Abstract
Dengue virus RNA-dependent RNA polymerase specifically binds to the viral genome by interacting with a promoter element known as stem-loop A (SLA). Although a great deal has been learned in recent years about the function of this promoter in dengue virus-infected cells, the molecular details that explain how the SLA interacts with the polymerase to promote viral RNA synthesis remain poorly understood. Using RNA binding and polymerase activity assays, we defined two elements of the SLA that are involved in polymerase interaction and RNA synthesis. Mutations at the top of the SLA resulted in RNAs that retained the ability to bind the polymerase but impaired promoter-dependent RNA synthesis. These results indicate that protein binding to the SLA is not sufficient to induce polymerase activity and that specific nucleotides of the SLA are necessary to render an active polymerase-promoter complex for RNA synthesis. We also report that protein binding to the viral RNA induces conformational changes downstream of the promoter element. Furthermore, we found that structured RNA elements at the 3′ end of the template repress dengue virus polymerase activity in the context of a fully active SLA promoter. Using assays to evaluate initiation of RNA synthesis at the viral 3′-UTR, we found that the RNA-RNA interaction mediated by 5′-3′-hybridization was able to release the silencing effect of the 3′-stem-loop structure. We propose that the long range RNA-RNA interactions in the viral genome play multiple roles during RNA synthesis. Together, we provide new molecular details about the promoter-dependent dengue virus RNA polymerase activity.
Keywords: Flavi Viruses, RNA, RNA Polymerase, RNA-Protein Interaction, RNA Viruses, RNA Synthesis, RNA-RNA Interaction, Dengue Virus, Viral RNA Replication
Introduction
Dengue virus (DENV)3 is a member of the Flavivirus genus in the Flaviviridae family, together with other important human pathogens such as yellow fever virus, West Nile virus (WNV), Saint Luis encephalitis virus, and Japanese encephalitis virus (1). DENV is the most significant mosquito-borne human viral pathogen worldwide and is responsible for the highest rates of disease and mortality among the members of the Flavivirus genus. The lack of vaccines and antivirals against DENV leaves two billion people at risk, mainly in poor countries.
DENV genome is a single-stranded RNA molecule of positive polarity of ∼11 kb in length that encodes a long polyprotein that is co- and post-translationally processed by host and viral proteases to yield three structural proteins (C, prM, and E), and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5). The coding sequence is flanked by highly structured 5′- and 3′-UTRs. In recent years, a number of cis-acting RNA elements have been identified in the viral 5′- and 3′-UTRs that are essential for viral RNA amplification (for review see Ref. 2). A model for DENV RNA synthesis has been proposed previously (3). This model involves binding of the viral RNA-dependent RNA polymerase (RdRp) to an RNA element present at the 5′ end of the genome known as stem-loop A (SLA). This element of 70 nucleotides folds into a Y-shaped structure that is conserved among different flaviviruses (4–8). Interaction of the SLA with the RdRp has been shown to be necessary for RNA synthesis in vitro and viral replication in transfected cells for DENV and WNV (3, 9–11). The SLA specifically binds the viral RdRp and promotes RNA synthesis 11,000 nucleotides away from the binding site, at the 3′ end of the genome. Genome cyclization mediated by long range RNA-RNA interactions was found to be crucial to reposition the SLA-RdRp complex near the 3′ end initiation site (3). Flaviviruses conserved both the SLA structure and the complementary sequences involved in RNA cyclization (12–19); therefore, it is likely that the proposed model is shared by different members of the genus.
The ∼100-nucleotide-long DENV 5′-UTR has a type 1 cap structure at the 5′ end and contains two RNA domains with distinct functions during viral RNA synthesis (6, 20). The first domain is the promoter SLA followed by an oligo(U) region that serves as spacer. The second domain is predicted to form a shorter stem-loop, named SLB. This element contains a 16-nucleotide-long sequence, known as 5′-UAR, which is complementary to a region present at the 3′ end of the viral genome (3′-UAR) (12, 13). Hybridization of 5′-3′-UAR together with other pair of complementary sequences, known as 5′ and 3′-CS, mediate the long range RNA-RNA interaction and genome cyclization (12, 13, 21–23). The ∼450-nucleotide-long DENV 3′-UTR lacks a poly(A) tail but ends in a very conserved 3′-stem-loop structure (3′-SL). Although a detailed functional analysis of the 3′-SL revealed its absolute requirement for genome replication (for review see Ref. 24), the mechanism by which this element controls RNA synthesis is still unclear. Specific bulges within the 3′-SL as well as nucleotides at the loop and the 3′ terminal sequence were found to be essential for RNA replication (25–28). The 5′ bottom part of the large stem of the 3′-SL contains the 3′-UAR sequence, complementary to the 5′-UAR region of the 5′-UTR (12, 13, 21). We have previously shown that 5′-3′-UAR complementarity, and not the UAR sequences per se, is essential for viral replication (12). Upstream of the 3′-SL there is the other essential cyclization element 3′-CS. This region is 11 nucleotides long and is complementary to the 5′-CS located in the coding sequence of the capsid protein at the 5′ end of the open reading frame (14, 15, 21, 29–32).
We have recently reported structural elements of the promoter SLA that are necessary for DENV replication in infected cells (10). However, the molecular details of how these elements interact with the viral polymerase to promote RNA synthesis are largely unknown. Here, we used the recombinant DENV RdRp and viral RNA molecules to investigate the requirements for polymerase binding and activity. Using footprinting, RNA binding studies, and polymerase activity assays, we defined two regions within the SLA that are important for protein binding and promoter function. Conformational changes of the RNA downstream of the promoter element were also observed upon RdRp binding. In addition, using different RNA molecules with mutations within the SLA, we found that mutations at the top loop of the SLA impaired polymerase activity without altering the dissociation constants of the complex RdRp-SLA, supporting the idea that recruitment of the viral polymerase to the SLA is not sufficient to promote RNA synthesis. Also, we demonstrate that structured 3′ end elements, including the viral 3′-SL, repress RNA synthesis. In this regard, the conformational change induced by 5′-3′ hybridization was sufficient to release the silencing effect of the 3′-SL on RNA synthesis.
EXPERIMENTAL PROCEDURES
RNA Preparation
RNAs for footprinting and in vitro RdRp assays were obtained by in vitro transcription using T7 RNA polymerase (90 min, 37 °C) and treated with DNase I RNase-free to remove templates. Except for the radiolabeled RNAs (see below), the RNAs were purified using the RNeasy mini kit (Qiagen) to remove free nucleotides, quantified spectrophotometrically, and their integrity was verified by electrophoresis on agarose gels. All of the numbers given in parentheses refer to nucleotide positions of a dengue virus type 2 strain 16681 infectious cDNA clone (GenBankTM accession number U87411) (33). Nucleotide sequences of primers used for PCR are listed in supplemental Table S1. The purified PCR products were directly used as templates for in vitro transcription. The sequence corresponding to the 5′-DV (nucleotides 1–160) was amplified by PCR from the 16681 DENV cDNA with the sense primer AVG 1 and the antisense primer AVG 130. Mutations, insertions, and deletions within 5′-DV (Mut 338, Mut 340, Mut 1–7, templates II–VI, and 5′-DV-CSN) were performed by PCR or overlapping PCR using 5′-DV as template. The forward and reverse oligonucleotides used to introduce the specific mutations are listed in supplemental Table S2. The sequence corresponding to the 3′-UTR (nucleotides 10269–10723) was amplified by PCR from the 16681 DENV cDNA with the sense primer AVG 2 and the antisense primer AVG 5. The SLA-3′-SL RNA was obtained by overlapping PCR using for the first round of PCR the forward primer AVG 1, the reverse primer AVG 515, the forward primer AVG 514, and the reverse primer AVG 5. The overlapping PCR was performed with AVG 1 and AVG 5. Mutations and insertions within the 3′-UTR (SLA-3′-SL Mut K, 3′-UTR-tail, and 3′-UTR-Mut K) were generated by PCR or overlapping PCR using the oligonucleotides listed in supplemental Table S2.
Preparation of Recombinant Proteins of Full-length NS5, Methyltransferase (MTase) Domain, and RdRp Domain
The RdRp domain of DENV2 NS5 (amino acids 268–909) was expressed in Escherichia coli (Rosettas pLac) and purified as described previously (3). The preparation of the DENV2 full-length NS5 (amino acids 1–909) and the MTase (amino acids 1–267) was identical to that for the RdRp, except that after the HisTrap nickel-agarose column purification, we performed a size exclusion chromatography step using a Superdex 200 column. The recombinant purified proteins were stored at −80 °C in a buffer containing 40% glycerol.
Methylation Assay
The MTase assay was performed as described elsewhere (34). Briefly, the [3H]methyl incorporation was measured in a total volume of 20 μl in buffer containing 50 mm glycine, pH 10, 2 mm dithiothreitol, 80 μm unlabeled S-adenosylmethionine (New England Biolabs), 2 μCi of [3H-methyl]S-adenosylmethionine (Amersham Biosciences), 10 pmol of GpppA-5′-DV RNA, and 30 pmol of full-length NS5 or MTase protein. The reaction mixture was incubated at 22 °C for 1 h. The reaction was ended by phenol extraction followed by ethanol precipitation. The RNA products were resuspended in milli-Q water and purified using MicroSpin G-25 columns (Amersham Biosciences). Then the radioactivity of the eluted sample was measured.
RdRp in Vitro Assay
The standard in vitro RdRp assay was carried out in a total volume of 25 μl in buffer containing 50 mm Hepes (pH 8.0), 10 mm KCl, 5 mm MgCl2, 2 mm MnCl2, 10 mm dithiothreitol, 4 units of RNase inhibitor, 500 μm (each) ATP, CTP, and UTP, 10 μm [α-32P]GTP, 0.5 μg of template RNA, and 0.15 μg of recombinant purified full-length NS5 or RdRp protein. The reaction mixture was incubated at 30 °C for 30 min, and the reaction was stopped by adding a denaturing solution to give final concentrations of 7% (w/v) TCA and 50 mm H3PO4 at 0 °C. The TCA-precipitated RNAs were then collected by vacuum filtration using a V-24 apparatus, with the mixture carefully added onto the center of a Millipore filter (type HAWP, 0.45-μm pore size). The filters were washed eight times with 5 ml each of cold 7% (w/v) TCA, 50 mm H3PO4 and dried, and the radioactivity was measured. For polyacrylamide gel electrophoresis analysis of the RNA products and the trans-initiation assay, the standard mixture was the same as the one described above except that the reaction was ended by phenol extraction followed by ethanol precipitation. The RNA products of the polymerase assay were resuspended in Tris-EDTA containing formamide (80%) and heated for 5 min at 65 °C. The samples were then analyzed by electrophoresis on a 5% denaturing polyacrylamide gel, 6 m urea and visualized by autoradiography.
RNA Binding Assay
RNA-RdRp interactions were analyzed by EMSA. Uniformly 32P-labeled RNA probes were obtained by in vitro transcription using T7 RNA polymerase and purified on 5% polyacrylamide gels and 6 m urea. The 5′-DV probe and the probes for the mutants (Mut 338, Mut 340, and Mut 1–7) were obtained as described under RNA templates. The SLA 1, SLA 2, and 5′-DV ΔSLA RNA probes were obtained using the PCR products with the primers described in supplemental Table S2 and in vitro transcription. The unstructured/unrelated RNA corresponds to the coding region of HCV1b (nucleotides 8451–8678) and was obtained by PCR using primers AVG245-AVG246. The binding reaction contained 5 mm Hepes (pH 7.9), 25 mm KCl, 2 mm MgCl2, 3.8% glycerol, 0.12 mg/ml heparin, 0.1 nm 32P-labeled probe and increasing concentrations of the RdRp domain. RNA-RdRp complexes were analyzed by electrophoresis through native 5% polyacrylamide gels supplemented with 5% glycerol. The gels were prerun for 30 min at 4 °C at 120 V, and then 20 μl of sample was loaded, and electrophoresis was allowed to proceed for 5 h at constant voltage. The gels were dried and visualized by autoradiography.
RNA-RNA interactions were also analyzed by EMSA. Uniformly 32P-labeled 3′-SL was obtained by in vitro transcription using T7 RNA polymerase and purified on 5% polyacrylamide gels and 6 m urea. The binding reaction contained 5 mm Hepes (pH 7.9), 25 mm KCl, 5 mm MgCl2, 3.8% glycerol, 2.8 μg of tRNA, uniformly 32P-labeled 3′-SL (0.1 nm, 0.013 μCi), and increasing concentrations of 5′-DV or 5′-DV-CSN in a final volume of 25 μl. RNA samples were heat-denatured at 85 °C for 5 min and slow cooled to room temperature. RNA-RNA complexes were analyzed as described above.
Footprinting Analysis
In vitro transcribed 5′-DV RNA was heated for 5 min at 85 °C and slow cooled to room temperature. Increasing concentrations of the RdRp (0, 30, 60, and 120 nm) or the full-length NS5 (0 and 500 nm) were added to the RNA, and the reactions were incubated at room temperature for 30 min to allow RNA-protein complex formation. Footprinting reactions (a total volume of 10 μl) were performed at 25 °C for 15 min in a mixture containing 30 mm Tris-HCl (pH 7), 100 mm KCl, 10 mm MgCl2, 1 mm dithiothreitol, 10% glycerol, 0.5 μg of 5′-DV RNA, the RdRp, or the full-length NS5 and 0.4 unit of RNase PhyM (Pierce) and stopped by the addition of 20 μl of RNase inactivation/precipitation buffer (Ambion). The reaction products were purified by incubation at −20 °C for 15 min followed by centrifugation at 15,700 × g for 15 min. The pellets were washed in 70% ethanol and resuspended in 5 μl of Milli-Q water. Control reactions were carried out in parallel under the same conditions, without the addition of RNases. The digested RNA products were used for primer extension. As primer, we used a 5′ end fluorescently labeled oligonucleotide (5′-cy5-TCATCAGAGATCTGCTCTCTAATTAAAAA-3′) (purchased from Integrated DNA Technologies). The reactions were performed with 20 units of ArrayScript reverse transcriptase (Ambion) at 42 °C for 50 min with a 10-μl mixture containing 50 mm Tris-HCl (pH 8.3), 75 mm KCl, 3 mm MgCl2, 5 mm dithiothreitol, and 500 μm of each of the four deoxynucleoside triphosphates. cDNA products were ethanol-precipitated, resuspended in 90% formamide, and heat-denatured for 3 min at 95 °C immediately prior to electrophoresis. A dideoxyadenosine sequencing reaction was carried out on unmodified RNA and run in parallel with the primer extension products on 10% polyacrylamide, 7 m urea sequencing gels. The gels were then analyzed directly using a Storm 840 imager (Molecular Dynamics). In addition, we performed footprinting assays using a 5′-DV end-labeled probe. In vitro transcribed 5′-DV RNA molecules were dephosphorylated by calf intestinal phosphatase (15 units) at 37 °C for 1 h and 5′ end-labeled with 50 μCi of [γ-32P]ATP and T4 polynucleotide kinase (20 units) at 37 °C for 1 h. The 5′ end-labeled RNA was gel-purified (5% polyacrylamide-6 M urea), eluted, and ethanol-precipitated. The recombinant RdRp (120 nm) was added to the RNA solution, and the reactions were incubated at room temperature for 30 min to allow RNA-protein complex formation. The RNA digestion was performed with 0.1 unit of RNase T1 or 0.01 unit of RNase A under the conditions described above. Control reactions were carried out under the same conditions, without the addition of RNases. A sequencing reaction was performed in parallel by digestion of 5′-DV RNA with 0.1 unit of RNase T1 under denaturing conditions (RNA sequence buffer; Ambion). After RNase precipitation/inactivation, the reaction products were resuspended in 90% formamide, heat-denatured, and directly resolved by electrophoresis on 10% polyacrylamide, 7 m urea sequencing gels.
RESULTS
RNA Binding of the RdRp, the MTase, and the Full-length NS5 Proteins
The DENV RdRp corresponds to the last 641 amino acids of NS5 and is physically linked to the N-terminal MTase domain. According to previous reports, the RdRp domain is able to interact with the 5′ end of the viral RNA in the absence of other viral or cellular proteins (3). However, the contribution of the MTase domain to this RNA binding has not been previously examined. In the case of WNV NS5, the MTase provides specificity for RNA binding (9). To investigate the role of the MTase domain in NS5 binding to the 5′ end of the DENV RNA, we expressed and purified the full-length NS5 and the RdRp and the MTase domains (Fig. 1A). To confirm that these recombinant proteins were properly folded and active, both MTase and RdRp activities were analyzed. The full-length NS5 and the RdRp domain displayed high levels of RNA synthesis (Fig. 1B, left panel). In addition, the full-length NS5 and the MTase showed [3H]methyl incorporation using [3H-methyl]S-adenosylmethionine and GpppA-RNA as substrates (Fig. 1B, right panel). These results indicate that the three proteins were active enzymes. Then we examined the ability of the recombinant proteins to bind the viral RNA. To this end, we performed EMSA assays using the purified proteins and a radiolabeled RNA corresponding to the first 160 nucleotides of the viral genome (5′-DV). A well defined and stable heparin-resistant complex was observed when the probe was incubated with the full-length NS5 or with the RdRp (Kd = 14 and 12 nm, respectively), whereas only a weak interaction was detected with the MTase domain alone (Kd > 240 nm) (Fig. 1C). This result suggests that DENV NS5 interacts with the viral RNA mainly by the polymerase domain, whereas the MTase domain does not contribute significantly to the observed interaction.
FIGURE 1.
Expression, purification, and characterization of DENV full-length NS5 and the two domains RdRp and MTase. A, SDS-PAGE analysis of purified recombinant proteins expressed in E. coli. Mobilities of molecular mass markers (MW) are indicated on the left (in kDa). B, in vitro polymerase and methyltransferase activities of recombinant proteins. The RdRp assay was carried out as described under “Experimental Procedures,” using an in vitro transcribed 5′-DV RNA as template. The RdRp activity is expressed in fmol of [α-32P]GMP incorporated into acid-insoluble RNA/min and per μg of protein. The MTase assay was carried out as described under “Experimental Procedures,” using as template an in vitro transcribed GpppA-5′-DV RNA or an uncapped 5′-DV RNA. The MTase activity is expressed in cpm of [3H]methyl-incorporated. C, binding of the recombinant proteins to the 5′-DV RNA. The interaction of the three proteins with the 5′-DV was evaluated by EMSA. The RNA corresponding to the first 160 nucleotides of the viral genome was uniformly 32P-labeled and titrated (0.1 nm, 30,000 cpm) with increasing concentrations of RdRp, MTase, or full-length NS5 (0, 0.2, 0.5, 1, 5, 10, 30, and 240 nm).
Binding of the Dengue Virus RNA-dependent RNA Polymerase to the SLA Promoter
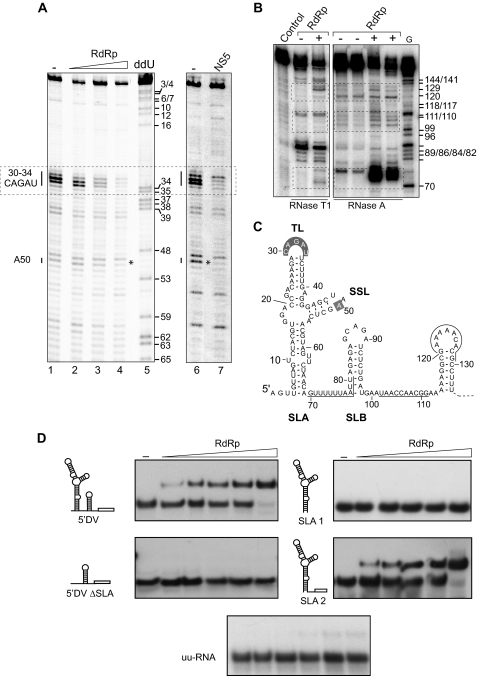
To investigate the mechanism by which the SLA promotes DENV RNA synthesis, we analyzed the interaction between the SLA and the viral RdRp or the full-length NS5 using footprinting analysis. The RNA corresponding to the 5′ end of the viral genome (5′-DV) was incubated with the purified recombinant viral proteins. After incubation, the RNA was digested with the PhyM RNase and used as a template for primer extension using a fluorescent probe that hybridized at the 3′ end of the SLA. The nucleotide sequences of the SLA protected by the viral proteins were identified in denaturing polyacrylamide gels (Fig. 2A). In both cases, we observed a strong protection in a region corresponding to nucleotides 30–34. In addition, a second protected region was detected at nucleotide 50. The region of nucleotides 30–34 mapped at the top loop (TL) of the SLA, which was highly sensitive to the RNase and became almost insensitive when the RNA was incubated with increasing amounts of protein. The other region mapped in the side stem-loop (SSL) and corresponded to 1 nucleotide of the loop, which was previously shown to be sensitive to single-stranded specific RNases (10). These results suggest that nucleotides of the TL and the SSL could be involved in RdRp binding.
FIGURE 2.
Interaction of the dengue virus RdRp with the viral RNA. A, footprinting assay indicates the interaction of the viral RdRp or full-length NS5 with elements of the SLA. The viral RNA was incubated with increasing concentrations of the RdRp (0, 30, 60, and 120 nm; left panel) or the full-length NS5 (0 and 500 nm; right panel) and subjected to RNase PhyM treatment. The cleaved RNAs were analyzed after primer extension in a sequencing gel along with a sequencing ladder (ddU). Regions showing protection from RNase cleavage upon protein binding are indicated on the left. B, the interaction of the RdRp with the viral RNA alters RNA structures downstream of the promoter element. A 5′ end-labeled RNA of the first 160 nucleotides of the viral genome was incubated with 120 nm RdRp and subjected to RNase T1 or RNase A treatment (as indicated at the bottom). The cleaved RNAs were analyzed along with a control sample without treatment (Control). The labeled RNA was also digested with RNase T1 under denaturing conditions to generate a G ladder (G). Regions showing exposure to RNase cleavages upon RdRp binding are indicated with dashed boxes. C, MFOLD-predicted RNA secondary structure of 5′-DV that includes a summary of protected (shaded gray) and exposed (underlined) nucleotides upon RdRp binding. The relevant elements are indicated: SLA, SLB, TL, and SSL. D, binding of the RdRp to different RNA molecules corresponding to the viral 5′-UTR. The interaction of four RNA probes with the RdRp was evaluated by EMSA. The RNA corresponding to the first 160 nucleotides of the viral genome (5′DV), the first 70 nucleotides (SLA 1), the 5′-DV RNA with a deletion of the SLA (5′DV ΔSLA), the SLA 1 followed by unstructured 32 nucleotides (SLA 2), or an unstructured unrelated RNA (uu-RNA) were uniformly 32P-labeled and titrated (0.1 nm, 30,000 cpm) with increasing concentrations of the RdRp (0, 5, 10, 15, 20, and 240 nm).
To investigate other possible interactions of the RdRp with the viral 5′ end of the genome, footprinting assays using a radiolabeled molecule corresponding to the first 160 nucleotides of the viral RNA was used. The RNA was end-labeled with [γ-32P]ATP, incubated with the viral RdRp, and digested with single-stranded specific RNases. No protected nucleotides were observed downstream of the promoter element; however, incubation of the RNA with the RdRp caused enhanced cleavage in three regions (Fig. 2B). The first one was a strong cleavage between nucleotides 70 and 80, which corresponded to a sequence located between the SLA and the SLB, and the other two regions corresponded to nucleotides 102–111 and 120–129. Residues with increased susceptibility to cleavage represent nucleotides that are more exposed to the RNases upon RdRp binding. We have previously shown that an oligo(U) sequence, present downstream of the promoter SLA (nucleotides 71–77; Fig. 2C), plays a role as spacer required for RNA synthesis in DENV-infected cells (10). The new observations indicate that the interaction of RdRp with its promoter also induces a rearrangement of RNA structures located downstream of the SLA.
To investigate the role of nucleotides present downstream of the promoter in RdRp binding, we performed EMSA assays using the first 160 nucleotides of the viral genome (5′-DV) or the first 70 nucleotides corresponding only to the SLA (Fig. 2D, SLA 1). The results indicated that although the 5′-DV RNA bound the viral protein with high affinity (3), the SLA alone was incapable of forming a stable ribonucleoprotein complex even at the highest concentration of protein (240 nm) (Fig. 2D). These results were unexpected because we had previously demonstrated that the SLA fused to a nonviral sequence was sufficient to promote polymerase activity (3). To analyze the requirement of downstream nucleotides for protein binding, the SLA was fused to a nonrelated sequence (Fig. 2D, SLA 2) and found that this RNA was an efficient binder of the RdRp. As control, an RNA probe containing the sequence of the 5′-DV with a deletion of the SLA (Fig. 2D, 5′-DV ΔSLA) and an unstructured unrelated RNA (Fig. 2D, uu-RNA) showed no protein binding. These results indicate that the SLA is the only viral specific sequence necessary for polymerase binding; however, nonspecific contacts with nucleotides downstream of the SLA are also necessary for a stable RNA-protein complex formation.
Recruitment of the RdRp to the SLA Is Not Sufficient to Promote RNA Synthesis
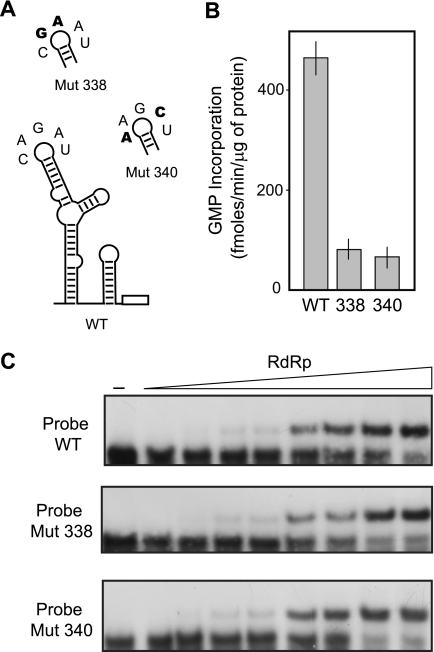
We have previously shown that mutations within the TL of the SLA impair viral replication in infected cells and that revertant viruses can be rescued with spontaneous changes that restore the TL sequence (3, 10). Based on the results shown in Fig. 2, we investigated the contribution of the TL sequence in the binding affinity of the RdRp to the promoter element. Radiolabeled RNAs carrying the wild type or mutated TL sequences (WT, Mut 338, or Mut 340; Fig. 3A) were used for EMSA assays to evaluate the RdRp binding. Although the mutations 338 and 340 almost abolished the promoter-dependent in vitro RNA synthesis by the viral RdRp (Fig. 3B), the estimated dissociation constants for the complexes Mut 338-RdRp and Mut 340-RdRp were indistinguishable from that observed with the wild type RNA. The Kd values were 11, 13, and 10 nm for the WT, Mut 338, and Mut 340, respectively. These results indicate that recruitment of the polymerase to the SLA is not sufficient to promote RNA synthesis. The mutants 338 and 340 are incompetent as promoters for RNA synthesis, but they bind the polymerase efficiently, suggesting that specific nucleotides in the TL participate in a post-binding step that is required to promote polymerase activity.
FIGURE 3.
Specific nucleotides at the top loop of the SLA promoter impair RNA synthesis without altering the RNA-RdRp complex formation. A, schematic representation of specific mutations within the TL of SLA. The nucleotide sequence of the WT and the mutants Mut 338 and Mut 340 are shown. The nucleotide changes are indicated in bold. B, in vitro polymerase activity using WT and mutated RNA templates. The activity is expressed in fmol of [α-32P]GMP incorporated into acid-insoluble RNA/min and per μg of protein. The reaction was carried out as described under “Experimental Procedures.” The error bars indicate the standard deviations of results from three experiments. C, RNA mobility shift assays showing the interaction between the purified RdRp and the WT or mutated RNA probes. Uniformly 32P-labeled RNAs corresponding to the first 160 nucleotides of the viral genome (0.1 nm, 30,000 cpm) were titrated with increasing concentrations of RdRp (0, 0.5, 1, 3, 5, 10, 15, 50, and 160 nm).
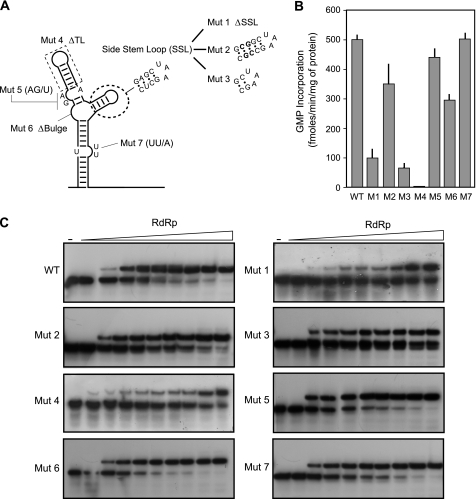
To further investigate the requirement of the TL, the SSL, and other elements of the SLA for polymerase binding and activity, different deletions and mutations were designed as described in Fig. 4A. The binding ability of the RNA molecules to the RdRp was tested for each mutant by EMSA. To this end, seven different radiolabeled probes were obtained. In addition, we analyzed whether protein binding resulted in an RNA-protein complex active for RNA synthesis.
FIGURE 4.
RNA elements of the SLA promoter required for RdRp binding and activity. A, schematic representation of specific mutations in the viral RNA. The nucleotide changes are indicated for each case: in Mut 1, the SSL was deleted, and a UUC sequence was inserted; in Mut 2, the stem of the SSL was stabilized by 5 GC base pairs; in Mut 3, a 2-base pair deletion in the stem of the SSL was included; in Mut 4, the TL was deleted; in Mut 5, the sequence of the GA bulge was replaced by a single U residue; in Mut 6, the GGA bulge was deleted; and in Mut 7 the sequence of the UU bulge was replaced by a single A residue. B, in vitro polymerase activity using WT and mutated RNA templates shown in A. The activity is expressed in fmol of [α-32P]GMP incorporated into acid-insoluble RNA/min and per μg of protein. The reaction was carried out as described under “Experimental Procedures.” The error bars indicate the standard deviations of results from three experiments. C, RNA mobility shift assays showing the interaction between the WT or the mutated RNA probes and the purified RdRp. Uniformly 32P-labeled RNA probes (0.1 nm, 30,000 cpm) were titrated with increasing concentrations of RdRp (0, 1, 5, 10, 15, 20, 25, 80, 160, and 240 nm).
First, we analyzed the requirement of the SSL for RdRp binding and activity. Deletion of the complete SSL (Mut 1) greatly decreased RNA synthesis, and the affinity for the RdRp was reduced (Kd > 240 nm) (Fig. 4, B and C). In the case in which the stem of the SSL was designed to be more stable than wild type (Mut 2), 80% of the polymerase activity and a Kd similar to that for the wild type (Kd = 9 nm) was observed. In contrast, using an RNA carrying an unstable stem of the SSL (Mut 3), RNA synthesis and polymerase binding was affected (Kd > 240 nm). These observations are in agreement with our previous in vivo studies, in which viral replication tolerated variation of the SSL sequence, but the structure was found to be essential for infectivity (10). RNAs with mutations in three bulges of the SLA (Fig. 4A, Mut 5, Mut 6, and Mut 7) were also tested. All three displayed almost wild type levels of promoter-dependent polymerase activity and efficient binding to the RdRp (Kd = 8, 9, and 8 nm, respectively). It is worth noting that Mut 7, which carries a deletion of a bulge of two Us that we have previously shown to be lethal in vivo (10), is fully functional in binding the RdRp and promoting RNA synthesis in vitro, suggesting an additional role of this element in the infected cell. Finally, a mutant RNA carrying the complete deletion of the TL was tested (Fig. 4A, Mut 4). In agreement with a possible role of this element in protein binding, RdRp activity was almost undetectable, and the Kd for complex formation was reduced (Kd > 240 nm). In summary, using structural and functional studies, we defined the TL and the SSL as important determinants for RdRp binding and activity. In addition, we found promoter mutants that retained the ability to bind the RdRp as the wild type RNA but were not competent to promote RNA synthesis. We conclude that binding of the RdRp to the SLA must be followed by an additional step that renders this complex competent for RNA synthesis.
3′ End Structures in the RNA Interfere with SLA-dependent RdRp Activity
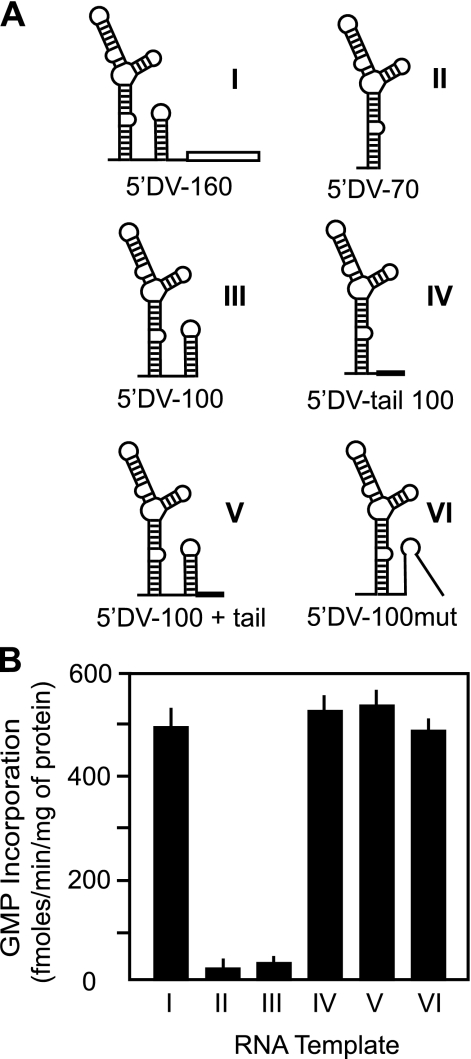
To define the minimal unit for promoter activity, we generated different RNAs carrying the SLA promoter and tested them as templates for in vitro polymerase assays. The RNA corresponding to the first 160 nucleotides of the viral genome (Fig. 5A, template I), showed high levels of RNA synthesis (Fig. 5B). In contrast, an RNA corresponding to the complete SLA (template II) was inactive when incubated with the viral enzyme. This result is in agreement with the lack of binding of the SLA to the RdRp observed in the EMSA assay (Fig. 2D). The 3′ end sequence of template II forms the stem of the SLA. In addition, template II is shorter than template I. Thus, we asked whether the sequence, the structure, or the length of the RNA was responsible for the lack of binding and the low levels of RNA synthesis observed with template II. To study these possibilities, we designed two 100-nucleotide-long RNAs, one that contained the complete SLA and SLB, and the other one in which the SLA was fused to an unstructured sequence (Fig. 5A, templates III and IV, respectively). When these templates were used for polymerase activity assays, we observed that template III was inactive, whereas template IV showed high polymerase activity. Because both RNAs were 100 nucleotides long, we rule out that the length could be the cause of the undetectable RNA synthesis with template III. To determine whether the structure at the 3′ end of the RNA was responsible of blocking RNA synthesis, we designed two new templates based on the inactive template III (Fig. 5A, templates V and VI). The unstructured sequence of template IV was fused to template III following the SLB to generate template V. To generate template VI, three point mutations were introduced in template III that open the stem of SLB, maintaining the same nucleotide sequence at the 3′ end of the RNA. Both templates were active to promote RNA synthesis. The results indicate that templates with structured 3′ ends blocked RNA synthesis, regardless of the presence of an active SLA promoter.
FIGURE 5.
Template activity of RNA molecules carrying different structures at the 3′ end. A, schematic representation of the structures of the 5′-DV RNAs used as templates. Template I (5′DV-160), template II (5′DV-70), and template III (5′DV-100) correspond to the first 160, 70, and 100 nucleotides of the viral genome, respectively. In templates IV (5′DV-tail 100) and V (5′DV-100 + tail), the first 77 or 100 nucleotides of the genome, respectively, were fused to a 26-nucleotide-long unstructured sequence. Template VI (5′DV-100mut) was generated introducing three point mutations in template III that opens the stem of SLB. B, in vitro polymerase activity for templates I–VI, expressed in fmol of [α-32P]GMP incorporated into acid-insoluble RNA/min and per μg of protein.
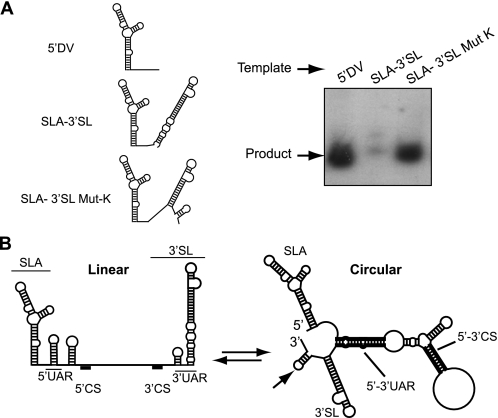
The SLA binds the viral RdRp and promotes RNA synthesis at the 3′ end of the viral genome, which ends in a highly structured 3′-SL. Therefore, we asked whether the 3′-SL can be used as template for RNA synthesis providing the SLA in the same molecule (Fig. 6A, SLA-3′-SL). In agreement with the results shown above, this RNA was inactive as template for polymerase activity (Fig. 6A).
FIGURE 6.
Effect of the 3′-SL structure on the SLA-dependent RdRp activity. A, analysis of radiolabeled RNA products in a denaturing 5% polyacrylamide gel synthesized by the recombinant RdRp. Schematic representations of the RNAs used are indicated on the left: 5′DV, 5′ end 160 nucleotides of DENV genome; SLA-3′SL, RNA containing the first 76 nucleotides fused to the last 107 nucleotides of DENV genome; SLA-3′SL Mut-K, SLA-3′-SL with seven point mutations at the 3′-SL, mimicking the long range interaction. B, schematic representation of linear and circular conformations of DENV genome. Relevant cis-acting elements are indicated: SLA, the cyclization sequences (5′-3′UAR and 5′-3′CS), and the 3′-SL. Note that in the circular conformation, the predicted SLA and the upper part of the 3′-SL are maintained after 5′-3′ hybridization, whereas the bottom part of the 3′-SL is disrupted to form an unstable hairpin (indicated by an arrow).
The current model for DENV RNA synthesis proposes that long range RNA-RNA interactions reposition the SLA-RdRp near the 3′ end of the RNA (3). In this regard, hybridization of complementary sequences present at the ends of the genome induces conformational changes including the opening of the bottom half of the 3′-SL, which leads to the formation of a less stable hairpin (indicated by an arrow in Fig. 6B). Experimental evidence of conformational changes within the 3′-SL upon 5′-3′ interaction have been previously provided using probing analysis with WNV RNA (9). To investigate whether the proposed conformational changes around the 3′-SL allow the polymerase to initiate RNA synthesis, we designed a molecule that mimics the long range interaction and formation of the terminal unstable hairpin by incorporating point mutations at the 5′ end of the 3′-SL (Fig. 6A, SLA-3′-SL Mut-K). Interestingly, this mutation was sufficient to allow efficient RNA synthesis by the viral RdRp. These results suggest that the conformational change induced by 5′-3′ hybridization could release the silencing effect of the 3′-SL.
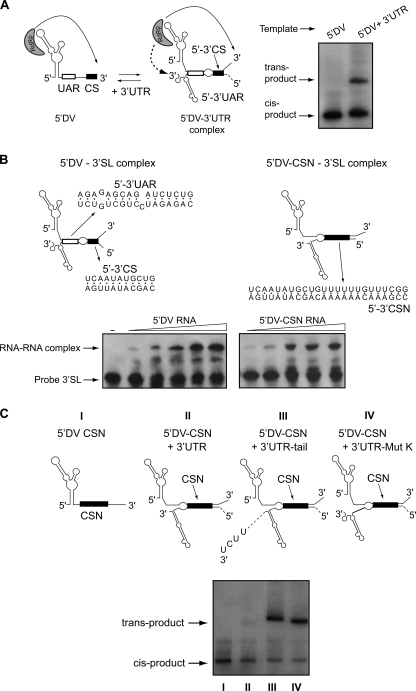
5′-3′-UAR Hybridization Allows Utilization of the Viral 3′-UTR as Template for RNA Synthesis
To further investigate the role of UAR hybridization on the initiation of RNA synthesis at the authentic 3′ end of the genome, we used a trans-initiation assay that resembles viral RNA synthesis in regard to the requirement of both the SLA promoter and the 5′-3′ RNA-RNA interaction (3, 12). In this assay, two RNA molecules (the first 160 nucleotides of the genome and the viral 3′-UTR of 454 nucleotides) are incubated with the viral polymerase (Fig. 7A). In these conditions, we have previously shown that the SLA promotes RNA synthesis copying from the 3′ end of both molecules (in cis and in trans), being the trans-initiation absolutely dependent on the hybridization of the two molecules (3, 12). It has been shown that 5′-DV molecules with point mutations that impaired 5′-3′-UAR hybridization abolished polymerase activity in trans, whereas substitution of these sequences by foreign nucleotides that restored base pairing restored polymerase function (12). This previous observation indicates that the hybridization of UAR and not the sequence per se is important for polymerase activity at the 3′ end. We hypothesized that 5′-3′-UAR interaction plays two roles: (a) contributes in bringing the SLA-RdRp near the 3′ end of the genome and (b) induces the conformational changes required to release the 3′ end sequence from the highly structure 3′-SL. To dissociate the two possible roles of UAR, we designed a 5′-DV molecule carrying a deletion of 5′-UAR together with the insertion of 10 nucleotides downstream of 5′-CS. These nucleotides were complementary to the 10 nucleotides that are preset just upstream of the 3′-CS, outside of the 3′-SL. Thus, the new complementary sequence, called CSN, is an extended 5′-3′-CS (Fig. 7B, top panel). To confirm that CSN was sufficient to mediate RNA-RNA complex formation, we performed EMSA assays incubating a uniformly labeled 3′-SL with increasing concentrations of 5′-DV-CSN or 5′-DV. An RNA-RNA complex was formed mediated by CSN complementarity (Fig. 7B, bottom panel). When the 5′-DV CSN molecule, which was an active template in cis, was incubated with the viral 3′-UTR and the RdRp, RNA synthesis in trans was undetectable (Fig. 7C, bottom panel), suggesting that the 3′-SL represses the utilization of the 3′-UTR as template. To confirm this possibility, we designed two different molecules: (a) the 3′-UTR was extended by a duplication of the last 10 nucleotides of the viral genome (Fig. 7C, 3′UTR tail) and (b) a mutation in the 3′-UTR was designed to open the bottom half of the 3′-SL (Fig. 7C, 3′UTR-Mut K). Both 3′-UTR molecules became templates for trans-activation by the SLA in the context of the 5′-DV. Polymerase activity was observed using the templates in cis and trans, confirming the requirement of unstructured 3′ ends for RNA synthesis. These studies confirm that the utilization of the authentic 3′-UTR as template for RNA synthesis requires a change in the 3′-SL structure and that UAR hybridization could facilitate this change.
FIGURE 7.
The SLA and changes in the 3′-SL structure are necessary for the RdRp activity using the 3′-UTR as template. A, trans-initiation assay. Left panel, schematic representation of the interaction of two RNA molecules representing the 5′-DV (160 nucleotides) and 3′-UTR (454 nucleotides) and the viral polymerase initiating at the 3′ end of the two RNA molecules. Right panel, a denaturing 5% polyacrylamide gel showing the radiolabeled products from an in vitro trans-initiation assay. B, schematic representation of the hybridized 5′-DV/3′-SL and the 5′-DV-CSN/3′-SL RNAs. The interacting sequences for both complexes are indicated below the structures. On the bottom, EMSA assays showing the interaction between the 3′-SL WT probe, corresponding to the last 106 nucleotides of the viral genome (nucleotides 10617–10723), and the 5′ DV or the 5′ DV-CSN RNAs. The mobility of the 3′-SL probe is indicated on the left. C, analysis of radiolabeled RNA products in a denaturing 5% polyacrylamide gel using as templates the molecules schematized at the top of the gel: 5′DV-CSN, 5′ end of DENV genome carrying a deletion of the SLB and an insertion of 16 nucleotides downstream 5′-CS; and the combination of 5′DV-CSN with the complete 3′-UTR of 454 nucleotides (5′DV-CSN + 3′-UTR), with the 3′-UTR carrying a duplication of the last 10 nucleotides of the viral genome (5′DV-CSN + 3′UTR-tail) or with the 3′-UTR with seven point mutations at the 3′-SL that mimic the long range interaction (5′DV-CSN + 3′-UTR-Mut K).
DISCUSSION
The mechanisms by which viral RNA polymerases specifically recognize viral genomes are still poorly understood. Here, we dissected structural elements present in the promoter SLA of DENV RNA that are necessary for polymerase binding and activity. Mutations of conserved sequence/structures of the SLA, which were previously found to be necessary for viral replication in infected cells (3, 10), were now identified as elements that contribute to an stable RNA-polymerase complex and polymerase activity in vitro. In addition, we provide evidence that supports the idea that recruitment of the RdRp to the viral RNA is not sufficient to promote RNA synthesis and that specific contacts, which do not contribute significantly in the promoter-RdRp affinity, are necessary for polymerase activity.
DENV RdRp is encoded in the NS5 protein, which also has an MTase domain in its N terminus. Previous studies using WNV reported that NS5 binds the viral RNA mainly through the MTase domain (9), whereas the RdRp showed weak binding by itself. In the case of DENV, in the present and in previous studies (3), we found that the RdRp domain interacts with the viral RNA with high affinity and that the SLA structure provides specificity for the interaction. This difference between WNV and DENV is also supported by a recent report showing that both viruses have different NS5 requirements. In this regard, chimeric DENVs containing the WNV MTase, RdRp, or full-length NS5 were found to be impaired in their replication, indicating that these proteins are not exchangeable (35). These recent observations together with our studies support the idea that, although flaviviruses share general strategies for viral RNA synthesis, they bear differences in regard to RNA recognition by NS5.
Using footprinting, we found that the top loop and nucleotides of the side stem-loop of the SLA element are protected by the RdRp. In addition, EMSA assays demonstrated that these two elements are necessary to form ribonucleoprotein complexes of high affinity (Figs. 2 and 4). However, although mutations or deletions of these elements decrease the affinity for the RdRp (Kd > 240 nm), they did not abolish protein binding, suggesting that other RdRp-RNA contacts are also involved in complex formation.
Within the SLA there is a UU or U bulge conserved in all flavivirus genomes (5–8). Our previous studies, in which this bulge was deleted in the context of the full-length viral RNA, resulted in revertant viruses that restore a U in this position, indicating an absolute requirement of this element for viral replication (10). However, in the present study, deletion of the bulge did not alter polymerase binding or RNA synthesis. This observation indicates that this UU element of the SLA must interact with other RNA sequence or protein that is crucial for viral replication. Furthermore, we observed that polymerase binding changes the structure of the RNA downstream of the SLA. In this regard, the oligo(U) track located at positions 70–77 became more exposed after protein binding. We have previously found that the oligo(U) track functions as a spacer, modulating RNA synthesis in cells transfected with full-length DENV genome (10). The previous observation is in agreement with the in vitro results obtained in the present study, which demonstrates that the RdRp requires a nonstructured nucleotide sequence downstream of the promoter element for interaction. Based on the footprinting exposure of the oligo(U) track upon RdRp binding, we speculate that this region acts as a hinge providing flexibility to the promoter element. Future three-dimensional studies of the structure of the ribonucleoprotein complex will be necessary to better define how the polymerase interacts with the viral RNA.
In addition to the SLA and the nonstructured sequences downstream of this element, we found that RNA synthesis by the DENV recombinant RdRp requires nucleotides at the 3′ end of the template that are not involved in stable secondary structures. Using in vitro assays, RNA templates with fully active SLAs but carrying a highly structured 3′ end element were inactive for polymerase activity. In this regard, the authentic 3′ end of the genome (3′-SL) was found to repress RNA synthesis. This observation could be explained by features of the DENV RdRp three-dimensional structure. It has been proposed that the template channel of this protein is narrow and would not accommodate the 3′ end of the RNA in a double-stranded form (36).
It is still unknown how interactions between the 5′ and 3′ ends of flavivirus genomes provide the correct conformation of the RNA for viral polymerase initiation at the 3′ end. Our in vitro polymerase assays indicate that hybridization of 5′-3′-UAR region is able to release the inhibitory effect of the 3′-SL. Interestingly, a common feature in mosquito- and tick-borne flavivirus cyclization sequences is the location of a complementary sequence within the 3′-SL. In all the genomes, hybridization of this essential element results in the release of the 3′ end of the bottom half of the 3′-SL sequence. Here, we found that changing the location of 3′-UAR outside of the 3′-SL impairs polymerase activity (Fig. 7). However, point mutations that mimic the structure generated by UAR hybridization results in a template that supports RdRp activity. Therefore, it is possible that the long range RNA-RNA interaction and genome cyclization facilitate the accessibility of the polymerase to the 3′ end nucleotides. cis-Acting elements acting as RNA replication silencers have been described at the 3′ end of the genome in other plus-strand RNA viruses. In this regard, structural changes around the 3′ terminal nucleotides of these RNA viruses were found to be a prerequisite for polymerase initiation of RNA synthesis (37–40). These observations, together with our studies, suggest common strategies for the regulatory function of cis-acting elements in diverse plus-strand RNA viruses.
We also analyzed here the requirement of a possible rearrangement of the 3′-SL using the full-length DENV infectious clone in transfected cells. In these studies, we relocated the 3′-UAR sequence outside the 3′-SL, which impaired viral replication, and introduced numerous unstructured elements or sequences at the 3′ of the viral genome to rescue the viral function. Unfortunately, all of the modifications introduced at the 3′ end of the genome were lethal (data not shown). The multiple functions of the 3′-SL in viral replication and the pleiotropic effect of the mutations introduced could explain our observations. In this regard, the results are in agreement with previous studies that have shown an essential function of the last nucleotides of flavivirus genomes (25, 26, 41). Therefore, to further understand the complex process of the structural changes around the 3′-SL during viral replication in infected cells, more information about the multiple functions of the 3′-SL is necessary. Importantly, different host proteins have been reported to interact with the 3′-SL, which could also play a role modulating the availability and the structure of the 3′-SL in vivo (42–46). It is also possible that 5′-3′-UAR hybridization provides the correct orientation of the promoter SLA respect to the 3′ end sequences during initiation of RNA synthesis in vivo. In this regard, mutations of the oligo(U) track (altering the SLA-UAR spacing) were previously shown to impair polymerase activity (10), suggesting a requirement of an adequate orientation of the multiple cis-acting elements.
We believe that understanding molecular aspects of DENV replication will aid the search for antiviral strategies against this important human pathogen. The results presented here provide new information about structural elements of the SLA involved in RdRp interaction and activation. To our knowledge, this is the first report showing that recruitment of the polymerase to the flavivirus RNA is not sufficient for RNA synthesis. Our data provide evidence of a specific activation of the RdRp, which can be dissociated from the binding requirements. Further three-dimensional structural studies of the complex formed between the promoter and the viral RdRp will be necessary to better define the conformational changes in the protein and/or the viral RNA that render an active polymerase for viral multiplication.
Supplementary Material
Acknowledgments
We are grateful to Richard Kinney for dengue virus cDNA clone. We also thank members of the Gamarnik laboratory for helpful discussions.
This work was supported by grants from the Howard Hughes Medical Institute and L'Oreal-UNESCO-CONICET (to A. V. G.) and by CONICET fellowships (to C. V. F., N. G. I., S. M. V., and D. E. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
- DENV
- dengue virus
- RdRp
- RNA-dependent RNA polymerase
- MTase
- methyltransferase
- SLA
- stem-loop A
- SLB
- stem-loop B
- 3′-SL
- 3′-stem-loop
- CS
- conserved sequence
- UAR
- upstream AUG region
- CSN
- new conserved sequence
- Mut
- mutant(s)
- TL
- top loop
- SSL
- side stem-loop
- WNV
- West Nile virus.
REFERENCES
- 1. Gubler D., Kuno G., Markoff L. (2007) in Fields Virology, Vol. 1, pp. 1153–1252, Lippincott-Raven, Philadelphia [Google Scholar]
- 2. Gamarnik A. V. (2010) in Frontiers in Dengue Virus Reserach (Hanley K. A., Weaver S. C. eds) pp. 55–78, Caister Academic Press, Norfolk, UK [Google Scholar]
- 3. Filomatori C. V., Lodeiro M. F., Alvarez D. E., Samsa M. M., Pietrasanta L., Gamarnik A. V. (2006) Genes Dev. 20, 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guzmán Tirado M. G. (1980) Rev. Cubana Med. Trop. 32, 123–130 [PubMed] [Google Scholar]
- 5. Gritsun T. S., Gould E. A. (2007) Virology 366, 8–15 [DOI] [PubMed] [Google Scholar]
- 6. Brinton M. A., Dispoto J. H. (1988) Virology 162, 290–299 [DOI] [PubMed] [Google Scholar]
- 7. Leyssen P., Charlier N., Lemey P., Billoir F., Vandamme A. M., De Clercq E., de Lamballerie X., Neyts J. (2002) Virology 293, 125–140 [DOI] [PubMed] [Google Scholar]
- 8. Thurner C., Witwer C., Hofacker I. L., Stadler P. F. (2004) J. Gen. Virol. 85, 1113–1124 [DOI] [PubMed] [Google Scholar]
- 9. Dong H., Zhang B., Shi P. Y. (2008) Virology 381, 123–135 [DOI] [PubMed] [Google Scholar]
- 10. Lodeiro M. F., Filomatori C. V., Gamarnik A. V. (2009) J. Virol. 83, 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X. F., Jiang T., Yu X. D., Deng Y. Q., Zhao H., Zhu Q. Y., Qin E. D., Qin C. F. (2010) J. Gen. Virol. 91, 1218–1223 [DOI] [PubMed] [Google Scholar]
- 12. Alvarez D. E., Filomatori C. V., Gamarnik A. V. (2008) Virology 375, 223–235 [DOI] [PubMed] [Google Scholar]
- 13. Alvarez D. E., Lodeiro M. F., Ludueña S. J., Pietrasanta L. I., Gamarnik A. V. (2005) J. Virol. 79, 6631–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Corver J., Lenches E., Smith K., Robison R. A., Sando T., Strauss E. G., Strauss J. H. (2003) J. Virol. 77, 2265–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khromykh A. A., Meka H., Guyatt K. J., Westaway E. G. (2001) J. Virol. 75, 6719–6728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kofler R. M., Hoenninger V. M., Thurner C., Mandl C. W. (2006) J. Virol. 80, 4099–4113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Villordo S. M., Gamarnik A. V. (2009) Virus Res. 139, 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu L., Nomaguchi M., Padmanabhan R., Markoff L. (2008) Virology 374, 170–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang B., Dong H., Stein D. A., Iversen P. L., Shi P. Y. (2008) Virology 373, 1–13 [DOI] [PubMed] [Google Scholar]
- 20. Cahour A., Pletnev A., Vazielle-Falcoz M., Rosen L., Lai C. J. (1995) Virology 207, 68–76 [DOI] [PubMed] [Google Scholar]
- 21. Polacek C., Foley J. E., Harris E. (2009) J. Virol. 83, 1161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. You S., Padmanabhan R. (1999) J. Biol. Chem. 274, 33714–33722 [DOI] [PubMed] [Google Scholar]
- 23. You S., Falgout B., Markoff L., Padmanabhan R. (2001) J. Biol. Chem. 276, 15581–15591 [DOI] [PubMed] [Google Scholar]
- 24. Markoff L. (2003) Adv. Virus Res. 59, 177–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Khromykh A. A., Kondratieva N., Sgro J. Y., Palmenberg A., Westaway E. G. (2003) J. Virol. 77, 10623–10629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tilgner M., Shi P. Y. (2004) J. Virol. 78, 8159–8171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu L., Markoff L. (2005) J. Virol. 79, 2309–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tilgner M., Deas T. S., Shi P. Y. (2005) Virology 331, 375–386 [DOI] [PubMed] [Google Scholar]
- 29. Hahn C. S., Hahn Y. S., Rice C. M., Lee E., Dalgarno L., Strauss E. G., Strauss J. H. (1987) J. Mol. Biol. 198, 33–41 [DOI] [PubMed] [Google Scholar]
- 30. Men R., Bray M., Clark D., Chanock R. M., Lai C. J. (1996) J. Virol. 70, 3930–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lo M. K., Tilgner M., Bernard K. A., Shi P. Y. (2003) J. Virol. 77, 10004–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alvarez D. E., De Lella Ezcurra A. L., Fucito S., Gamarnik A. V. (2005) Virology 339, 200–212 [DOI] [PubMed] [Google Scholar]
- 33. Kinney R. M., Butrapet S., Chang G. J., Tsuchiya K. R., Roehrig J. T., Bhamarapravati N., Gubler D. J. (1997) Virology 230, 300–308 [DOI] [PubMed] [Google Scholar]
- 34. Ray D., Shah A., Tilgner M., Guo Y., Zhao Y., Dong H., Deas T. S., Zhou Y., Li H., Shi P. Y. (2006) J. Virol. 80, 8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dong H., Chang D. C., Xie X., Toh Y. X., Chung K. Y., Zou G., Lescar J., Lim S. P., Shi P. Y. (2010) Virology 405, 568–578 [DOI] [PubMed] [Google Scholar]
- 36. Yap T. L., Xu T., Chen Y. L., Malet H., Egloff M. P., Canard B., Vasudevan S. G., Lescar J. (2007) J. Virol. 81, 4753–4765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang G., Zhang J., George A. T., Baumstark T., Simon A. E. (2006) RNA 12, 147–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang G., Zhang J., Simon A. E. (2004) J. Virol. 78, 7619–7633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Olsthoorn R. C., Mertens S., Brederode F. T., Bol J. F. (1999) EMBO J. 18, 4856–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pogany J., Fabian M. R., White K. A., Nagy P. D. (2003) EMBO J. 22, 5602–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Teramoto T., Kohno Y., Mattoo P., Markoff L., Falgout B., Padmanabhan R. (2008) RNA 14, 2645–2656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Blackwell J. L., Brinton M. A. (1997) J. Virol. 71, 6433–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. De Nova-Ocampo M., Villegas-Sepúlveda N., del Angel R. M. (2002) Virology 295, 337–347 [DOI] [PubMed] [Google Scholar]
- 44. García-Montalvo B. M., Medina F., del Angel R. M. (2004) Virus Res. 102, 141–150 [DOI] [PubMed] [Google Scholar]
- 45. Paranjape S. M., Harris E. (2007) J. Biol. Chem. 282, 30497–30508 [DOI] [PubMed] [Google Scholar]
- 46. Yocupicio-Monroy M., Padmanabhan R., Medina F., del Angel R. M. (2007) Virology 357, 29–40 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.