Abstract
Mammalian Sterile 20-like kinase 1 (MST1) protein kinase plays an important role in the apoptosis induced by a variety of stresses. The MST1 is a serine/threonine kinase that is activated upon apoptotic stimulation, which in turn activates its downstream targets, JNK/p38, histone H2B and FOXO. It has been reported that overexpression of MST1 initiates apoptosis by activating p53. However, the molecular mechanisms underlying MST1-p53 signaling during apoptosis are unclear. Here, we report that MST1 promotes genotoxic agent-induced apoptosis in a p53-dependent manner. We found that MST1 increases p53 acetylation and transactivation by inhibiting the deacetylation of Sirtuin 1 (Sirt1) and its interaction with p53 and that Sirt1 can be phosphorylated by MST1 leading to the inhibition of Sirt1 activity. Collectively, these findings define a novel regulatory mechanism involving the phosphorylation of Sirt1 by MST1 kinase which leads to p53 activation, with implications for our understanding of signaling mechanisms during DNA damage-induced apoptosis.
Keywords: Apoptosis, p53, Serine/Threonine Protein Kinase, Signal Transduction, SIRT
Introduction
The protein kinase mammalian Sterile 20-like kinase 1 (MST1)4 contains a Ste20-related kinase catalytic domain in the amino-terminal segment followed by a regulatory domain at the COOH terminus (1). It has been implicated in diverse biological functions, including cell proliferation, differentiation, morphogenesis, and cytoskeletal rearrangements. Previous studies have indicated that the noncatalytic tail of MST1 is cleaved by caspase-3 in response to a number of apoptotic stimuli such as death receptor triggering by CD95/FasL, staurosporine or ceramide, and heat shock and arsenite. The amino-terminal fragment of cleaved MST1 translocates into the nucleus where it contributes to chromatin condensation and then apoptosis (2–4). Furthermore, when MST1 is overexpressed, cleavage and subsequent induced apoptosis can also be observed (5). It has been reported that the two major cleavage sites are Asp-326 and Asp-349, and kinase activation, nuclear translocation, and the ability of MST1 to induce cell death are apparently attenuated by mutating these cleavage sites (4, 6). Recently threonine 183 has been identified as a crucial phosphoactivation site in subdomain VIII of MST1, and it is essential for kinase activation. Autophosphorylation of threonine 183 within the MST1 kinase domain is required for its activation (7). Hippo, a mammalian homolog of MST1/2 in Drosophila, has been extensively shown to restrain cell growth and proliferation through inhibition of the transcription and/or degradation of cyclin E and Drosophila Inhibitor of Apoptosis proteins (8, 9) or the phosphorylation and inhibition of Yorkie (10). In mammals, it has been shown that MST1 can activate c-Jun amino-terminal kinase (JNK) and p38 MAPK kinase signaling pathways through MKK4/MKK7 and MKK3/MKK6, respectively (3). Recently, it has been suggested that JNK is essential and sufficient for MST1 activation and MST1-mediated apoptosis via phosphorylation of serine 82 in MST1 (11, 12). In addition, MST1-induced caspase activation and apoptosis are inhibited by dominant negative JNK, but not by dominant negative p38 or the p38 inhibitor (13).
MST1 induces apoptosis by phosphorylating histone H2B on a relatively conserved site, Ser-14 in mammalian cells and Ser-10 in Saccharomyces cerevisiae (5, 14). We have reported previously that MST1 is implicated in the control of FOXO-dependent neuronal cell death via phosphorylating FOXO3a at Ser-207 and the corresponding site of FOXO1 at Ser-212 (15, 16). Recently, we have shown that phosphorylation of threonine 120 by phosphoinositide 3-kinase/Akt can inhibit the MST1-mediated proapoptotic signaling pathway (17).
Sirt1 is an NAD+-dependent deacetylase that has many substrates that participate in various cellular processes (18). Sirt1-dependent deacetylation of its targets can lead to either an increase or a decrease in the biological activities of these proteins, thus influencing a plethora of cellular processes including transcriptional silencing, genetic control of aging, cell metabolism, energy homeostasis, DNA repair, and cell survival (19–23). p53 functions as a key tumor suppressor and plays a vital role in invoking cellular responses to numerous stresses, including DNA damage, hypoxia, and aberrant proliferation (24, 25). The function of p53 in maintaining genome stability is mainly achieved via p53-dependent apoptosis during tumor suppression or tumor clearance. In general, the biological activity of p53 in response to DNA damage is tightly regulated by its post-translational modification status, particularly by site-specific phosphorylation, acetylation, and ubiquitination (26, 27). Sirt1 has been reported to bind strongly to its substrate p53 and can deacetylate p53 at lysine 382 (28). Sirt1-mediated deacetylation antagonizes p53-dependent transcriptional activation and specifically inhibits p53-dependent apoptosis in response to DNA damage and oxidative stress (29, 30).
Previous studies have shown that MST1 promotion of cell death is p53-dependent (31), but the molecular mechanisms underlying MST1-p53 signaling during apoptosis are still largely unknown. In this work, we found that MST1 promotes genotoxic agent-induced apoptosis in a p53-dependent manner. We also found that MST1 augments p53 acetylation through repressing the deacetylation activity of Sirt1 and its interaction with p53. Sirt1 can be phosphorylated by MST1 and results in the inhibition of Sirt1 activity. Collectively, these observations help us gain further insight into the connection between MST1 kinase and p53 activation in cellular processes and define a novel regulatory mechanism between MST1 and Sirt1, with implications for our understanding of signaling mechanisms during apoptosis.
EXPERIMENTAL PROCEDURES
Reagents and Cell Culture
DMEM and FBS were purchased from Invitrogen. Murine embryonic fibroblast (MEF) cells from MST1+/+ and MST1−/− mice, U2OS, HCT116, H1299, 293T cells, and several tumor cell lines were cultured at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS. Anti-FLAG antibody (M2) was obtained from Sigma. Anti-p53 acetylation and anti-Ser(P)/Thr(P) antibodies were purchased from Cell Signaling Technology (Beverly, MA). Monoclonal anti-MST1 antibody was from Zymed Laboratories Inc..(South San Francisco, CA). Anti-HA, anti-FLAG, anti-GFP, anti-GST, anti-14-3-3β, and anti-p53 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Expression Constructs
3×FLAG-tagged MST1 and 3×FLAG-tagged Sirt1 were cloned, respectively, into a pCMV5 expression vector. HA-tagged p53 was cloned into a pcDNA3 expression vector. The hpRNA targeting sequences used included an MST1 siRNA targeting sequence, GGGCACTGTCCGAGTAGCAGC; a Sirt1 siRNA targeting sequence, GATGAAGTTGACCTCCTCA; and p53 hpRNA, GGAGTCTTCCAGTGTGATGAT. All of these complementary hairpin sequences were commercially synthesized and p53 hpRNA was cloned into pSilencer 2.0 under promoter U6 (Ambion).
Western Blotting
Western blotting was performed according to standard procedures. The appropriate antibodies are noted in the figure legends. Detection of band intensity was carried out with an ECL Western Blotting Analysis System.
Immunoprecipitation and Immunoblotting
Immunoprecipitation and immunoblotting were carried out as described previously (16). Briefly, protein lysates were incubated with appropriate antibodies as indicated in the figure legends in the presence of 15 μl of protein A-protein G (2:1)-agarose beads for 2 h at 4 °C. After washing four times, the immunoprecipitates were subjected to immunoblotting. Protein expression was examined by probing Western blots of total cell lysates or immunoprecipitates with the appropriate antibodies as noted in the figure legends. Detection of band intensity was carried out with an ECL Western Blotting Analysis System.
In Vitro Kinase Assays
Protein kinase assays were carried out as described previously (16). Briefly, reactions were carried out in the presence of 10 μCi of [γ-32P]ATP (Amersham Biosciences) and 3 μm cold ATP in 30 μl of buffer containing 20 mm HEPES (pH 7.4), 10 mm MgCl2, 10 mm MnCl2, and 1 mm dithiothreitol, and the proteins indicated in the figure legends were used as the exogenous substrates. After incubation at room temperature for 30 min, the reaction was stopped by adding protein loading buffer and then separated by SDS-PAGE. Each experiment was repeated three times, and the relative amounts of radioactivity incorporated were determined by autoradiography and quantified with a PhosphorImager (Amersham Biosciences) or immunoblotting using the appropriate antibody as noted in the figure legends.
Luciferase Reporter Assay
H1299 cells were cultured in 24-well plates. 14*p53 binding site artificial luciferase reporter, p21 luciferase reporter, and the plasmids indicated were co-transfected. 36 h after transfection, cells were harvested, and luciferase activity was measured by using a luciferase assay kit (Promega) according to the manufacturer's guidelines. All luciferase activities were normalized to Renilla.
Apoptosis Analysis
Assays were performed according to the manufacturer's guidelines (BD Pharmingen). Briefly, cells were cultured in 6-well plates and grown in DMEM supplemented with 10% FBS for 24 h, then treated with etoposide (37.5 μm) for 36 h. Both floating and attached cells were collected for analysis. Cells were washed with PBS and resuspended in binding buffer containing 5 μl of annexin V followed by flow cytometry (Beckman Coulter). The experiments were performed three times.
RESULTS AND DISCUSSION
MST1 Promotion of Cell Death Is p53-dependent
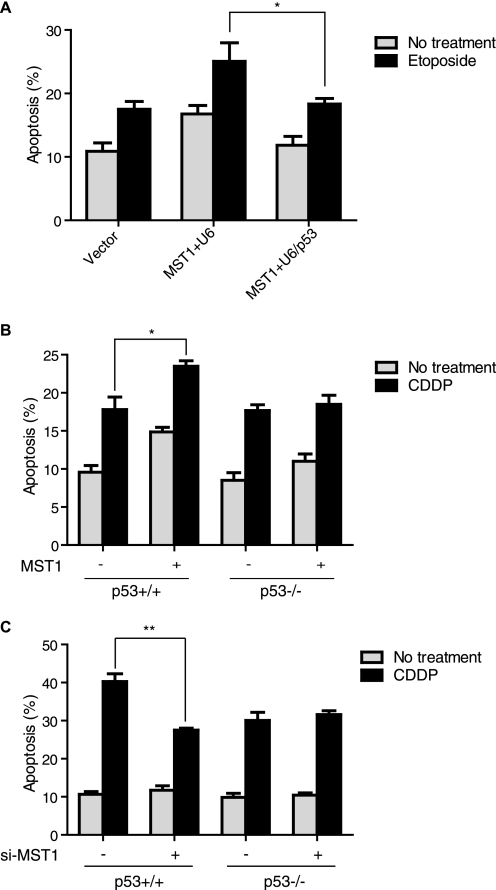
Previous studies have indicated that MST1 induces cell apoptosis upon DNA damage. p53 has been well documented as a major cell death initiator (31, 32). We first investigated whether the induction of cell death by MST1 is p53-dependent. p53 knockdown in U2OS cells reduced MST1 overexpression-induced cell death upon etoposide treatment (Fig. 1A). We also found that MST1 increased cisplatin-induced cell death in HCT116 p53+/+ cells and that MST1 expression failed to induce cisplatin-triggered cell death in HCT116 p53−/− cells (Fig. 1B). Accordingly, knockdown of MST1 mitigated cisplatin-induced cell death in p53+/+ HCT116 cells but not in p53−/− cells (Fig. 1C). Together, these experiments support our conclusion that MST1 induction of cell death under DNA damage is p53-dependent. p53 activity can be regulated through phosphorylation by a panel of kinases, such as CK2, CDK9, and p38 MAPK (33–35). Because MST1 is a serine/threonine kinase and has been shown to promote cell death through phosphorylation of its substrates, such as histone H2B (5, 14) and FOXO (15, 16), we then asked that whether MST1 can phosphorylate p53 and directly regulate p53 function. Using in vitro MST1 kinase assays with recombinant p53 or histone H2B as the substrates, we found that MST1 failed to phosphorylate p53 in vitro (supplemental Fig. S1), indicating that MST1 may regulate p53 indirectly.
FIGURE 1.
MST1 promotion of cell death is p53-dependent. A, U2OS cells were transfected with plasmids encoding MST1 and p53 shRNA or the control vector, then treated with etoposide (36 h) before analyzing apoptosis with annexin V staining and flow cytometry (t test; n = 3, p < 0.05). Error bars represent the mean ± S.E. (error bars). p53 knockdown significantly attenuates MST1-induced cell apoptosis after DNA damage. B, both HCT116 p53+/+ and p53−/− cells stably transfected with MST1 or control vectors were treated with cisplatin (CDDP), and cell death analysis was performed as in A (t test; n = 3, p < 0.05). MST1-induced apoptosis is p53-dependent. C, HCT116 p53+/+ and p53−/− cells stably transfected with si-MST1 or control vectors were treated with cisplatin, and cell death analysis was performed as in A (t test; n = 3, p < 0.01).
MST1 Inhibits Sirt1-mediated Deacetylation of p53
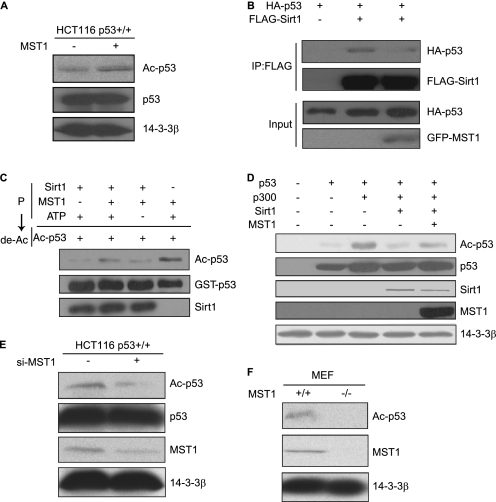
It has been shown that the acetylation and deacetylation of p53 dynamically regulate p53 transactivation and p53-dependent apoptosis (36, 37). Sirt1 is an NAD+-dependent deacetylase and is known to be involved in cell aging and the response to DNA damage (19, 20). Sirt1 binding and deacetylation of p53 protein occur in its COOH-terminal lysine 382 residue, leading to p53 inactivation. Expression of Sirt1 attenuates both transcriptional activity of p53 and p53-dependent apoptosis upon DNA damage (29, 30). On the other hand, in Sirt1-deficient mice p53 hyperacetylation after DNA damage is accompanied by a significant increase in p53-dependent apoptosis in thymocytes (38). As shown in Fig. 2A, we stably transfected MST1 in HCT116 p53+/+ cells and characterized the acetylation status of p53. Western blotting using a p53-specific acetylation antibody revealed that the acetylation of endogenous p53 was up-regulated in MST1-overexpressing cells. We then asked whether MST1 increases the acetylation of p53 through inhibition of the deacetylation activity of Sirt1. First, we confirmed that MST1 and Sirt1 interact in cells (supplemental Fig. S2, A and B), and results suggested that they may interact functionally. We then observed that MST1 overexpression inhibited the interaction between p53 and Sirt1 (Fig. 2B and supplemental Fig. S2C). In addition, we carried out in vitro phosphorylation and deacetylation reactions. In the presence of MST1 and ATP, Sirt1 did indeed decrease Sirt1-induced p53 deacetylation (Fig. 2C), indicating that MST1 inhibits Sirt1-mediated p53 deacetylation through phosphorylating Sirt1. We also found that MST1 reduces Sirt1-mediated FOXO3 deacetylation in vitro (supplemental Fig. S3), indicating that MST1 might regulate the biological function of other Sirt1 substrates.
FIGURE 2.
MST1 inhibits Sirt1-mediated deacetylation of p53. A, lysates of HCT116 p53+/+ cells stably transfected with MST1 or the control vector were immunoblotted with anti-p53-K382Ac or anti-p53 antibody. Gel loading was normalized by using the protein 14-3-3β. Overexpression of MST1 augments the acetylation level of p53. B, FLAG-Sirt1 immunoprecipitates from cells transfected with HA-p53 or GFP-MST1 were immunoblotted with anti-HA or anti-FLAG antibody. MST1 inhibits the interaction between Sirt1 and p53. C, in vitro phosphorylation was performed by incubating active MST1 and Sirt1 in the presence of cold ATP. A deacetylation reaction was then performed by incubating the products of the in vitro phosphorylation reaction with acetylated p53. The reaction was analyzed by SDS-PAGE followed by immunoblotting with anti-p53-K382Ac, anti-GST, or Sirt1 antibodies. MST1 negatively regulates the deacetylation activity of Sirt1 by phosphorylation. D, lysates of 293T cells transfected with plasmids encoding p53, p300, Sirt1, and MST1 as indicated were immunoblotted with anti-p53-K382Ac antibody and other antibodies as indicated. MST1 inhibits Sirt1-mediated repression of acetylation of p53 in vivo. E, lysates of HCT116 p53+/+ cells transfected with si-MST1 and the control vector were immunoblotted with anti-p53-K382Ac antibody and other antibodies as indicated. F, lysates of MST1+/+ or MST1−/− cells were blotted with anti-Ac-p53 antibody, MST1, or 14-3-3β antibody.
Consistent with these observations, in HCT116 cells, Sirt1-mediated deacetylation of p53 was inhibited in the presence of MST1 (Fig. 2D). The endogenous acetylation level of p53 was dramatically reduced by using si-MST1 to knock down endogenous MST1 in HCT116 p53+/+ cells (Fig. 2E). Furthermore, the level of p53 acetylation was lower in MST1−/− MEFs than MST1+/+ MEF cells (Fig. 2F). Taken together, these results indicate that MST1 inhibits Sirt1-mediated p53 deacetylation by inhibiting the interaction between Sirt1 and p53 and the deacetylation activity of Sirt1.
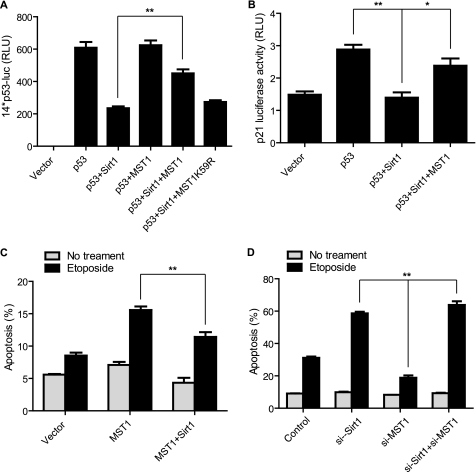
MST1 Increases p53 Transcriptional Activity and DNA Damage-mediated Apoptosis by Inhibiting Sirt1 Activity
To evaluate the functional consequence of interactions among MST1, Sirt1, and p53 proteins, we investigated whether MST1 can influence Sirt1-mediated repression of p53 transcriptional activation. As shown in Fig. 3A, Sirt1 dramatically inhibits p53-mediated expression of the 14*p53 reporter. Wild-type MST1 significantly rescued Sirt1-induced p53 repression, but the kinase-dead MST1 in which the ATP binding site was mutated (MST1 K59R) failed to do so. Similarly, Sirt1 inhibited p53-dependent p21 luciferase activity and MST1 could reverse Sirt1-mediated p21 repression (Fig. 3B). Because we found that MST1 promotes DNA damage-induced apoptosis in U2OS cells (Fig. 1A), we next investigated the functional interaction of MST1 and Sirt1 in the etoposide-induced U2OS cell death. Fig. 3C shows that Sirt1 overexpression decreases MST1-mediated apoptosis. We also found that knockdown of MST1 significantly mitigated the etoposide-induced apoptosis in U2OS cells, whereas Sirt1 knockdown increased cell death (Fig. 3D). Importantly, with knockdown of Sirt1 and MST1 together, the etoposide-induced cell death is similar to that of Sirt1 knockdown, suggesting that Sirt1 functions downstream of MST1 in the DNA damage-induced apoptosis (Fig. 3D). Taken together, these results indicate that MST1 promotes p53 activation through inhibiting Sirt1.
FIGURE 3.
MST1 enhances p53 transcriptional activity. A, H1299 cells were co-transfected with a 14*p53 artificial luciferase construct and p53, MST1, MST1-K59R, or Sirt1 as indicated. Lysates were assayed for dual luciferase activity (t test; n = 4, p < 0.01). Error bars represent the mean ± S.E. MST1 expression decreases Sirt1-mediated repression of p53 transcriptional activation. B, H1299 cells were co-transfected with a p21-luciferase promoter construct and p53, MST1, or Sirt1 plasmids as indicated. Lysates were assayed for dual luciferase activity as in A (t test; n = 3, p < 0.05). Sirt1 reduces p53-mediated p21 expression, and MST1 expression can reverse Sirt1-mediated repression of p53-induced p21 expression. C, U2OS cells were transfected with plasmids encoding MST1 together with Sirt1 or an empty control vector. Apoptosis was analyzed after 36-h etoposide treatment by annexin V staining followed by flow cytometry (t test; n = 3, p < 0.01). D, U2OS cells were transfected with either si-Sirt1, si-MST1, or both si-Sirt1 and si-MST1, or the control vector. Apoptosis was analyzed after 36-h etoposide treatment by annexin V staining followed by flow cytometry (t test; n = 3, p < 0.01).
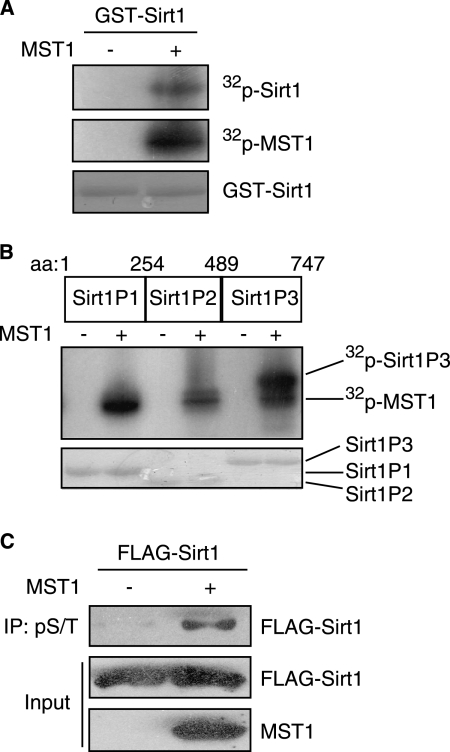
MST1 Phosphorylates Sirt1
Because MST1 kinase activity is required for the inhibition of Sirt1-mediated p53 deacetylation and that MST1 failed to phosphorylate p53 directly, we tested whether MST1 might phosphorylate Sirt1. In vitro MST1 kinase assays using recombinant Sirt1 as the substrate indicated that Sirt1 can be phosphorylated by MST1 (Fig. 4A). We then identified the phosphorylated region within Sirt1 using recombinant GST fusion proteins encoding three nonoverlapping Sirt1 domains (peptides P1–P3). In vitro kinase assays showed that the P3 fragment containing the C-terminal 489–747 amino acids is the major region of phosphorylation (Fig. 4B). We next determined whether MST1 can phosphorylate Sirt1 in cells and found that MST1 expression increased Sirt1 phosphorylation (Fig. 4C). Together, these results support the conclusion that MST1 phosphorylates Sirt1.
FIGURE 4.
MST1 phosphorylates Sirt1 in vitro and in vivo. A. In vitro MST1 kinase assays were performed by incubating recombinant active MST1 with GST-Sirt1 as substrate in the presence of [32P]ATP. The reaction was analyzed by SDS-PAGE followed by autoradiography. Autophosphorylation of MST1 and expression of GST-Sirt1 are shown in the middle panel and bottom panel, respectively. MST1 can phosphorylate Sirt1 in vitro. B, in vitro MST1 kinase assay was performed by incubating the recombinant active MST1 with different Sirt1 fragments (P1, P2, and P3) in the presence of [32P]ATP. The reaction was analyzed by SDS-PAGE followed by autoradiography. The Sirt1 site phosphorylated by MST1 kinase is in the P3 fragment. C, lysates of 293T cells transfected with FLAG-Sirt1 together with MST1 or control vector were immunoprecipitated with anti-Ser(P)/Thr(P) antibody followed by immunoblotting with anti-FLAG antibody. Total lysates were immunoblotted with FLAG or MST1 antibody. MST1 induces Sirt1 phosphorylation in cells.
To summarize, we have identified a new signaling link between MST1 and p53 (supplemental Fig. S4). Our major findings are: (i) MST1 promotion of cell death is p53-dependent; (ii) MST1 cannot phosphorylate p53 directly; (iii) MST1 phosphorylation of Sirt1 inhibits the deacetylation ability of Sirt1; (iv) MST1 increases p53 acetylation and DNA damage-induced apoptosis through inhibiting Sirt1-mediated p53 deacetylation and the interaction between p53 and Sirt1.
It has been reported that Sirt1 is regulated by Ser/Thr kinases, such as CK2 (39), cyclinB/Cdk1 (40) and DYRK1A/DYRK3 (41) at different phosphorylation sites. For example, DYRK1A/DYRK3 promotion of cell survival upon DNA damage through phosphorylation of Sirt1 at Thr-522 results in the enhancement of p53 deacetylation (41), suggesting that phosphorylation of Sirt1 by different kinases confers a variety of biological responses. It will thus be interesting to define the precise MST1 phosphorylation site(s) in the COOH terminus of Sirt1 and to characterize the molecular mechanisms of the phosphorylation that underlies the functional regulation of Sirt1 in the context of DNA damage.
Supplementary Material
Acknowledgments
We thank Dr. Y. Zhang for providing the p21 and 14*p53 luciferase constructs, Dr. J. Ren for the HCT116 cells, and members of the Yuan laboratory for discussion and critical reading of the manuscript.
This work was supported by the National Science Foundation of China Grants 30870792 and 81030025 and the Ministry of Science and Technology of China Grants 973-2009CB918704 and 2006CB910903.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MST1
- mammalian Sterile 20-like kinase 1
- hp
- hairpin
- MEF
- murine embryonic fibroblast
- Sirt1
- Sirtuin 1.
REFERENCES
- 1. Creasy C. L., Ambrose D. M., Chernoff J. (1996) J. Biol. Chem. 271, 21049–21053 [DOI] [PubMed] [Google Scholar]
- 2. Taylor L. K., Wang H. C., Erikson R. L. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 10099–10104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Graves J. D., Gotoh Y., Draves K. E., Ambrose D., Han D. K., Wright M., Chernoff J., Clark E. A., Krebs E. G. (1998) EMBO J. 17, 2224–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee K. K., Murakawa M., Nishida E., Tsubuki S., Kawashima S., Sakamaki K., Yonehara S. (1998) Oncogene 16, 3029–3037 [DOI] [PubMed] [Google Scholar]
- 5. Cheung W. L., Ajiro K., Samejima K., Kloc M., Cheung P., Mizzen C. A., Beeser A., Etkin L. D., Chernoff J., Earnshaw W. C., Allis C. D. (2003) Cell 113, 507–517 [DOI] [PubMed] [Google Scholar]
- 6. Graves J. D., Draves K. E., Gotoh Y., Krebs E. G., Clark E. A. (2001) J. Biol. Chem. 276, 14909–14915 [DOI] [PubMed] [Google Scholar]
- 7. Glantschnig H., Rodan G. A., Reszka A. A. (2002) J. Biol. Chem. 277, 42987–42996 [DOI] [PubMed] [Google Scholar]
- 8. Harvey K. F., Pfleger C. M., Hariharan I. K. (2003) Cell 114, 457–467 [DOI] [PubMed] [Google Scholar]
- 9. Wu S., Huang J., Dong J., Pan D. (2003) Cell 114, 445–456 [DOI] [PubMed] [Google Scholar]
- 10. Huang J., Wu S., Barrera J., Matthews K., Pan D. (2005) Cell 122, 421–434 [DOI] [PubMed] [Google Scholar]
- 11. Ura S., Nishina H., Gotoh Y., Katada T. (2007) Mol. Cell. Biol. 27, 5514–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bi W., Xiao L., Jia Y., Wu J., Xie Q., Ren J., Ji G., Yuan Z. (2010) J. Biol. Chem. 285, 6259–6264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ura S., Masuyama N., Graves J. D., Gotoh Y. (2001) Genes Cells 6, 519–530 [DOI] [PubMed] [Google Scholar]
- 14. Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., Allis C. D. (2005) Cell 120, 25–36 [DOI] [PubMed] [Google Scholar]
- 15. Lehtinen M. K., Yuan Z., Boag P. R., Yang Y., Villén J., Becker E. B., DiBacco S., de la Iglesia N., Gygi S., Blackwell T. K., Bonni A. (2006) Cell 125, 987–1001 [DOI] [PubMed] [Google Scholar]
- 16. Yuan Z., Lehtinen M. K., Merlo P., Villén J., Gygi S., Bonni A. (2009) J. Biol. Chem. 284, 11285–11292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan Z., Kim D., Shu S., Wu J., Guo J., Xiao L., Kaneko S., Coppola D., Cheng J. Q. (2010) J. Biol. Chem. 285, 3815–3824 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18. Imai S., Armstrong C. M., Kaeberlein M., Guarente L. (2000) Nature 403, 795–800 [DOI] [PubMed] [Google Scholar]
- 19. Gottlieb S., Esposito R. E. (1989) Cell 56, 771–776 [DOI] [PubMed] [Google Scholar]
- 20. Haigis M. C., Guarente L. P. (2006) Genes Dev. 20, 2913–2921 [DOI] [PubMed] [Google Scholar]
- 21. Matsushita N., Takami Y., Kimura M., Tachiiri S., Ishiai M., Nakayama T., Takata M. (2005) Genes Cells 10, 321–332 [DOI] [PubMed] [Google Scholar]
- 22. Alcendor R. R., Kirshenbaum L. A., Imai S., Vatner S. F., Sadoshima J. (2004) Circ. Res. 95, 971–980 [DOI] [PubMed] [Google Scholar]
- 23. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 24. Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 25. Riley T., Sontag E., Chen P., Levine A. (2008) Nat. Rev. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 26. Maya R., Balass M., Kim S. T., Shkedy D., Leal J. F., Shifman O., Moas M., Buschmann T., Ronai Z., Shiloh Y., Kastan M. B., Katzir E., Oren M. (2001) Genes Dev. 15, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bode A. M., Dong Z. (2004) Nat. Rev. 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 28. Zhao Y., Lu S., Wu L., Chai G., Wang H., Chen Y., Sun J., Yu Y., Zhou W., Zheng Q., Wu M., Otterson G. A., Zhu W. G. (2006) Mol. Cell. Biol. 26, 2782–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 30. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 31. Lin Y., Khokhlatchev A., Figeys D., Avruch J. (2002) J. Biol. Chem. 277, 47991–48001 [DOI] [PubMed] [Google Scholar]
- 32. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 33. Hupp T. R., Meek D. W., Midgley C. A., Lane D. P. (1992) Cell 71, 875–886 [DOI] [PubMed] [Google Scholar]
- 34. Huang C., Ma W. Y., Maxiner A., Sun Y., Dong Z. (1999) J. Biol. Chem. 274, 12229–12235 [DOI] [PubMed] [Google Scholar]
- 35. Radhakrishnan S. K., Gartel A. L. (2006) Cell Cycle 5, 519–521 [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi H., Woods N. T., Piluso L. G., Lee H. H., Chen J., Bhalla K. N., Monteiro A., Liu X., Hung M. C., Wang H. G. (2009) J. Biol. Chem. 284, 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Luo J., Su F., Chen D., Shiloh A., Gu W. (2000) Nature 408, 377–381 [DOI] [PubMed] [Google Scholar]
- 38. Cheng H. L., Mostoslavsky R., Saito S., Manis J. P., Gu Y., Patel P., Bronson R., Appella E., Alt F. W., Chua K. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 10794–10799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zschoernig B., Mahlknecht U. (2009) Biochem. Biophys. Res. Commun. 381, 372–377 [DOI] [PubMed] [Google Scholar]
- 40. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) PloS One 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo X., Williams J. G., Schug T. T., Li X. (2010) J. Biol. Chem. 285, 13223–13232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.