Abstract
Melleolides and related fungal sesquiterpenoid aryl esters are antimicrobial and cytotoxic natural products derived from cultures of the Homobasidiomycetes genus Armillaria. The initial step in the biosynthesis of all melleolides involves cyclization of the universal sesquiterpene precursor farnesyl diphosphate to produce protoilludene, a reaction catalyzed by protoilludene synthase. We achieved the partial purification of protoilludene synthase from a mycelial culture of Armillaria gallica and found that 6-protoilludene was its exclusive reaction product. Therefore, a further isomerization reaction is necessary to convert the 6–7 double bond into the 7–8 double bond found in melleolides. We expressed an A. gallica protoilludene synthase cDNA in Escherichia coli, and this also led to the exclusive production of 6-protoilludene. Sequence comparison of the isolated sesquiterpene synthase revealed a distant relationship to other fungal terpene synthases. The isolation of the genomic sequence identified the 6-protoilludene synthase to be present as a single copy gene in the genome of A. gallica, possessing an open reading frame interrupted with eight introns.
Keywords: Drug Design, Enzyme Purification, Enzymes, Isoprenoid, Metabolism, Basidiomycete, Melleolide, Protoilludene, Terpene Synthases, Fungal Natural Products
Introduction
Terpenes are the largest and most diverse group of natural products, with more than 50,000 known structures (1). The biosynthesis of some terpenes has been studied in detail, particularly those from plants (e.g. menthol, artemisinin, and taxol), but less attention has been paid to terpenes from other sources. Fungi are a rich source of physiologically active natural products, and mushroom-forming fungi and other Basidiomycota are particularly well known for the synthesis of numerous bioactive sesquiterpenoids and, to a lesser extent, diterpenoids (2, 3). The biosynthesis of all terpenes begins with the cyclization and rearrangement of one of three universal precursors, geranyl diphosphate, farnesyl diphosphate, or geranylgeranyl diphosphate, to yield monoterpenoids, sesquiterpenoids, or diterpenoids, respectively. These cyclization reactions are among the most complex chemical reactions known in nature, and they are catalyzed by terpene synthases (also known as terpene cyclases). Following the terpene synthase reaction, which establishes the complete terpene carbon scaffold, additional reactions typically catalyzed by cytochrome P450-dependent mono-oxygenases generate further structural diversity (4–6).
Several terpene synthases have been isolated from plants (7, 8), but only a few microbial and fungal terpene synthases have been reported (9–21). A limited number of fungal sesquiterpenoid synthases have also been cloned and functionally characterized, including trichodiene synthase, aristolochene synthase, presilphiperfolan-8β-ol synthase and, recently, six sesquiterpene synthases from Coprinopsis cinerea (Coprinus cinereus) that yield germacrene A, α-muurolene, δ-cadinene, and α-cuprenene as major products (11, 13, 19–26). Generally, plant and fungal terpene synthases show only low level sequence identity, and whereas genes related to plant terpene biosynthesis pathways are often scattered throughout the genome, fungal terpene synthase genes are frequently clustered together, along with genes encoding terpene-modifying enzymes such as cytochrome P450 mono-oxygenases.
The Basidiomycota, a subdivision of the Dicarya, comprises >30,000 known species and represents approximately one-third of the phylum Eumycota (fungi) (27). This is a particularly rich source of complex, structurally diverse, and bioactive sesquiterpenoids (2, 3). Semisynthetic derivatives of pleuromutilin such as tiamulin, valnemulin, and retapamulin are already used as drugs to treat mycoplasma infections and bacterial pathogens that affect livestock (28).
Many known sesquiterpenoids such as illudane, marasmane, cerapicane, and, to a lesser extent, lactarane and sterpurane (29) are structurally related to the protoilludane skeleton. Although some of these compounds are thought to be synthesized uniquely in the basidiocarps of symbiotic fungi, others are produced in large quantities by the mycelial cultures of saprotrophic or facultative parasitic species, which are easier to culture under laboratory conditions. To study fungal terpenoids and investigate the genetic background that has led to the phenotypic diversity in the synthesis of such compounds, we studied the important cosmopolitan genus Armillaria (commonly known as honey mushrooms and currently classified in the family Physalaciaceae). Armillaria species are not only regarded as edible mushrooms, but many species are also notorious forest parasites, with a host range of the most virulent species, primarily Armillaria mellea and Armillaria ostoyae, comprising >600 species worldwide (30). This is also reflected by their ability to form rhizomorphs that allow them to grow across nutrient-poor areas located between large food sources such as tree stumps and, finally, to infect entire forests. Smith et al. (31) have even postulated that Armillaria bulbosa (current name: Armillaria gallica) may be among the oldest and largest organisms on Earth.
The characteristic secondary metabolites produced by Armillaria species are protoilludene-type sesquiterpenoids, potent antimicrobial molecules also known as melleolides. Some of the examined melleolides, like melleolide B, C, and D, possess remarkable antibacterial activity against Bacillus cereus, Bacillus subtilis, and Escherichia coli (32). For armillaric acid, antimicrobial activity against Micrococcus luteus, B. subtilis, Candida albicans, and Staphylococcus aureus has been reported (33). There are ∼50 known melleolides, produced almost exclusively by this fungal genus. Each molecule comprises a tricyclic sesquiterpenoid skeleton linked to an orsellinic acid-like polyketide side chain via an ester bond (34).
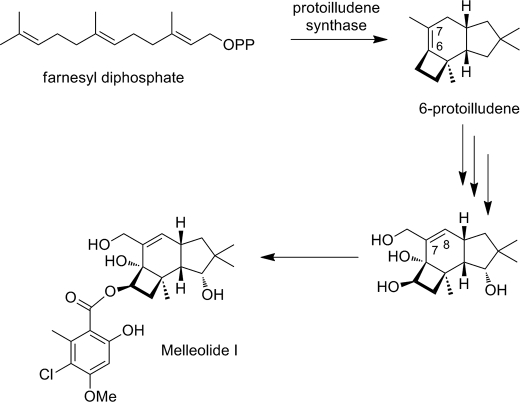
The biosynthesis of protoilludene is thought to involve cyclization of the universal sesquiterpenoid precursor farnesyl diphosphate to protoilludene followed by further modification by cytochrome P450 mono-oxygenases and subsequent attachment of the polyketide side chain (Fig. 1). In this study, we describe the partial purification and characterization of A. gallica protoilludene synthase and the cloning and expression of the corresponding cDNA. We found that A. gallica protoilludene synthase exclusively produces 6-protoilludene as a reaction product, requiring a double bond rearrangement from the 6–7 to the 7–8 position encountered in the melleolides.
FIGURE 1.
Plausible scheme of melleolide I biosynthesis involving the cyclization of farnesyl diphosphate to 6-protoilludene, oxygenation reactions, and the side chain attachment.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Armillaria gallica strain FU02472 was established from basidiocarps collected near Traunsee, Austria and was propagated in submerged culture in 500-ml Erlenmeyer shake flasks containing 200 ml of YMG medium at 23 °C with agitation at 140 rpm, as described previously (35). Mycelia were harvested from the culture broth by filtration, shock-frozen with liquid nitrogen, and stored at −80 °C. E. coli strain TOP10 (Invitrogen) was used for cloning, and strain BL21(DE3) CodonPlus (Agilent) was used for heterologous protein expression, along with the GatewayTM-compatible vector pDEST14 (Invitrogen).
Protein Purification and Biochemical Characterization
Shock-frozen A. gallica mycelia were powdered using a mortar and pestle and transferred into 5 volumes (w/v) of extraction buffer (50 mm MES, 20 mm MgCl2, 5 mm 2-mercaptoenthanol, 10% (v/v) glycerol, and 0.1 g/g mycelia poly(vinylpolypyrrolidone), pH 6.5). After additional treatment with an Ultraturrax, the protein extract was cleared by centrifugation. Protein concentration was determined using the Quick Start Bradford Protein Assay (Bio-Rad). E. coli protein extracts were prepared by pelleting overnight cultures and resuspending the cell pellet in extraction buffer (see above), followed by lysis under constant cooling by two rounds of microfluidizer treatment. Protoilludene synthase activity was determined using [1-3H]-farnesyl diphosphate (20 Ci/mmol) (Biotrend, Cologne) in assay buffer (50 mm MOPS, 20 mm MgCl2, 5 mm 2-mercaptoethanol, pH 7.2). Standard protoilludene synthase activity measurements were performed with 500 nm of [1-3H]-farnesyl diphosphate for 2 min followed by quenching with ethyl acetate. Part of the organic extract was then spotted onto silica-gel TLC plates and separated using 9:1 cyclohexane:ethyl acetate as the solvent, prior to analysis in a radio-TLC reader (Raytest). To determine Km values, the reactions were stopped by quenching with 100 mm EDTA (final concentration) followed by extraction with n-pentane, purification by silica gel column chromatography and quantitation by liquid scintillation counting. All kinetic activity assays were performed in triplicate. Mass spectrometry analysis of solvent extracts was performed on a QP2010S quadrupole mass spectrometer (Shimadzu, Duisburg) equipped with an RxiTM-5 ms (0.25-mm inner diameter and 30-m length) column (Restek, Bad Homburg) using the following temperature program: 80 °C for 20 min, followed by heating the column at a rate of 15 °C/min to 300 °C with a final constant temperature of 300 °C for 4 min. Fragmentation was achieved by electric ionization at 1 keV.
cDNA Library Construction and Sequencing
An A. gallica CloneMinerTM cDNA library (Invitrogen) was constructed according to the manufacturer's protocol using a cesium chloride density gradient and A. gallica strain FU02472 mRNA purified by Oligotex (Qiagen). Recombinant E. coli were selected on 2YT-agar plates containing 50 μg/ml kanamycin, and 2800 randomly picked colonies were transferred to 96-well microtiter plates containing 200 μl 2YT medium per well with 50 μg/ml kanamycin for selection. The plates were incubated at 37 °C with continuous shaking at 160 rpm for ∼12 h. A. gallica cDNAs were amplified directly from the culture using forward primer (5′-CTC GCG TTA ACG CTA GCA TGG ATG-3′) and reverse primer (5′-GTG AGT CGT ATT ACA TGG TCA TAG CTG-3′). PCR products were cleaned and sequenced by Raphael Soeur (Fraunhofer IME Aachen, Functional and Applied Genomics Group) using primer (5′-CGA CGG CCA GTC TTA AGC TCG GGC-3′) on an Applied Biosystems 3730 DNA Analyzer. Sequence data were analyzed using CLC Combined Workbench software (version 3; CLC bio), the Lasergene Package (DNASTAR), NCBI BLASTx, and Local BLAST.
Heterologous Expression of A. gallica Protoilludene Synthase in E. coli
Where cDNA sequencing identified potential terpene synthase clones, the corresponding pENTRY vectors were used in LR recombination reactions involving the pDEST14 destination vector. The resulting expression constructs were then introduced into E. coli BL21(DE3) CodonPlus cells (Stratagene) for heterologous expression. Recombinant bacteria were cultivated in Fernbach-baffled flasks and were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside when the A600 reached 0.5. The induced bacteria were maintained at 28 °C for 8 h with constant shaking at 160 rpm. Cells were then harvested by centrifugation, resuspended in protoilludene assay buffer, and lysed using a microfluidizer.
Genomic DNA Isolation and Southern Blot Hybridization
A. gallica genomic DNA was isolated using the cetyltrimethylammonium bromide method, and 120 μg was digested with 50 units of BamHI, EcoRI, or HindIII (New England Biolabs), as appropriate, for 8 h. The digested DNA was fractionated by 0.7% agarose gel electrophoresis at a constant 50 V overnight, transferred to a positively charged nylon membrane (Roche Applied Science) and prehybridized with Roti®-Hybri-Quick (Roth) containing single-stranded salmon sperm DNA. Two nucleic acid probes (∼400 bp) were synthesized by PCR using forward primer (5′-CCT TCC TGA TAC TCT TGC CAA CTG-3′) and reverse primer (5′-CCT CCT CCG TCG AGA CGT CCG AGT AC-3′) for probe 1 and forward primer (5′-GTC ATC AAT CAT CCG GTT ATC AAA G-3′) and reverse primer (5′-CTT GGG CAT CAG CGT TAT CCA CCT C-3′) for probe 2. These products were labeled with [α-32P]dATP (Hartmann Analytic, Braunschweig) using the DecaLabelTM DNA Labeling kit (Fermentas) according to the manufacturer's recommendations.
Isolation of an A. gallica Protoilludene Synthase Genomic Clone
The genomic clone encompassing the A. gallica protoilludene synthase gene was isolated by amplifying 100 ng of A. gallica genomic DNA using forward primer (5′-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CGA AGG AGA TAG AAC CAT GTC TCA ACG CAT CTT CCT TCC TG-3′), reverse primer (5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTT TAG AGA TGA AAT CCG TCA ACA ATT TGA GG-3′) and Herculase® II Fusion DNA Polymerase (Stratagene) in a 50-μl reaction. The PCR product was purified and sequenced as described above.
RESULTS
Characterization of Melleolide Biosynthesis by A. gallica Cell Culture
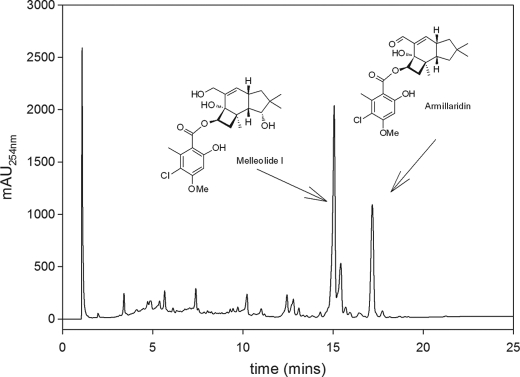
To isolate and characterize a putative honey mushroom (A. gallica) protoilludene synthase, we established a cell culture (FU02472) from a mushroom specimen collected near Traunsee, Austria. To examine secondary metabolite formation, the culture was cultivated by fermentation in liquid YM6.3 medium for 500 h to promote melleolide production. The culture was then harvested, and the melleolide product profile was characterized by LC-UV-MS using appropriate reference substances. The major melleolides produced by FU02472 were identified as melleolide I and armillaridin (Fig. 2). We also investigated the melleolide accumulation profile and changes in protoilludene synthase activity over time by sampling the culture at different time points (Fig. 3).
FIGURE 2.
HPLC analysis of A. gallica strain FU02472 submerged-culture organic extract. Using available standards, two of the observed compound peaks were identified as melleolide I and armillaridin. mAu, milli absorption units.
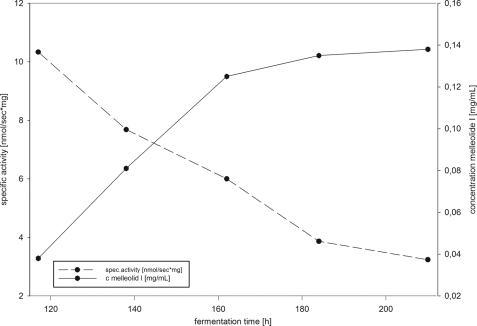
FIGURE 3.
Time course of protoilludene synthase activity and accumulation of melleolide I during liquid fermentation. The graph shows the accumulation of melleolide I and the detected protoilludene synthase activity over a 220-h fermentation of A. gallica strain FU2472.
Protoilludene Synthase Activity
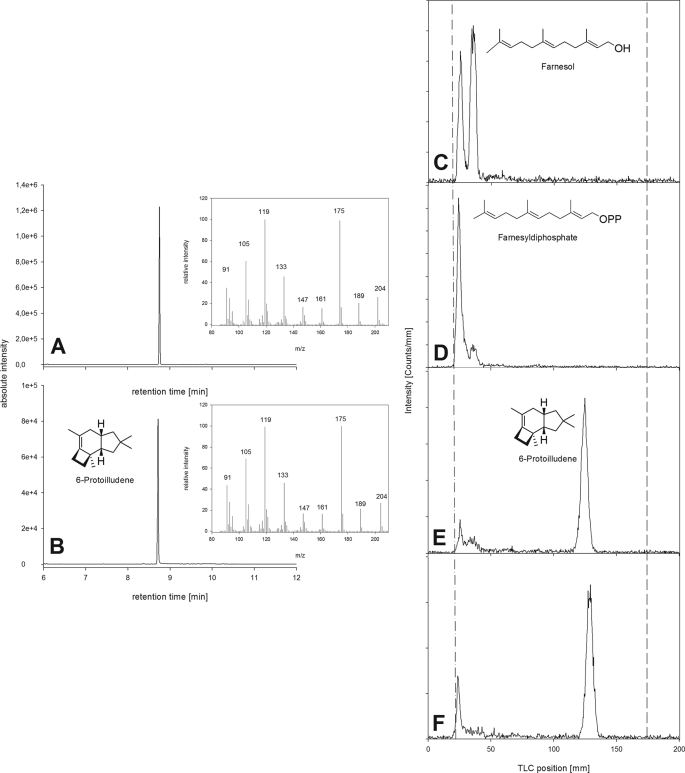
Enzyme activity was tested by incubating soluble enzyme extracts from the culture with radioactively labeled farnesyl diphosphate. Organic extracts from these reactions were then analyzed by radio-TLC (Fig. 3) Variable levels of protoilludene synthase activity were observed in all the crude protein extracts we tested, generating a strongly nonpolar product with an RF value of 0.7 (tentatively identified as 6-protoilludene) and a product with an RF value of 0.1 (tentatively identified as farnesol). The identity of both products was later confirmed by GC-MS (see below). Thermally inactivated control fractions were unable to convert farnesyl diphosphate into nonpolar products. The highest protoilludene synthase activity was observed after 185 h in culture, and this time point was therefore chosen for enzyme purification. To confirm that the putative protoilludene synthase activity in the soluble protein fraction was indeed due to a protoilludene synthase enzyme present in the fungal extract, cold farnesyl diphosphate was spiked with tritium-labeled material and incubated with the FU02472 soluble protein extract for 12 h. The radioactive fraction was then analyzed by radio-TLC and GC-MS to confirm the RF values and identities of the products. The mass spectrum of the extracted product yielded ions at an m/z of 175 (100%), 119 (91%), 105 (59%) 133 (35%), 91 (40%), 189 (17%), 161 (15%), and 147 (14%), with the molecular parent ion at an m/z of 204 (24%), which matched the fragmentation pattern previously reported for 6-protoilludene (Fig. 4) (36, 38). Surprisingly, there was no evidence indicating the presence of 7-protoilludene among the reaction products.
FIGURE 4.
Nonradioactive and radioactive assays to confirm that the Pro1 clone expressed in E. coli has protoilludene synthase activity. A and B, gas chromatograms showing a peak at retention time 8.77 min with characteristic mass spectra. A, extract from E. coli clone expressing Pro1. B, extract from A. gallica mycelia. C–F, radio-TLC assays: the dashed/dotted line is the start, and the dashed line is the solvent front. C, E. coli pUC19 negative control. D, E. coli Pro1 clone incubated with [1-3H]GGPP, a substrate for diterpene synthases. E, E. coli Pro1 clone incubated with [1-3H]farnesyl diphosphate. F, A. gallica positive control.
Enzymatic Characterization of A. gallica Protoilludene Synthase
Further characterization of the enzyme was carried out using partially purified A. gallica protoilludene synthase, tritium-labeled farnesyl diphosphate and radio-TLC analysis. These experiments revealed a Km for farnesyl diphosphate of 0.53 μm. As expected, we found that the enzyme activity was absolutely dependent on divalent metal ions. The highest protoilludene synthase activity was achieved in the presence of 5 mm MgCl2, and it fell by 75% when MgCl2 was replaced with MnCl2. The temperature optimum was 22 °C, with nearly complete loss of activity at temperatures >35 °C. Several buffering systems were tested at 50 mm, with optimal activity at pH 7.2 (MOPS buffer). The activity fell by 50% at pH 5.8 (MES buffer) and pH 8.5 (Tris buffer). The enzyme was also sensitive to the presence of ethanol, with concentrations as low as 5% causing a dramatic reduction in activity. Attempts to purify the A. gallica protoilludene synthase to homogeneity and determine the N-terminal amino acid sequence by Edman sequencing proved unsuccessful, but they showed that the enzyme was likely a 45-kDa monomer.
Cloning of A. gallica Protoilludene Synthase
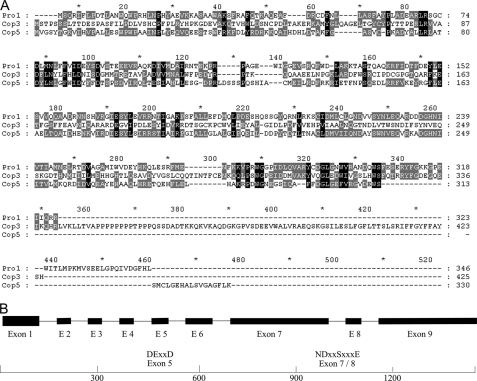
A cDNA library was constructed from the A. gallica FU02472 mycelial culture, and the analysis of 2592 randomly chosen clones led to the identification of six partial sequences with homology to fugal terpene synthases, e.g. from C. cinerea (Coprinus cinereus) (20). Five of these sequences (two full-length cDNAs, one partial cDNA, and two clones containing introns) represented a single putative protoilludene synthase gene, designated Pro1. Further analysis of Pro1 revealed an open reading frame 1041 bp in length encoding a protein with 347 amino acid residues and a predicted molecular mass of 40 kDa. The Pro1 polypeptide contained DEXXD and NDxxSxxxE motifs in the appropriate orientation, as is characteristic for other terpene synthases (40). The Pro1 amino acid sequence was used to search GenBankTM, revealing close relationships with the Cop3 and Cop5 sesquiterpene synthases from the basidiomycete Coprinopsis cinerea (32.6 and 33% identity, respectively) (Fig. 4). Cop3 is an α-muurolene synthase, whereas the precise function of Cop5 remains to be determined (20). No significant homology was observed between Pro1 and terpene synthases from ascomycetes or plants.
Heterologous Expression of Pro1 in E. coli
Heterologous expression of the Pro1 gene in E. coli resulted in a crude soluble protein extract possessing sufficient sesquiterpene synthase activity to convert 80% of the 0.5 μm tritium-labeled farnesyl diphosphate substrate into a product matching the properties of 6-protoilludene (RF = 0.7) within 5 min. Incubation of the same lysate with geranylgeranyl diphosphate did not produce significant amounts of a less polar product (<1%), and neither farnesyl diphosphate nor geranylgeranyl diphosphate was converted into less polar products when using E. coli control lysates derived from the empty vector control (Fig. 4). Characterization of the heterologous terpene synthase using cold farnesyl diphosphate, after pentane extraction and analysis of the organic extract by GC-MS, revealed the formation of a product with an identical retention time and mass spectrum as the native protoilludene synthase.
Isolation of Genomic Protoilludene Synthase Gene Sequence
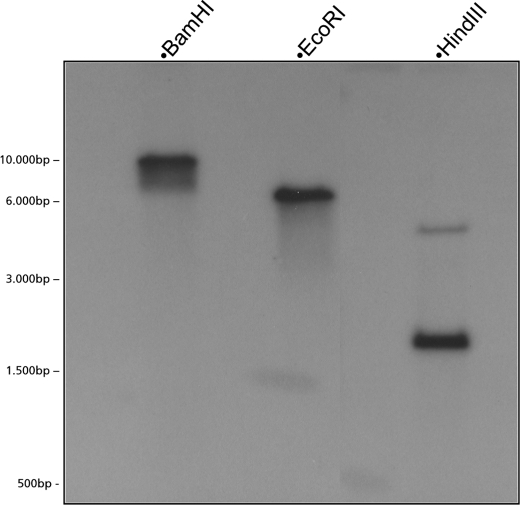
Secondary metabolite biosynthetic pathways in microorganisms are often arranged in biosynthetic gene clusters. Thus, the isolation of the protoilludene synthase genomic clone would facilitate the isolation of a potential melleolide biosynthetic gene cluster in A. gallica. By using the 1270-bp full-length Pro1 cDNA clone, we designed primers to amplify the corresponding genomic DNA sequence (Fig. 5). Analysis of the 1645-bp product revealed the presence of eight introns and nine exons, with some as short as 50 bp. The observed pattern of intron/exon structures differed significantly from the patterns observed in a previous analysis of plant terpene synthase genes (7). Southern blot hybridization, using oligonucleotide probes representing the 5′ and 3′ ends of the Pro1 clone, respectively, revealed only a single copy of the Pro1 gene in the A. gallica genome (Fig. 6).
FIGURE 5.
A, deduced amino acid sequence alignment of fungal sesquiterpene synthases. The sequences of A. gallica protoilludene synthase (clone Pro1), Coprinopsis cinerea α-muurolene synthase (Cop3) and Cop5 (a sesquiterpene synthase with yet unknown product) are compared. Black boxes indicate identical residues for the three sequences; gray boxes indicate identical residues for two of the three sequences. B, a schematic presentation of the A. gallica protoilludene synthase genomic sequence with exons (shown as black boxes) and conserved DEXXD and NDxxSxxxE motifs.
FIGURE 6.
Determination of A. gallica protoilludene synthase gene copy number by Southern blot hybridization. One clear band is visible in the BamHI and EcoRI lanes (neither enzyme has a target site in the genomic clone), whereas two bands are visible in the HindIII lane (which has two target sites in the clone, 85 bp apart).
DISCUSSION
The emergence of multi-drug-resistant microbial pathogens, such as methicillin-resistant strains of S. aureus, has triggered a resurgence of interest in novel antibiotics with new mechanisms of action. Natural products traditionally represent a good source of antibiotic substances. Although terpenoids represent the largest as well as most diverse group of natural products, only two antibiotics based on a fungal sesquiterpenoid are currently in use.
Melleolides from the Armillaria species have long been known for their potent antibacterial activity (31, 32, 37). However, limited supplies of individual melleolides have prevented them from being more extensively examined and clinically developed. Metabolic pathway engineering in easily culturable microbial hosts such as Saccharomyces cerevisiae or Aspergillus nidulans may provide an efficient method of producing advanced melleolide precursors for semisynthesis or the desired product by total fermentation.
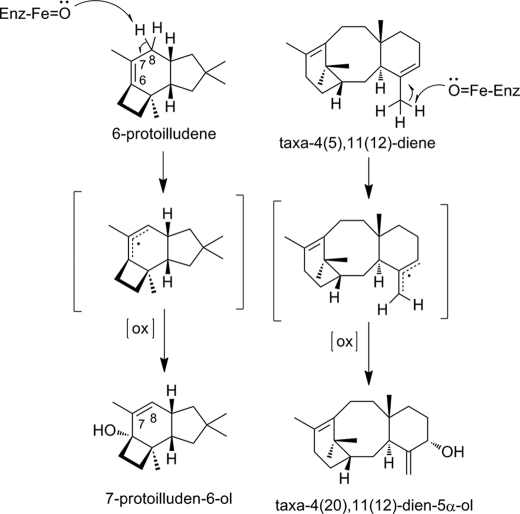
For successful metabolic engineering, the functional characterization and molecular cloning of the underlying enzymatic steps is a key prerequisite. The cyclization of the universal sesquiterpenoid building block franesyl diphosphate to 6-protoilludene constitutes the initial pathway-committing step in the biosynthesis of all melleolides. Phytochemical analysis of Formitopsis insularis has led to the isolation of 6-protoilludene and 7-protoilluden-6-ol (38). Using a chemically synthesized reference, the analysis of the organic extract of Formitopsis insularis also yielded no indication of the formation of 7-protoilludene. Thus, the observed product 6-protoilludene instead of 7-protoilludene requires a subsequent rearrangement of the double bond from the 6–7 to the 7–8 position to generate the melleolide end products (Fig. 7).
FIGURE 7.
Proposed mechanism for double bond rearrangement and hydroxylation of 6-protoilludene to 7-protoilluden-6-ol involving the abstraction of a hydrogen at the C7, yielding an allylic radical, followed by the stereoselective insertion of an oxygen. A similar reaction, involving double bond rearrangement and stereoselective oxygen insertion, is found in the biosynthesis of the diterpenoid taxol, for which the underlying enzymatic activity has been identified as a cytochrome P450-dependent mono-oxygenase (39).
A similar rearrangement has been described for the biosynthesis of taxol (paclitaxel), where taxa-4(5),11(12)-diene is converted to taxa-4(5),11(12)-diene-5α-ol by a cytochrome P450-dependent mono-oxygenase (39). On basis of the isolated intermediate 7-protoilludene-6-ol, we postulate as a subsequent biosynthesis step a similar concomitant double bond rearrangement followed by an oxidation reaction at the C6 position catalyzed by a cytochrome P450-dependent mono-oxygenase.
To further elucidate the melleolide biosynthetic pathway in A. gallica, we cloned the genomic sequence of the identified 6-protoilludene synthase, assuming a potential clustering of the genes involved in the melleolide biosynthesis. Sequencing of the genomic clone identified a 1645-bp sequence with eight introns interrupting the open reading frame. A comparison of angiosperm and gymnosperm terpene synthases has identified distinct classes of plant terpene synthases based on intron/exon pattern, suggesting an ancestral terpenoid synthase containing 12 introns and 13 exons as a common evolutionary precursor (7). Sequence comparisons at the amino acid and genomic levels underscore the distant evolutionary relatedness of fungal terpene synthases to the more extensively studied plant terpene synthases.
In summary, our analysis of a partially purified A. gallica protoilludene synthase and the expression of a corresponding cDNA in E. coli have provided important insight into the melleolide biosynthesis pathway in fungi and may contribute to the development of new strategies for the synthesis of novel antimicrobial compounds.
Acknowledgments
We are grateful for expert technical assistance from Raphael Soeur (Fraunhofer IME), Beate Schmieschek, Dirk Müller, and Stefan Heke (InterMed Discovery GmbH). We thank professor Ruth Seeger for providing the A. gallica culture.
Footnotes
This work was supported by a grant from Arbeitsgemeinschaft industrielle Forschungsvereinigungen (Otto von Guericke) Pro Inno II.
REFERENCES
- 1. Conolly J. D., Hill R. A. (1991) Dictionary of Terpenoids, Chapman & Hall, London [Google Scholar]
- 2. Ayer W. A., Browne L. M. (1981) Tetraedron 37, 2199–2248 [Google Scholar]
- 3. Lorenzen K., Anke T. (1998) Curr. Org. Chem. 2, 329–364 [Google Scholar]
- 4. Sacchettini J. C., Poulter C. D. (1997) Science 277, 1788–1789 [DOI] [PubMed] [Google Scholar]
- 5. Davis E. M., Croteau R. (2000) Topics Curr. Chem. 209, 53–95 [Google Scholar]
- 6. Christianson D. W. (2006) Chem. Rev. 106, 3412–3442 [DOI] [PubMed] [Google Scholar]
- 7. Trapp S. C., Croteau R. B. (2001) Genetics 158, 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tholl D. (2006) Curr. Opin. Plant. Biol. 9, 297–304 [DOI] [PubMed] [Google Scholar]
- 9. Kawaide H., Imai R., Sassa T., Kamiya Y. (1997) J. Biol. Chem. 272, 21706–21712 [DOI] [PubMed] [Google Scholar]
- 10. Lesburg C. A., Zhai G., Cane D. E., Christianson D. W. (1997) Science 277, 1820–1824 [DOI] [PubMed] [Google Scholar]
- 11. Caruthers J. M., Kang I., Rynkiewicz M. J., Cane D. E., Christianson D. W. (2000) J. Biol. Chem. 275, 25533–25539 [DOI] [PubMed] [Google Scholar]
- 12. Dairi T., Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H. (2001) J. Bacteriol. 183, 6085–6094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rynkiewicz M. J., Cane D. E., Christianson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13543–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hamano Y., Kuzuyama T., Itoh N., Furihata K., Seto H., Dairi T. (2002) J. Biol. Chem. 277, 37098–37104 [DOI] [PubMed] [Google Scholar]
- 15. Toyomasu T., Nakaminami K., Toshima H., Mie T., Watanabe K., Ito H., Matsui H., Mitsuhashi W., Sassa T., Oikawa H. (2004) Biosci. Biotechnol. Biochem. 68, 146–152 [DOI] [PubMed] [Google Scholar]
- 16. Toyomasu T., Tsukahara M., Kaneko A., Niida R., Mitsuhashi W., Dairi T., Kato N., Sassa T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3084–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kawaide H. (2006) Biosci. Biotechnol. Biochem. 70, 583–590 [DOI] [PubMed] [Google Scholar]
- 18. Shishova E. Y., Di Costanzo L., Cane D. E., Christianson D. W. (2007) Biochemistry 46, 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinedo C., Wang C. M., Pradier J. M., Dalmais B., Choquer M., Le Pêcheur P., Morgant G., Collado I. G., Cane D. E., Viaud M. (2008) ACS Chem. Biol. 3, 791–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agger S., Lopez-Gallego F., Schmidt-Dannert C. (2009) Mol. Microbiol. 72, 1181–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez-Gallego F., Agger S. A., Abate-Pella D., Distefano M. D., Schmidt-Dannert C. (2010) ChemBioChem. 11, 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hohn T. M., Beremand P. D. (1989) Gene 79, 131–138 [DOI] [PubMed] [Google Scholar]
- 23. Hohn T. M., Plattner R. D. (1989) Arch. Biochem. Biophys. 272, 137–143 [DOI] [PubMed] [Google Scholar]
- 24. Cane D. E., Shim J. H., Xue Q., Fitzsimons B. C., Hohn T. M. (1995) Biochemistry 34, 2480–2488 [DOI] [PubMed] [Google Scholar]
- 25. Hohn T. M., Desjardins A. E., McCormick S. P. (1995) Mol. Gen. Genet. 248, 95–102 [DOI] [PubMed] [Google Scholar]
- 26. Cane D. E., Kang I. (2000) Arch. Biochem. Biophys. 376, 354–364 [DOI] [PubMed] [Google Scholar]
- 27. Kirk P. F., Cannon P. F., Minter D. W., Stalper J. A. (eds) (2008) Dictionary of the Fungi, 10th Ed., CABI Publishing, Egham, United Kingdom [Google Scholar]
- 28. Heinzl S. (2006) Chemother. J. 15, 40–44 [Google Scholar]
- 29. Abraham W. R. (2001) Curr. Med. Chem. 8, 583–606 [DOI] [PubMed] [Google Scholar]
- 30. Tainter F. H., Baker F. A. (1996) Principles of Forest Pathology, Wiley, Chichester, United Kingdom [Google Scholar]
- 31. Smith M., Bruhn J., Anderson J. (1992) Nature 356, 428–431 [Google Scholar]
- 32. Arnone A., Cardillo R., Nasini G. (1986) Phytochemistry 25, 471–474 [Google Scholar]
- 33. Obuchi T., Kondoh H., Watanabe N., Tamai M., Omura S., Yang J. S., Liang X. T. (1990) Planta Med. 56, 198–201 [DOI] [PubMed] [Google Scholar]
- 34. Laatsch H. (2010) AntiBase 2010: The Natural Compound Identifier, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 35. Stadler M., Tichy H. V., Katsiou E., Hellwig V. (2003) Mycol. Progr. 2, 95–122 [Google Scholar]
- 36. Joulain D., König W. A. (1998) The Atlas of Spectral Data of Sesquiterpene Hydrocarbons, EB-Verlag, Berlin [Google Scholar]
- 37. Oduro K. A., Munnecke D. E., Sims J. J., Keen N. T. (1976) Trans. Br. Mycol. Soc. 66, 195–199 [Google Scholar]
- 38. Nozoe S., Kobayashi H., Urano S., Furukawa J. (1977) Tetrahedron Lett. 18, 1381–1384 [Google Scholar]
- 39. Jennewein S., Long R. M., Williams R. M., Croteau R. (2004) Chem. Biol. 11, 379–387 [DOI] [PubMed] [Google Scholar]
- 40. Vedula S. L., Jiang J., Zakkarian T., Cane D. E., Christianson D. W. (2008) Arch. Biochem. Biophys. 469, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]