Abstract
We demonstrate that the levels of native as well as transfected prion protein (PrP) are lowered in various cell lines exposed to phosphorothioate oligodeoxynucleotides (PS-DNA) and can be rapidly reverted to their normal amounts by removal of PS-DNA. This transient modulation was independent of the glycosylation state of PrP, and in addition, all three PrP glycoforms were susceptible to PS-DNA treatment. Deletion of the N-terminal domain (amino acids 23–99), but not of the other domains of PrP, abrogated its PS-DNA-mediated down-regulation. PrP versions localized in the mitochondria, cytoplasm, or nucleus were not modulated by PS-DNA, indicating that PrP surface exposure is required for executing this effect. Proteins that in their native forms were not responsive to PS-DNA, such as thymocyte antigen 1 (Thy1), Doppel protein (Dpl), green fluorescent protein (GFP), and cyan fluorescent protein (CFP), became susceptible to PS-DNA-mediated down-regulation following introduction of the N terminus of PrP into their sequence. These observations demonstrate the essential role of the N-terminal domain for promoting oligonucleotide-mediated reduction of the PrP level and suggest that transient treatment of cultured cells with PS-DNA may provide a general method for targeted modulation of the levels of desired surface proteins in a conditional and reversible manner.
Keywords: Membrane Proteins, Prions, Protein Degradation, Protein DNA-Interaction, siRNA, Oligotherapy
Introduction
Prion diseases are fatal neurodegenerative disorders caused by the conversion of the cellular prion protein (PrPC)4 into its infectious conformation, PrPSc (1). The accumulation of PrPSc in the central nervous system results in neurodegeneration and lethal disease progression by a mechanism that is still unknown (2, 3). PrPC is a phylogenetically conserved protein (4, 5), which is present in a wide range of cell types besides neuronal cells and is localized mainly at the plasma membrane, although cytosolic and nuclear localizations were described (6). Nevertheless, prion disease pathogenesis seems to depend on its membrane localization (7). Neuronal PrP was demonstrated to play a role in neuroprotection (8) and to be involved in cell signaling (9, 10). The important role attributed to PrP based on its prevalence is complicated by the observation that mice lacking PrP display normal development and behavior (11). Recently, it was demonstrated that axonal prion protein is required for peripheral myelin maintenance (12). Thus, the physiological role of PrP is yet to be fully elucidated.
PrPC proteins possess high ability to bind short nucleic acid molecules regardless of their sequence, and it was suggested that this DNA binding aptitude may be involved in the pathogenicity of prion diseases (for a review, see Ref. 13). The use of oligodeoxynucleotides as a modality of affecting PrPSc levels was suggested by in vivo studies reported by Sethi et al. (14), who have documented that co-inoculation of mice with prions and oligodeoxynucleotides significantly prolonged survival times. This effect was suggested to reflect the elimination of PrPSc following activation of the innate immune response by the CpG motif in the oligodeoxynucleotides. Other studies showed that prion pathogenesis can be prevented by such treatments in the absence of the receptor that specifically binds CpG oligonucleotides (15), suggesting that the suppression of disease was not mediated by the activation of the immune system through the CpG motif but more likely by direct interaction of PrP with nucleic acids.
Modified oligodeoxynucleotides such as phosphorothioate oligodeoxynucleotides (PS-DNA), which exhibit reduced enzymatic degradation, were shown to down-regulate levels of both PrPSc and PrPC in mouse-scrapie cells and animal models (16, 17). The ability to prevent or eliminate scrapie infectivity in cell culture correlated with the size of PS-DNA (>18-mer) and was sequence-independent. Single-stranded and double-stranded PS-DNA, but not PS-RNA, were equally able to promote a drop in PrPSc levels and did not involve inhibition of the translation or transcription machinery but rather an effect on preexisting PrP (16). Examination of the interaction of PS-DNA with PrP established that PS-DNA may directly bind PrP and that PS-DNA may co-localize with internalized PrP in both infected and uninfected cells (18). This suggested that the anti-scrapie effect of PS-DNA might not be through interaction with PrPSc but rather with PrPC. PrP degradation in the presence of PS-DNA is probably occurring at the lysosome (14), and binding of PS-DNA to PrPC leads to its increased internalization (18).
The studies documented in the present report further examine PrP down-regulation in response to PS-DNA exposure. We demonstrate that the N terminus domain of PrP is essential for the susceptibility to PS-DNA-mediated diminution of plasma membrane PrP levels in a variety of cell lines. Chimeric heterologous proteins, engineered to contain the PrP N terminus, could be endowed with PS-DNA sensitivity, suggesting that the PS-DNA induced down-regulation may serve as the basis for a general method for modulation of protein levels.
EXPERIMENTAL PROCEDURES
Plasmids
The following constructs were described previously: g1, g2, and g12 (19); mutants 1, 2, 3, 5, 12, and 25 (20, 21); CD4-PrP, NT-Thy1, and Thy1 (22); MoXenPrP (23); and PrP targeted to the cytoplasm (CytoPrP) (24), the nucleus (NucPrP), and the mitochondria (MitoPrP) (25). For cloning of PrP-Thy1, a PCR fragment encompassing the GPI attachment signal of murine Thy1 protein (amino acids 127–162) was fused by standard cloning techniques to a truncated 3F4-PrP comprising residues 1–231. All amino acid numbers refer to the mouse PrP sequence (GenBankTM accession number M18070). All plasmid constructs were sequenced using the primers for sequence verification, and amplified in Escherichia coli (DH10B), kindly provided by Dr. Saul Burdman.
Treatment of Cells with Oligodeoxynucleotides
The following purified PS oligodeoxynucleotides were purchased from TriLink Biotechnologies (San Diego, CA): CpG 22-mer CpG-PS-DNA, 5′-TGACTGTGAACGTTCGAGTGA-3′; and Scrambled 22-mer SCR-PS-DNA, 5′-CAGTGATAGCTATGTGAGCTAG-3′. In all experiments, 10 μm PS-DNA was added to the medium for 24 h unless otherwise noted. Following treatment of cells with oligodeoxynucleotides, cells were washed with PBS, and 100 μl of lysis buffer (10 mm Tris-HCl, pH 8; 100 mm NaCl; 0.5% Nonidet P-40; and 0.5% deoxycholate) was added to the 60-mm plates for 5 min on ice. DNA aggregates were collected from the lysate using a sterile tip.
Cell Cultures and Transfections
Murine neuroblastoma cell lines (N2a), Chinese hamster ovary cells (CHO), and hypothalamic cell line infected with scrapie and stably transfected with prion protein (ScGT1-MHM2) were kindly provided by Dr. Albert Taraboulos (16, 26). Human embryonic kidney cells (HEK293) were purchased from ATCC. Cells were grown, treated with oligodeoxynucleotides, harvested, and maintained as described (13), unless otherwise noted. Cell lines were grown in DMEM medium enriched with 10% v/v FBS (Kibbutz, Beth Ha'Emek, Israel), 1% v/v l-glutamine (Kibbutz, Beth Ha'Emek, Israel), and 1% v/v penicillin-streptomycin (Sigma); ScGT1 cells were grown in 50% v/v Opti-MEM medium (Invitrogen), 43.5% DMEM, 5% v/v FCS, 0.5 v/v l-glutamine, and 1% v/v penicillin-streptomycin. Transient transfection of the cells was performed using the FuGENE kit with 2 μg of DNA construct per transfection. Seventy-two hours after transfection, cells were exposed to PS-DNA. For stable transfections, 72 h after transfection, cells were exposed to selective medium containing 1 g/liter G418. Stably transfected cells were established after a period of 3–4 weeks of selection.
Antibodies and Immunoblots
Protein concentration in each sample was determined using a Bradford reagent (Bio-Rad). Samples were loaded onto 12% SDS-PAGE gels. In all experiments, 100 μg/well of total protein lysates was loaded, unless otherwise noted. Prestained protein molecular weight markers were purchased from Fermentas. Western blots were generated by standard procedures (16). PrP was detected using Fab D13 (27) (InPro Biotechnology). Goat anti-human Fab was used as secondary antibody (Pierce). Levels of transfected PrP or PrP protein fusions containing the hamster PrP epitope 109–112 (see Fig. 2c for location of this epitope) were detected using the 3F4 (Sigma). Anti-tubulin and anti-actin monoclonal antibodies were from Sigma. Blots probed with monoclonal antibodies were developed using a goat anti-mouse IgG. Levels of MoDpl, NT-Thy1, and Thy1 were detected as described in Ref. 22. Levels of CytoPrP, NucPrP, and MitoPrP were detected as described (24, 25). All secondary antibodies were peroxidase-conjugated, enabling ECL visualization of proteins.
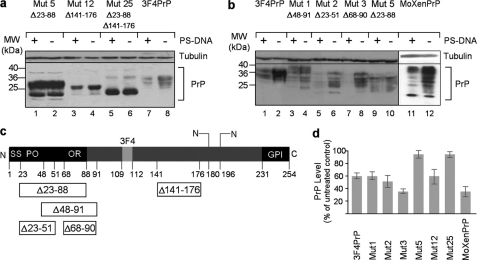
FIGURE 2.
PrP N terminus is essential for PrP down-regulation in the presence of PS-DNA. a and b, Western blot analysis of PS-DNA-treated N2a cells transiently transfected with various deletion-mutated forms of PrP. MW, molecular mass markers; Mut, mutation. c, schematic representation of PrP. SS, signal sequence; PO, pre-octarepeat domain; OR, octarepeat domain; GPI, glycosylphosphatidylinositol anchor. Deletions are indicated as open boxes containing the aa positions of their boundaries. d, densitometric quantification of Western blot analyses (n = three independent studies, p = 0.009 for mutants 1 and 12, p = 0.0005 for MHM2, mutant 3, and mutant 2, and p = 0.0008 for MoXenPrP).
Tunicamycin and Phosphatidylinositol Phospholipase C (PIPLC) Treatments
Twenty-four hours following 1:10 splitting, the cells were washed, and 1.5 mg/ml tunicamycin was added. PS-oligodeoxynucleotides were then added to the dish for variable periods of time. PIPLC was diluted to a final concentration of 200 μm with 0.05% BSA, 144 mm NaCl, 10 mm Tris-HCl, pH 7.4. N2a cells were grown as described for oligodeoxynucleotide treatment. Oligodeoxynucleotides were then added to the dish (10 μm) for 72 h. Following extensive washing, fresh serum-free medium in the presence or absence of PIPLC (200 mm) was added for 6 h at 37 °C. PrP in the supernatant was concentrated by TCA precipitation.
Densitometric Quantification and Statistics
Protein quantification of Western blots was done using the NIH ImageJ software. PrP signal was normalized to total protein present on Coomassie Blue-stained gels, to tubulin, or to actin signals. The data represent the average and S.D. of three independent experiments for each treatment. Student's t tests were performed for two-group comparisons. For comparisons involving more than two groups, post hoc least squares difference one-way analysis of variance was used. Treatments were considered to be significant when p < 0.05.
RESULTS
PrP Is Efficiently Degraded upon PS-DNA Treatment
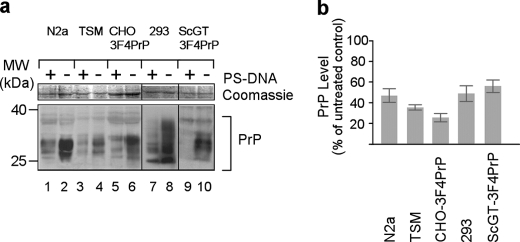
The phenomenon of native PrP down-regulation following PS-DNA treatment was demonstrated in N2a and GT1 neuronal mouse cell lines (16). To explore the possibility that this phenomenon is of a broader nature, CHO cells transfected with 3F4PrP, a murine PrP containing hamster PrP epitope 109-112 (CHO-3F4PrP), mouse neurocortical neuroblast cells (TSM), and human kidney 293 cells, were exposed to 10 μm PS-DNA for 24 h, and the level of PrP in each case was inspected by Western blot analysis. Transfected murine PrP contained the 3F4 epitope enabling specific immunodetection of the transfected proteins but not of the endogenous mouse PrP (see “Experimental Procedures”). Treatment resulted in a significant decrease in the amount of native or recombinant PrP in all cell lines (Fig. 1). PS-DNAs exhibiting two different DNA sequences were used, CpG-PS-DNA and Scr-PS-DNA. Treatment with either of these sequences generated identical results verifying our initial observation that the PS-DNA-mediated PrP down-regulation is independent of the PS-DNA sequence (16). We observed that shorter incubations (2 h) are sufficient to promote significant lowering of PrP levels (>50% as compared with untreated cells, supplemental Fig. 3, compare lane 2 with lane 1). Exposure to PS-DNA for 8 h was more potent than the 4-h treatment (80% PrP level diminution, supplemental Fig. 1, compare lane 2 with lane 4). We observed rapid restoration of the PrP levels following termination of the treatment, to 80% of the initial levels (supplemental Fig. 1, lane 3). A longer recovery time of 24 h fully restored the PrP levels (supplemental Fig. 1, lane 6).
FIGURE 1.
Native or transfected PrP proteins are down-regulated in various cell lines exposed to PS-DNA. a, Western blot analysis of N2a (mouse neuroblastoma), TSM (mouse neurocortical neuroblasts), CHO-3F4PrP (CHO cells stably transfected with mouse PrP containing the 3F4 PrP epitope), ScGT-3F4PrP (scrapie-infected mouse hypothalamic neuronal cell line stably transfected with 3F4PrP), and HEK 293 cells (human embryonic kidney). PrP was detected using the D13 (lanes 1–4, 7, and 8) or the 3F4 (lanes 5, 6, 9, and 10) anti-PrP antibody. MW, molecular mass markers. b, densitometric quantification of Western blot analyses (n = three independent experiments, p = 0.001).
Glycosylation Is Not Essential for PS-DNA-mediated PrP Down-regulation That Involves Existing as Well as Newly Synthesized PrP
PrP is a glycosylated protein and therefore generates three electrophoretically distinct isoforms distinguished by molecular weight (mouse PrP harbors two N-glycosylation sites at positions 180 and 196). To assess whether this post-translation modification affects PrP down-regulation mediated by PS-DNA, we examined N2a cells transfected with three different mouse PrP recombinant versions (supplemental Fig. 2). In these PrP forms, each glycosylation site or both of them were mutated, resulting in an altered state of glycosylation. We observed that significant down-regulation (50–80% reduction in the level of recombinant PrP) occurs in all cases (supplemental Fig. 2), clearly indicating that PrP glycosylation has no impact on PS-DNA-mediated PrP down-regulation. Next, we addressed the question whether de novo synthesis of PrP is involved in the observed PS-DNA-mediated reduction in the level of PrP. To distinguish between newly synthesized PrP and mature pre-existing PrP, cells were exposed to tunicamycin, which inhibits the N-glycosylation of newly synthesized proteins (28). Accordingly, mature glycosylated PrP forms can be detected as upper higher molecular weight bands on Western blots, whereas the newly synthesized nonglycosylated PrP constitutes mainly the low molecular band. Both newly synthesized as well as mature PrP were down-regulated in the presence of PS-DNA (supplemental Fig. 3).
The N Terminus of PrP Is Essential for the PS-DNA-mediated Down-regulation
To determine whether any specific domain of PrP is essential for the reduction in the level of PrP, PrP mutated versions entailing various deletions were transfected into N2a cells (Fig. 2). The mutated versions of PrP inspected were: mutant 5, exhibiting a deletion between aa positions 23 and 88; mutant 12, exhibiting a deletion between aa positions 141 and 176; and mutant 25, exhibiting two deletions between aa positions 23 and 88 and positions 141 and 176. The data demonstrate that N terminus deletion, specifically residues 23–88, abolished the down-regulation of PrP in response to PS-DNA treatment (Fig. 2a). To further map the domain essential for this effect, three different versions of PrP harboring different deletions within the N terminus were tested: mutant 1, exhibiting a deletion between aa positions 48 and 91; mutant 2, exhibiting a deletion between aa positions 23 and 51; and mutant 3, exhibiting a deletion between aa positions 68 and 90. These partial deletions did not affect the reduction of PrP level, strongly indicating that an intact N terminus is necessary for this phenomenon. The resistance of the N-deleted PrP mutant could be reversed by fusing it with the N terminus domain of Xenopus laevis PrP (Fig. 2b). We note that some deletions conferred a higher susceptibility to PS-DNA treatment as compared with the full-length PrP such as in the case of the aa 68–90 domain removal (Fig. 2b, mutant 3 as compared with mutant 1). This issue needs further investigation, and we conclude that the N terminus integrity is essential for the PS-DNA-mediated down-regulation.
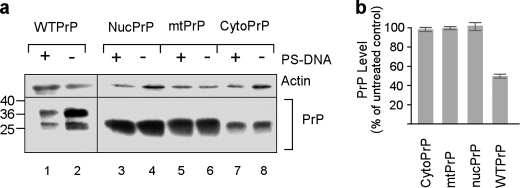
Plasma Membrane Localization Is Essential for Reduction in the Level of PrP
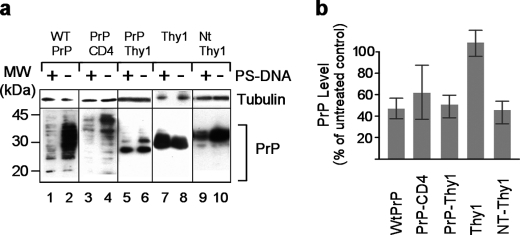
Subcellular localization of PrP can be controlled by exploiting recombinant versions of PrP, which are targeted specifically to the nucleus, mitochondria, cytoplasm, or cell surface of the cells. Accordingly, PrP targeting to the nucleus or mitochondria may be achieved by replacing its endoplasmic reticulum signal sequence with the nuclear localization signal of SV40 large T antigen or the mitochondrial signal sequence of subunit 1 of the ubiquinol cytochrome c reductase complex, respectively (25). Deletion of the endoplasmic reticulum signal sequence leads to cytoplasmic PrP expression (24). Using such PrP versions, we addressed the question whether a specific cellular localization of PrP is essential for its down-regulation in the presence of PS-DNA. CytoPrP, MitoPrP, and NucPrP of N2a cells, which are not imported into the endoplasmic reticulum/Golgi, can be distinguished by the fact that they generate only one electrophoretic band of unglycosylated PrP (Fig. 3a). Neither one of these PrP versions were affected by the presence of PS-DNA, as compared with positive controls entailing plasma membrane-localized PrP (Fig. 3b). The notion that membrane localization is necessary and sufficient was substantiated by experiments employing various membrane attachment modalities, which may alter the PrP microenvironment on the plasma membrane. This issue was examined by substituting the GPI anchor attachment signal (aa 231–254) with either the transmembrane mouse CD4 domain (PrP-CD4) or the GPI of thymocyte antigen 1 (PrP-Thy1). The first modification shifts PrP from the rafts/caveolae-like domain to a clathrin-coated pit trafficking environment (29, 30). The second modification directs PrP to a different domain than PrP within the lipid rafts (31, 32). These two replacements did not affect the PS-DNA-mediated diminution, although the results for PrP-CD4 construct were not statistically significant (Fig. 4). Therefore, the modality by which the protein is attached to the membrane did not affect the reduction in PrP levels following exposure to PS-DNA.
FIGURE 3.
Nuclear, cytosolic, and mitochondrial PrP are not down-regulated in cells exposed to PS-DNA. a, Western blot analysis of N2a cells transiently transfected with plasmid constructs directing PrP into different cellular compartment. mtPrP, MitoPrP. b, densitometric quantification of Western blot analyses (n = three independent studies).
FIGURE 4.
Different membrane-anchoring modalities do not affect PS-DNA-induced PrP down-regulation. a, Western blot analysis of N2a cells transiently transfected with the indicated plasmid constructs. PrP-CD4, fusion of PrP aa 1–230 and the transmembrane domain of mouse CD4 (aa 369–431); PrP-Thy1, fusion of PrP (aa 1–231) and the GPI anchor of Thy1 (aa 127–163); NT-Thy1, fusion of aa 1–22 of PrP, the FLAG epitope, PrP N terminus (aa 23–88), and Thy1. Thy1 was detected using anti-HA antibodies, and NT-Thy1 was detected using anti-FLAG antibodies. All other PrP forms were detected using the 3F4 monoclonal antibody. MW, molecular mass markers. b, densitometric quantification of Western blot analyses (n = three independent studies, p = 0.0001, p = 0.005, p = 0.0004 for 3F4PrP, PrP-Thy1, and for NT-Thy1, respectively).
Taken together, these experiments indicate that only PrP localized at the outer leaflet of the plasma membrane interacts with PS-DNA, leading to its level reduction. This conclusion was strengthened by the observation that cells treated with PS-DNA and subsequently exposed to PIPLC, which cleaves all GPI membrane-associated molecules, released into the medium significantly lower amounts of PrP. These results indicate that the levels of membrane PrP were indeed reduced in the presence of PS-DNA prior to the PIPLC-mediated cleavage (supplemental Fig. 4, compare lane 1 with lane 3).
Attaching PrP at the N Terminus or C Terminus of Other Proteins Leads to Their Down-regulation in the Presence of PS-DNA in Cell Culture
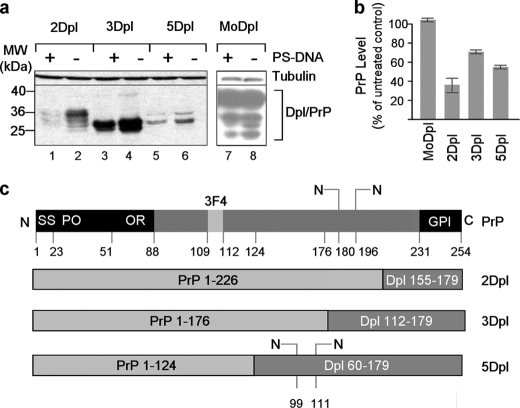
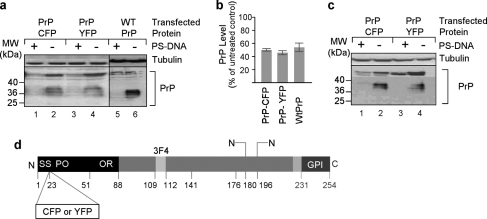
We explored the possibility to promote reduction in the levels of other membrane proteins, which are not affected by PS-DNA, by attaching the PrP N terminus domain into their sequence. One of these proteins previously shown to be resistant to PS-DNA treatment is Dpl (16), despite the high degree of spatial homology that it shares with PrP (33). We therefore used fusion products of different portions of the PrP N terminus with different segments of the C terminus of Dpl and addressed their modulation in N2a cells upon PS-DNA exposure (Fig. 5c). The results indicate that the levels of all PrP-Dpl protein fusions were lowered in the presence of PS-DNA as compared with unaffected Dpl levels (Fig. 5a). We then examined another recombinant protein fusion generated by addition of the N-terminal portion of PrP to the protein Thy1 (PrP-Thy1), which does not share any structural homology with PrP yet is a GPI-anchored membrane protein. The data reveal that the chimeric form of Thy1 became susceptible to PS-DNA treatment due to the PrP N terminus (Fig. 4a, compare lane 8 with lane 7). Furthermore, we inquired whether the levels of proteins that are resistant to PS-DNA and are not anchored to the cell surface in their native form could be lowered in the presence of PS-DNA if tagged with the N terminus of PrP and anchored to the membrane. Accordingly, we generated two chimeric molecules; the first is a cyan fluorescent protein (CFP) recombinant form in which CFP is inserted into PrP downstream of the signal sequence (Fig. 6c, SS) and upstream of the PrP N-terminal degradation-mediating domain. The second molecule involved similar modifications of the yellow fluorescent protein (YFP) (Fig. 6c). Both PrP-CFP and PrP-YFP fusion proteins were susceptible to PS-DNA treatment. This phenomenon was observed both in N2a and in CHO cells (Fig. 6). We therefore conclude that the levels of proteins with no structural homology to PrP can be reduced by PS-DNA treatment when tagged with the N terminus of PrP as long as they are anchored to the cell membrane.
FIGURE 5.
Attachment of PrP at the N terminus of the Dpl protein results in susceptibility to PS-DNA. a, Western blot analysis of N2a cells transiently transfected with different fusion products of PrP and Dpl. MW, molecular mass markers. b, densitometric quantification of Western blot analyses (n = three independent studies, p = 0.04). c, schematic representation of the fusion products examined. The aa positions of the boundaries of PrP (gray boxes) and Dpl (dark boxes) are indicated in each case. SS, signal sequence; PO, pre-octarepeat domain; OR, octarepeat domain; GPI, glycosylphosphatidylinositol anchor.
FIGURE 6.
Attachment of PrP at the C terminus of the cytosolic proteins YFP and CFP promotes PS-DNA susceptibility. a, Western blot analysis of CHO cells transiently transfected with CFP-PrP or YFP-PrP fusion products and treated with PS-DNA. (n = two independent studies). MW, molecular mass markers. b, densitometric quantification of Western blot analyses in panel a. c, Western blot analysis of N2a cells transiently transfected with CFP-PrP or YFP-PrP fusion products and treated with PS-DNA. d, positions of YFP or CFP insertion in PrP. SS, signal sequence; PO, pre-octarepeat domain; OR, octarepeat domain; GPI, glycosylphosphatidylinositol anchor.
DISCUSSION
We and others documented in the past that PS-DNA treatment of cells in culture reduces the levels of PrPSc and PrPC and suggested that PS-DNA may serve as the basis for the development of a future potent countermeasure for treatment of prion diseases (16, 18, 34). The current study aims to assess the essential factors needed for the observed modulatory effect of PS-DNA treatment on PrPC levels.
The current major observations are the essential role of the N-terminal domain and the membranal localization of PrP for the PS-DNA-mediated reduction. Testing different deletion-mutated versions of PrP established that removal of its N terminus (aa 23–90) resulted in loss of susceptibility to PS-DNA treatment, whereas other deletions such as that of an essential internal domain for prion infectivity (aa 141–176) had no effect. Furthermore, proteins that are resistant to PS-DNA exposure became susceptible when tagged only with PrP N terminus. Thus, the proteins Dpl, Thy1, CFP, and YFP, which do not share sequence homology with PrP and which in their native form are not affected by PS-DNA, were reduced when anchored to the cell surface and fused either to full-length PrP or only to a portion of the N terminus of PrP. Moreover, preliminary data suggest that PrP can be attached to the C terminus of CFP and YFP proteins for PS-DNA susceptibility, without any effect on their fluorescence. These observations are in good agreement with previous studies demonstrating that nucleic acid binding properties of PrP are confined to its N-terminal region (35, 36).
Apart from DNA binding, the N terminus of PrP may bind copper via an octarepeat peptide domain (37, 38), which resides at position 48–90. We note that PS-DNA treatment of cells transfected with a recombinant version of murine PrP in which its native N-terminal domain (aa 1–90) was replaced with the homologous sequence from X. laevis PrP (aa 1–69), and therefore devoid of the copper-binding domain, resulted in increased susceptibility to reduction in PrP levels. This observation may reflect the previously reported increased degradation of PrP devoid of the copper-binding domain (39). On the other hand, one may speculate that the copper-binding domain may interfere with PS-DNA binding or that the structure of the N terminus devoid of the copper domain is more amenable to interaction with PS-DNA. This issue needs to be further investigated.
We demonstrate that PrP or heterologous proteins tailored with the N-terminal domain of PrP have to be anchored to the cell membrane for efficient reduction in response to PS-DNA treatment of the cells. PrP proteins directed to the cytosol, mitochondria, or nucleus became resistant to PS-DNA exposure. The mode of anchoring to the cell surface and the microenvironment of PrP on the cell surface does not influence its susceptibility to PS-DNA, as shown by efficient reduction in the levels of PrP versions in which the PrP GPI anchor was replaced with the Thy1 GPI or with the CD4 transmembrane domain. Alteration of a different PrP post-modification such as N-glycosylation does not seem to influence the PS-DNA effect, which efficiently occurred with non-, mono-, and diglycosylated isoform, as well as following tunicamycin treatment. These observations are somewhat contradictory to the reported failure of partially glycosylated PrP to be efficiently implanted in the cell surface (19, 40, 41), suggesting that the exact involvement of glycosylation for the migration of PrP to the surface and/or its stable localization at the membrane are yet to be fully understood. We note that Capellari et al. (42) suggested in the past that glycan occupancy of the first PrP glycosylation does not appear to be essential for its transport through the secretory pathway.
Based on the current study, we may only speculate on the mechanism responsible for PrP down-regulation upon PS-DNA exposure. One possible explanation invokes enhanced internalization of PrP promoted by PS-DNA in line with the reported internalization of PrP following exposure to polyamines (31, 43) and PS-DNA (18). Because no secreted forms of PrP could be detected in the cell medium following PS-DNA exposure, we suggest that PrP is internalized and degraded within the cells, as suggested previously (15). Preliminary results suggest that the polyanion suramin was not able to block PrP down-regulation in the presence of PS-DNA, indicating that these two negatively charged compounds are probably down-regulating PrP levels by different mechanisms and are not competing with the same binding domain on PrP (not shown). Suramin was shown to induce misfolding of plasma membrane PrP and depends on the proximity to the C-terminal domain of PrP (31). Further studies will be necessary to accurately distinguish between the different mechanisms mediating PrP down-regulation in response to PS-DNA (presumably involving internalization and degradation) as opposed to suramin (which involves misfolding). An alternative, yet not mutually exclusive, mechanism may involve the previously proposed unfolding of prion protein promoted by interaction with DNA resulting in enhanced accessibility to proteolytic degradation (44).
Previous studies demonstrated the in vivo anti-prion effect of PS-DNA containing the CpG motif, suggesting that the anti-prion effect may be due to the stimulation of the innate immune response (14, 45). Other studies suggested that down-regulation of PrPSc following exposure of PS-DNA is due to the direct interaction of PS-DNA to PrPC (18). Furthermore, Zou et al. (46) reported that an anti-DNA antibody or the bacterial DNA-binding protein g5p co-captures PrP from cell lysates in pulldown experiments, strengthening the notion that DNA and PrP closely interact. Although the current study cannot rule out the possibility that the observed effect involves an additional, yet to be identified, molecule, it provides strong evidence in support of the direct binding hypothesis. Most notably, this phenomenon is shown to occur equally efficiently in various cell lines of various histological and phylogenetic origins, strongly suggesting that the interaction of PS-DNA and PrP does not involve tissue-specific factors. Considering the possibility that PS-DNA treatment may represent the basis for a future therapeutic approach, it will be interesting to determine whether a similar diminution in the PrP levels may be observed in cultures of primary neuronal cells.
The study presented here suggests a novel method by which membrane-located proteins can be down-regulated in a controlled and transient manner in cell cultures. We detect significant reduction in the protein levels following 2 h of incubation in the presence of PS-DNA. This reduction may gradually progress for extended incubation periods. Moreover, the effect is reversible, and removal of PS-DNA from the medium results in restoration of PrP levels to normal values within 4 h.
The approach that may be pursued according to this methodology may involve the use of a PrP tag (28 amino acids of PrP) fused to a protein of interest expressed in cell cultures. Subsequently, similar to other specific gene-silencing experimental setups, the physiological significance of the down-regulation of the desired protein may be interrogated by a variety of approaches. The method proposed here may be superior to the siRNA approach for assaying physiological events occurring short times after the specific removal of a protein of interest (2–8 h) and especially by virtue of its rapid and full restoration to normal levels. Obviously the recovery time following cessation of the treatment will depend on the turnover of the protein under study, which may differ substantially from case to case. Nevertheless, other approaches, such as siRNA-mediated inhibition of gene expression, typically require days for reversal of the effect.
Apart from serving as the basis for an experimental approach for protein level transient modulation, the study documented in this report underscores the puzzling phenomenon of reduction in the PrP level in response to oligonucleotide contact. The biological significance of membrane-anchored PrP susceptibility to oligodeoxynucleotides is poorly understood. It is tempting to speculate that this universal and highly conserved process, as emerging from the present study, is relevant to physiological states in which PrP down-regulation is imperative and which implicate the contact of PrP with short DNA fragments. The presence of extracellular DNA may prevent the detrimental release of PrP. The quantitative approach adopted in this study may contribute to future elucidation of this phenomenon.
Supplementary Material
Acknowledgments
We thank Professor Stanley Prusiner for kindly providing PrP deletion mutants and PrP-Dpl chimeras. We thank David Shitrit, Eran Fink, and Marina Brumin for technical assistance. We are grateful to Professor Gil Segal, Professor Ruth Gabizon, Professor Albert Taraboulos, Professor Donald C. Lo, Dr. Saul Burdman, and Dr. Christian Essrich for providing reagents and for useful discussion and insights. Drs. Theodor Chitlaru, Edith Chitlaru, Liliana Bar, Brett Monia, and Shuly Cooper and Professor Fritz Eckstein are thanked for valuable comments in preparing the manuscript.
This work was supported by the Career Development Award Grant, Human Frontier Science Program (to M. V. K.), grants from the Deutsche Forschungsgemeinschaft (SFB 596), the Max Planck Society, and the BMBF (BioDisc, DIP5.1) (to J. T.), and grants from the Deutsche Forschungsgemeinschaft (SFB 596) and European Union NoE Neuroprion (to H. M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- PrP
- prion protein
- PrPC
- cellular PrP
- PS-DNA
- phosphorothioate oligodeoxynucleotides
- Dpl
- Doppel protein
- GPI
- glycosylphosphatidylinositol
- PIPLC
- phosphatidylinositol phospholipase c
- aa
- amino acids.
REFERENCES
- 1. Prusiner S. B. (1982) Science 216, 136–144 [DOI] [PubMed] [Google Scholar]
- 2. Chakrabarti O., Hegde R. S. (2009) Cell 137, 1136–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rane N. S., Kang S. W., Chakrabarti O., Feigenbaum L., Hegde R. S. (2008) Dev. Cell 15, 359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schätzl H. M., Da Costa M., Taylor L., Cohen F. E., Prusiner S. B. (1995) J. Mol. Biol. 245, 362–374 [DOI] [PubMed] [Google Scholar]
- 5. Calzolai L., Lysek D. A., Pérez D. R., Güntert P., Wüthrich K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gabus C., Derrington E., Leblanc P., Chnaiderman J., Dormont D., Swietnicki W., Morillas M., Surewicz W. K., Marc D., Nandi P., Darlix J. L. (2001) J. Biol. Chem. 276, 19301–19309 [DOI] [PubMed] [Google Scholar]
- 7. Radford H. E., Mallucci G. R. (2010) Curr. Issues Mol Biol. 12, 119–127 [PubMed] [Google Scholar]
- 8. Roucou X., LeBlanc A. C. (2005) J. Mol. Med. 83, 3–11 [DOI] [PubMed] [Google Scholar]
- 9. Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J. L., Lehmann S., Launay J. M., Kellermann O. (2000) Science 289, 1925–1928 [DOI] [PubMed] [Google Scholar]
- 10. Spielhaupter C., Schätzl H. M. (2001) J. Biol. Chem. 276, 44604–44612 [DOI] [PubMed] [Google Scholar]
- 11. Büeler H., Fischer M., Lang Y., Bluethmann H., Lipp H. P., DeArmond S. J., Prusiner S. B., Aguet M., Weissmann C. (1992) Nature 356, 577–582 [DOI] [PubMed] [Google Scholar]
- 12. Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A. D., Toyka K. V., Nave K. A., Weis J., Aguzzi A. (2010) Nat. Neurosci. 13, 310–318 [DOI] [PubMed] [Google Scholar]
- 13. Silva J. L., Lima L. M., Foguel D., Cordeiro Y. (2008) Trends Biochem. Sci. 33, 132–140 [DOI] [PubMed] [Google Scholar]
- 14. Sethi S., Lipford G., Wagner H., Kretzschmar H. (2002) Lancet 360, 229–230 [DOI] [PubMed] [Google Scholar]
- 15. Spinner D. S., Cho I. S., Park S. Y., Kim J. I., Meeker H. C., Ye X., Lafauci G., Kerr D. J., Flory M. J., Kim B. S., Kascsak R. B., Wisniewski T., Levis W. R., Schuller-Levis G. B., Carp R. I., Park E., Kascsak R. J. (2008) J. Virol. 82, 10701–10708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karpuj M. V., Giles K., Gelibter-Niv S., Scott M. R., Lingappa V. R., Szoka F. C., Peretz D., Denetclaw W., Prusiner S. B. (2007) Mol Med. 13, 190–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prusiner S. B. (2004) Prion Biology and Diseases, pp. 961–1014, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Kocisko D. A., Vaillant A., Lee K. S., Arnold K. M., Bertholet N., Race R. E., Olsen E. A., Juteau J. M., Caughey B. (2006) Antimicrob. Agents Chemother. 50, 1034–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korth C., Kaneko K., Prusiner S. B. (2000) J. Gen. Virol. 81, 2555–2563 [DOI] [PubMed] [Google Scholar]
- 20. Scott M. R., Köhler R., Foster D., Prusiner S. B. (1992) Protein Sci. 1, 986–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muramoto T., Scott M., Cohen F. E., Prusiner S. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15457–15462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilch S., Nunziante M., Ertmer A., Wopfner F., Laszlo L., Schätzl H. M. (2004) Traffic 5, 300–313 [DOI] [PubMed] [Google Scholar]
- 23. Nunziante M., Gilch S., Schätzl H. M. (2003) J. Biol. Chem. 278, 3726–3734 [DOI] [PubMed] [Google Scholar]
- 24. Heske J., Heller U., Winklhofer K. F., Tatzelt J. (2004) J. Biol. Chem. 279, 5435–5443 [DOI] [PubMed] [Google Scholar]
- 25. Rambold A. S., Miesbauer M., Rapaport D., Bartke T., Baier M., Winklhofer K. F., Tatzelt J. (2006) Mol. Biol. Cell 17, 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butler D. A., Scott M. R., Bockman J. M., Borchelt D. R., Taraboulos A., Hsiao K. K., Kingsbury D. T., Prusiner S. B. (1988) J. Virol. 62, 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hori H., Elbein A. D. (1981) Plant Physiol. 67, 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Taraboulos A., Scott M., Semenov A., Avrahami D., Laszlo L., Prusiner S. B., Avraham D. (1995) J. Cell Biol. 129, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Keller G. A., Siegel M. W., Caras I. W. (1992) EMBO J. 11, 863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kiachopoulos S., Heske J., Tatzelt J., Winklhofer K. F. (2004) Traffic 5, 426–436 [DOI] [PubMed] [Google Scholar]
- 32. Hollrigel G. S., Morris R. J., Soltesz I. (1998) Proc. Biol. Sci. 265, 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Whyte S. M., Sylvester I. D., Martin S. R., Gill A. C., Wopfner F., Schätzl H. M., Dodson G. G., Bayley P. M. (2003) Biochem. J. 373, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prusiner S. B. (2001) N. Engl. J. Med. 344, 1516–1526 [DOI] [PubMed] [Google Scholar]
- 35. Lima L. M., Cordeiro Y., Tinoco L. W., Marques A. F., Oliveira C. L., Sampath S., Kodali R., Choi G., Foguel D., Torriani I., Caughey B., Silva J. L. (2006) Biochemistry 45, 9180–9187 [DOI] [PubMed] [Google Scholar]
- 36. Cordeiro Y., Machado F., Juliano L., Juliano M. A., Brentani R. R., Foguel D., Silva J. L. (2001) J. Biol. Chem. 276, 49400–49409 [DOI] [PubMed] [Google Scholar]
- 37. Pauly P. C., Harris D. A. (1998) J. Biol. Chem. 273, 33107–33110 [DOI] [PubMed] [Google Scholar]
- 38. Rachidi W., Vilette D., Guiraud P., Arlotto M., Riondel J., Laude H., Lehmann S., Favier A.(2003) J. Biol. Chem. 278, 9064–9072 [DOI] [PubMed] [Google Scholar]
- 39. Haigh C. L., Edwards K., Brown D. R. (2005) Mol. Cell. Neurosci. 30, 186–196 [DOI] [PubMed] [Google Scholar]
- 40. Lehmann S., Harris D. A. (1997) J. Biol. Chem. 272, 21479–21487 [DOI] [PubMed] [Google Scholar]
- 41. Neuendorf E., Weber A., Saalmueller A., Schatzl H., Reifenberg K., Pfaff E., Groschup M. H. (2004) J. Biol. Chem. 279, 53306–53316 [DOI] [PubMed] [Google Scholar]
- 42. Capellari S., Zaidi S. I., Long A. C., Kwon E. E., Petersen R. B. (2000) Alzheimers Dis. 2, 27–35 [DOI] [PubMed] [Google Scholar]
- 43. Shyng S. L., Lehmann S., Moulder K. L., Harris D. A. (1995) J. Biol. Chem. 270, 30221–30229 [DOI] [PubMed] [Google Scholar]
- 44. Nandi P. K., Leclerc E., Nicole J. C., Takahashi M. (2002) J. Mol. Biol. 322, 153–161 [DOI] [PubMed] [Google Scholar]
- 45. Gilch S., Schmitz F., Aguib Y., Kehler C., Bülow S., Bauer S., Kremmer E., Schätzl H. M. (2007) FEBS J. 274, 5834–5844 [DOI] [PubMed] [Google Scholar]
- 46. Zou W. Q., Zheng J., Gray D. M., Gambetti P., Chen S. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.