PubMed] [Google Scholar]
- 104.Hendricks JC, Williams JA, Panckeri K, Kirk D, Tello M, Yin JC, Sehgal A. A non-circadian role for cAMP signaling and CREB activity in Drosophila rest homeostasis. Nat Neurosci. 2001;4:1108–1115. doi: 10.1038/nn743. [DOI] [PubMed] [Google Scholar]
- 105.Sakai T, Tamura T, Kitamoto T, Kidokoro Y. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc Natl Acad Sci USA. 2004;101:16058–16063. doi: 10.1073/pnas.0401472101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xia S, Miyashita T, Fu TF, Lin WY, Wu CL, Pyzocha L, Lin IR, Saitoe M, Tully T, Chiang AS. NMDA receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 2005;15:603–615. doi: 10.1016/j.cub.2005.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waltereit R, Weller M. Signaling from cAMP/PKA to MAPK and synaptic plasticity. Mol Neurobiol. 2003;27:99–106. doi: 10.1385/MN:27:1:99. [DOI] [PubMed] [Google Scholar]
- 108.Tully T. On the road to a better understanding of learning and memory in Drosophila melanogaster. In: Spatz GHaH-CH., editor. Modulation of Synaptic Transmission and Plasticity in Nervous Systems. Berlin: NATO ASI series, Springer Verlag; 1988. pp. 401–417. [Google Scholar]
- 109.Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- 110.Redmond L, Oh SR, Hicks C, Weinmaster G, Ghosh A. Nuclear Notch1 signaling and the regulation of dendritic development. Nat Neurosci. 2000;3:30–40. doi: 10.1038/71104. [DOI] [PubMed] [Google Scholar]
- 111.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 112.Franklin JL, Berechid BE, Cutting FB, Presente A, Chambers CB, Foltz DR, Ferreira A, Nye JS. Autonomous and non-autonomous regulation of mammalian neurite development by Notch1 and Delta1. Curr Biol. 1999;9:1448–1457. doi: 10.1016/s0960-9822(00)80114-1. [DOI] [PubMed] [Google Scholar]
- 113.Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25:549–561. doi: 10.1016/s0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- 114.Giniger E, Jan LY, Jan YN. Specifying the path of the intersegmental nerve of the Drosophila embryo: a role for Delta and Notch. Development. 1993;117:431–440. doi: 10.1242/dev.117.2.431. [DOI] [PubMed] [Google Scholar]
- 115.Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13:1348–1354. doi: 10.1016/s0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 116.Harris JG, Filley CM. CADASIL: neuropsychological findings in three generations of an affected family. J Int Neuropsychol Soc. 2001;7:768–774. doi: 10.1017/s135561770176612x. [DOI] [PubMed] [Google Scholar]
- 117.Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 118.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 119.Calignano A, Piomelli D, Sacktor TC, Schwartz JH. A phospholipase A2-stimulating protein regulated by protein kinase C in Aplysia neurons. Brain Res Mol Brain Res. 1991;9:347–351. doi: 10.1016/0169-328x(91)90083-a. [DOI] [PubMed] [Google Scholar]
- 120.Nishizaki T, Nomura T, Matsuoka T, Enikolopov G, Sumikawa K. Arachidonic acid induces a long-lasting facilitation of hippocampal synaptic transmission by modulating PKC activity and nicotinic ACh receptors. Brain Res Mol Brain Res. 1999;69:263–272. doi: 10.1016/s0169-328x(99)00117-5. [DOI] [PubMed] [Google Scholar]
- 121.Muzzio IA, Gandhi CC, Manyam U, Pesnell A, Matzel LD. Receptor-stimulated phospholipase A(2) liberates arachidonic acid and regulates neuronal excitability through protein kinase C. J Neurophysiol. 2001;85:1639–1647. doi: 10.1152/jn.2001.85.4.1639. [DOI] [PubMed] [Google Scholar]
- 122.Gerber B, Tanimoto H, Heisenberg M. An engram found? Evaluating the evidence from fruit flies. Curr Opin Neurobiol. 2004;14:737–744. doi: 10.1016/j.conb.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 123.Hammer M, Menzel R. Multiple sites of associative odor learning as revealed by local brain microinjections of octopamine in honeybees. Learn Mem. 1998;5:146–156. [PMC free article] [PubMed] [Google Scholar]
- 124.Menzel R. Searching for the memory trace in a mini-brain, the honeybee. Learn Mem. 2001;8:53–62. doi: 10.1101/lm.38801. [DOI] [PubMed] [Google Scholar]
- 125.Patterson TA, Gilbert DB, Rose SP. Pre- and post-training lesions of the intermediate medial hyperstriatum ventrale and passive avoidance learning in the chick. Exp Brain Res. 1990;80:189–195. doi: 10.1007/BF00228860. [DOI] [PubMed] [Google Scholar]
- 126.Gilbert DB, Patterson TA, Rose SP. Dissociation of brain sites necessary for registration and storage of memory for a one-trial passive avoidance task in the chick. Behav Neurosci. 1991;105:553–561. doi: 10.1037//0735-7044.105.4.553. [DOI] [PubMed] [Google Scholar]
- 127.Chen C, Kim JJ, Thompson RF, Tonegawa S. Hippocampal lesions impair contextual fear conditioning in two strains of mice. Behav Neurosci. 1996;110:1177–1180. doi: 10.1037//0735-7044.110.5.1177. [DOI] [PubMed] [Google Scholar]
- 128.Frankland PW, Cestari V, Filipkowski RK, McDonald RJ, Silva AJ. The dorsal hippocampus is essential for context discrimination but not for contextual conditioning. Behav Neurosci. 1998;112:863–874. doi: 10.1037//0735-7044.112.4.863. [DOI] [PubMed] [Google Scholar]
- 129.Milner B. Disorders of learning and memory after temporal lobe lesions in man. Clin Neurosurg. 1972;19:421–446. doi: 10.1093/neurosurgery/19.cn_suppl_1.421. [DOI] [PubMed] [Google Scholar]
- 130.Scoville WB, Milner B. Loss of recent memory after bilateral hippocampal lesions. 1957. J Neuropsychiatry Clin Neurosci. 2000;12:103–113. doi: 10.1176/jnp.12.1.103. [DOI] [PubMed] [Google Scholar]
- 131.Pascual A, Huang KL, Neveu J, Preat T. Neuroanatomy: brain asymmetry and long-term memory. Nature. 2004;427:605–606. doi: 10.1038/427605a. [DOI] [PubMed] [Google Scholar]