Abstract
The mammalian circadian oscillator is primarily driven by an essential negative feedback loop comprising a positive component, the CLOCK-BMAL1 complex, and a negative component, the PER-CRY complex. Numerous studies suggest that feedback inhibition of CLOCK-BMAL1 is mediated by time-dependent physical interaction with its direct target gene products PER and CRY, suggesting that the ratio between the negative and positive complexes must be important for the molecular oscillator and rhythm generation. We explored this idea by altering expression of clock components in fibroblasts derived from Per2Luc and Per mutant mice, a cell system extensively used to study in vivo clock mechanisms. Our data demonstrate that the stoichiometric relationship between clock components is critical for the robustness of circadian rhythms and provide insights into the mechanistic organization of the negative feedback loop. Our findings may explain why certain mutant mice or cells are arrhythmic, whereas others are rhythmic, and suggest that robustness of circadian rhythms can be increased even in wild-type cells by modulating the stoichiometry.
Keywords: Gene Regulation, Gene Transcription, Mouse Genetics, Neurological Diseases, Neuroscience, Neurospora, Protein-Protein Interactions, Circadian, Clock Proteins, Negative Feedback
Introduction
Sleep/wake cycles and other mammalian circadian rhythms are synchronized with changes in the local environment, most notably light/dark cycles, through endogenous circadian clocks (1–5). A master clock is located in the suprachiasmatic nuclei in the anterior hypothalamus; this clock adjusts itself based on light/dark information and synchronizes peripheral clocks present in most tissues. The molecular composition and operating mechanism of the clocks are very similar, if not identical, among suprachiasmatic nuclei and peripheral tissues (6, 7).
The cell-autonomous molecular clock consists of several interacting transcriptional/post-translational feedback loops (8, 9). However, as found in most organisms, including Neurospora, Drosophila, and mammals (1, 10–12), one negative feedback loop seems to be the primary driver of clock function; this loop is composed of positive elements and negative elements. In mammalian clock cells, CLOCK (or NPAS2) and BMAL1 are the positive elements, and they form a heterodimer that activates transcription of the negative components PER and CRY, which then constitute an inhibitory complex. The inhibitory complex closes the negative feedback loop by inhibiting the positive complex (CLOCK-BMAL1) through direct physical interaction (3, 4, 13–18). Although CLOCK and BMAL1 are dynamically regulated at the post-translational level in a circadian fashion (14, 19–22), their oscillations in abundance do not seem to be required for clock function (15, 23, 24). However, oscillations of the negative complex are critical for the clock, and PER seems to be rate-limiting for the rhythmic formation of the complex (14, 15). Constitutive overexpression of PER leads to constitutively elevated levels of the negative complex and constitutive down-regulation of CLOCK-BMAL1-controlled genes (15).
Although the precise mechanism of the inhibition by the negative complex is not known, the mode of the inhibition may vary from species to species. In Drosophila, stable and stoichiometric interaction between the negative and the positive complexes seems to be required for the inhibition, as Menet et al. (25) recently showed that Drosophila PER can sequester Drosophila CLOCK in a 1:1 stoichiometric complex with low DNA-binding affinity. However, the mode of the inhibition does not seem to depend on stoichiometric interaction between two complexes in Neurospora. Although the total levels of inhibitor (FRQ) and activator (WCC) complexes are similar in Neurospora, the levels of FRQ are significantly lower than those of WCC in the nucleus, and they do not form stable complexes (26, 27). Nuclear FRQ can overcome the abundance of nuclear WCC by catalytically inactivating nuclear WCC (27–29). Preliminary evidence suggests that the inhibitory mechanism in mammals is similar to that in Drosophila. In the mouse, both positive (CLOCK-BMAL1) and negative (PER1/2-CRY1/2) complexes are predominantly nuclear and about equal in abundance (14, 30). Furthermore, they are co-eluted in fractionation by gel filtration chromatography, and stable complex formation between CLOCK-BMAL1 and PER-CRY and the resulting negative feedback are tightly linked to the abundance of the rate-limiting component PER, supporting the importance of the stoichiometry (14, 15). If the time-dependent stoichiometric interaction between the positive and negative complexes drives the negative feedback inhibition, then the relative ratio between them must be critical for proper functioning of the circadian clock.
To test how varying ratios of negative to positive complexes affect circadian rhythms and the molecular clock in mammals, we modulated expression levels of the complexes in Per2Luc mouse embryonic fibroblasts (MEFs)4 using an adenoviral vector. Previous studies have demonstrated that bioluminescence rhythms from these cells grown in vitro reflect in vivo cell-autonomous circadian rhythms and can be genetically modulated in the same way as in vivo (7, 15, 24, 31–33). In this work, we show that circadian period and amplitude can be profoundly affected by modulation of the relative ratio of negative to positive complexes in the feedback loop. Furthermore, our studies demonstrate that PER1 and PER2 are indeed functionally redundant in the negative feedback loop. In Per1 mutant cells, exogenous expression of Per2 via a Per2 promoter could rescue arrhythmicity of the mutant cells, as could expression of Per1 via the Per2 promoter. Likewise, in Per2 mutant cells, exogenous expression of Per1 could rescue arrhythmicity. Our quantification experiments suggest that CLOCK-BMAL1 is significantly more abundant than PER-CRY in cultured mouse fibroblasts, unlike in liver in vivo. Consistent with our hypothesis, robustness of circadian rhythms in cultured fibroblasts was dramatically enhanced by equalizing the stoichiometry.
EXPERIMENTAL PROCEDURES
Cell Culture and Antibodies
Wild-type MEF protocols have been described previously (15). Per1 and Per2 mutant mouse fibroblasts were generously provided by Drs. Andrew C. Liu and Steve A. Kay. COS-7 cells were obtained from ATCCAmerican Type Culture Collection. Antibodies to clock proteins were described previously (14, 34). Rabbit anti-actin antibody was purchased from Sigma. Anti-V5 and anti-FLAG antibodies were from Invitrogen and Sigma, respectively.
Monitoring of Bioluminescence Rhythms and Immunoblots with MEFs
Bioluminescence rhythms were measured as described previously (15). To measure bioluminescence rhythms from fibroblasts expressing GFP and clock proteins, fibroblasts were infected with GFP- or clock protein-expressing adenovirus for 2 h and serum-shocked with 50% horse serum for 2 h. These fibroblasts were immediately placed into a LumiCycle luminometer (Actimetrics, Wilmette, IL). To detect protein rhythms in MEFs, MEFs in 60-mm dishes were serum-shocked for 2 h, harvested at selected intervals, and subjected to immunoblotting.
Quantification of in Vivo Clock Proteins in MEFs
MEF extracts (in vivo clock proteins) were prepared from MEFs harvested at 16 and 24 h after a 2-h serum shock (35). In vitro translated proteins were prepared using TnT rabbit reticulocyte extract (Promega, Madison, WI) and pcDNA-clock gene templates (see below) in the presence of l-[35S]methionine to allow quantification of the labeled product (14). Known amounts of in vitro translated proteins (∼1, 0.2, and 0.05 fmol) were resolved with MEF extracts on the same blot, and signal intensities were quantified using a film densitometer as has been done previously (14).
Immunoprecipitation, Immunocytochemistry, and Luciferase Reporter Assay
Immunoprecipitation, immunocytochemistry, and the luciferase reporter assay were performed as described previously (16, 36).
Recombinant Plasmids, Adenoviral Constructs, and Virus Production
pcDNA plasmids for Cry1HA, Cry2-HA, Per2-V5, Per1-V5, 3×FLAG-Clock, and 2×HA-Bmal1 were described previously (15, 16). The construction of the recombinant adenoviral vectors encoding various clock proteins and generation of recombinant adenovirus were performed using the procedures of He et al. (37). Adenoviral constructs for GFP, BMAL1, CRY1, Per2-PER2, and Per2-LUC have been described previously (15). For the adenoviral Clock construct, 3×FLAG-Clock was cloned into the XhoI and EcoRV/PmeI sites of pAd-Track-CMV using the following primers: forward, ATCCCTCGAGGCCACCATGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGGTGTTTACCGTAAGCTGTAGTAAAATGAGC; and reverse, TAGGGTTTAAACCTGTGGCTGGACCTTGGAAGGGTC. For the mutant Bmal1 adenoviral construct, 2×HA-mutant Bmal1 (amino acid 86 to the last amino acid) was cloned into the EcoRV site of pAd-Track-CMV using the following primers: forward, ATCCGTTTAAACGCCACCATGTACCCATACGATGTTCCAGATTACGCTTACCCATACGATGTTCCAGATTACGCTGACAAAATG; and reverse, TAGGGTTTAAACTCAATGGTGATGGTGATG. For the Per2-PER1-V5 adenoviral construct, the Per2 promoter and the Bmal1 3′-UTR described previously (15) were transferred into pcDNA3.1-Per1-V5, and then Per2 promoter-Per1-V5-Bmal1 3′-UTR from the pcDNA was cloned into the KpnI/PmeI sites of pAd-Track-CMV by two-step ligation. Complete adenoviral vectors were generated by in vivo recombination as described previously (15).
Period and Amplitude Calculation
The period was calculated with the first four peaks using the periodogram function in the ClockLab software. The amplitudes in Fig. 6C were calculated with five peaks after the fourth peak using the Fast Fourier transform function in the software. Significance levels were measured by Student's t test.
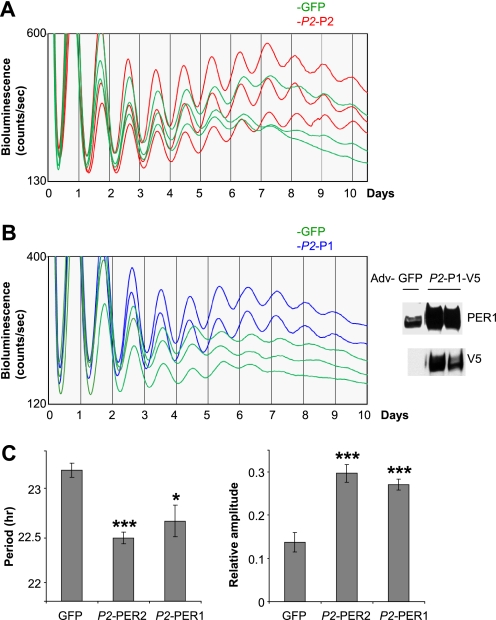
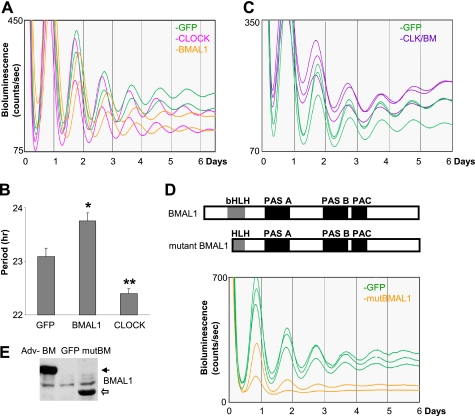
FIGURE 6.
Exogenous expression of PER under the control of the Per2 promoter enhances robustness of circadian rhythms in wild-type MEFs. A and B, robustness of bioluminescence rhythms is enhanced in PER2 (A) or PER1 (B) MEFs compared with GFP MEFs. Exogenous PER1-V5 was overexpressed relative to endogenous PER1 in GFP MEFs as shown in the right panel of B. More traces from separate experiments are shown in supplemental Fig. S5. Adv, adenovirus. C, quantification of A and B. Periods were calculated using the first four peaks, and amplitudes were calculated using five peaks after the fourth peak. For GFP, n = 12; for Per2-PER2 (P2-P2), n = 12; and for Per2-PER1 (P2-P1), n = 6. *, p < 0.05; ***, p < 0.001.
Real-time Quantitative PCR and ChIP
Quantitative PCR in Fig. 4A was performed as described previously (15). For ChIP, MEFs were grown to confluency in 100-mm dishes, infected with GFP or CLOCK/BMAL1 adenovirus for 2 h, and then treated with 1% formaldehyde 24 h after the infection. The reaction was stopped by treatment with 0.125 m glycine and washing with PBS three times. The cells were harvested and processed without SDS as described previously (14). Quantitative PCR was performed on ChIP-isolated DNA for Per1 E3, Per2 E2, and Dbp E2 (14, 38–40). For the top panels in Fig. 4C, PCR amplification was performed for 32 cycles using ChIP samples and 100-fold diluted input samples.
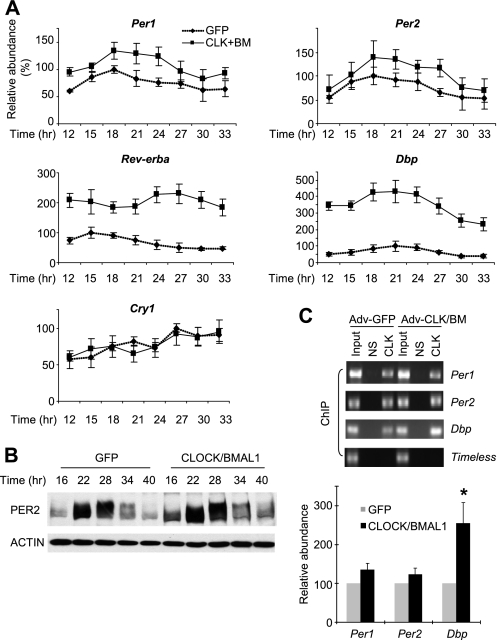
FIGURE 4.
Endogenous clock genes are modulated in a target-specific manner by increased levels of CLOCK/BMAL1. A, MEFs were harvested at the indicated times after GFP or CLOCK/BMAL1 (CLK+BM) adenoviral infection. mRNA levels were measured by quantitative real-time PCR as described previously (15). Levels of Rev-erbα and Dbp were elevated (∼2- and ∼4-fold, respectively, above GFP MEFs), and rhythm amplitudes were significantly reduced in CLOCK/BMAL1-expressing MEFs. Amplitudes of Rev-erbα and Dbp mRNA rhythms in GFP MEFs were ∼2- and ∼3-fold, respectively. Relative values were calculated with the highest number in GFP MEFs set as 100 in each experiment. Results are shown as means ± S.E. of three experiments. B, PER2-LUC protein rhythms in GFP or CLOCK/BMAL1 MEFs. C, CLOCK binding to E-box motifs is elevated in Dbp, but it is not significantly increased in Per1 and Per2 when measured by ChIP. MEFs were infected with GFP or CLOCK/BMAL1 adenovirus (Adv), harvested 24 h later, and subjected to ChIP for CLOCK-bound chromatin as described previously (14, 38). Co-immunoprecipitated DNA levels were measured by real-time quantitative PCR, and relative values were calculated against the value in GFP MEFs. Results are shown as means ± S.E. of three experiments. NS, nonspecific immunoprecipitation. *, p < 0.05.
RESULTS
CLOCK-BMAL1 Is Significantly More Abundant than PER-CRY in the Circadian Negative Feedback Loop in MEFs
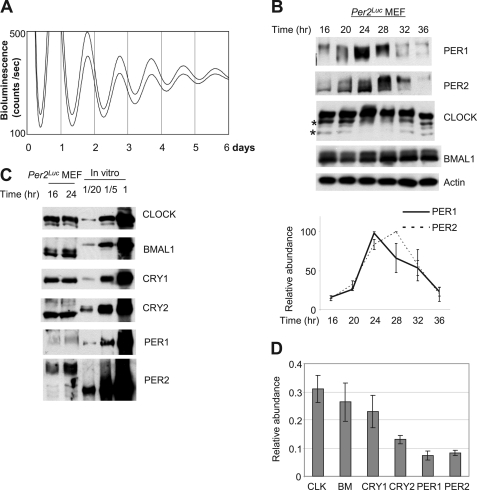
As suggested by circadian bioluminescence in Per2Luc MEFs, the proteins that compose the circadian feedback loop oscillate in cultured MEFs much as they do in vivo (Fig. 1, A and B) (14, 15, 19, 41). Both PER1 and PER2 showed robust oscillations in abundance and phosphorylation, as they do in mouse tissues. As in liver, four isoforms of CLOCK were observed and oscillated in abundance and phosphorylation. BMAL1 exhibited modest oscillations in abundance and phosphorylation. Although it has been suggested from previous studies that PER is limiting for formation of the inhibitory PER-CRY complex in liver and cultured fibroblasts (14, 15), the stoichiometric relationship between negative and positive components is not known in cultured fibroblasts; this knowledge is critical for using this system to test our hypothesis. Thus, we determined absolute concentrations of the clock components in the feedback loop by comparing peak levels of in vivo clock proteins with known amounts of in vitro translated proteins, as has been done previously (14). The comparison of clock proteins was performed with MEFs harvested 24 h after serum shock because our quantification results showed that the limiting clock components PER1 and PER2 are both near peak levels at this time (Fig. 1, B–D). In MEFs, CLOCK, BMAL1, and CRY1 were similarly abundant, which is different from liver, where BMAL1 is far less abundant than the other two (Fig. 1, C and D). CRY2, PER1, and PER2 were less abundant than the other core clock proteins. Unlike in mouse liver, where the levels of PER1/2 (the limiting component in the negative complex) to BMAL1 (the limiting component in the positive complex) are almost 1:1, in MEFs, the combined levels of PER1 and PER2 were only about half of those of CLOCK and BMAL1, implying that CLOCK-BMAL1 would be twice as abundant as the negative complex, PER-CRY, assuming that positive and negative heterodimers predominate over other possible complexes, including homodimers. Like PER, endogenous CLOCK and BMAL1 were predominantly nuclear in MEFs (supplemental Fig. S1) (41). Based on these data, it is tempting to speculate that the low amplitude in clock gene and clock-controlled gene mRNA rhythms in fibroblasts (e.g. Refs. 7, 15, and 42) could be due to inadequate levels of the negative complex, resulting in weaker inhibition of the positive complex as compared with liver tissue. On the other hand, it was intriguing that the negative feedback loop is still functioning in MEFs, although there is significantly less of the negative complex than the positive complex. This raised the possibility that the feedback inhibition may also be mediated at least in part by a catalytic activity of the negative complex, as described in Drosophila, where the inhibitor complex induces phosphorylation of Drosophila CLOCK and dissociation of the positive complex from DNA (43, 44).
FIGURE 1.
PER is the stoichiometrically limiting factor in the circadian negative feedback loop in MEFs. A, robust bioluminescence rhythms in Per2Luc MEFs. The bioluminescence rhythms were measured after a 2-h serum shock. B, circadian rhythms of endogenous clock proteins in MEFs. Per2Luc MEFs were harvested at the indicated times after serum shock and immunoblotted. Note that there are four CLOCK isoforms: two hyperphosphorylated isoforms and two non- or hypophosphorylated isoforms (indicated by two asterisks), as has been shown in liver (14). Relative abundance of PER1 and PER2 was calculated from three different experiments (bottom panel). C, PER is the limiting factor among the clock proteins. Endogenous clock proteins in MEFs harvested at 16 and 24 h after serum shock were compared with known amounts of in vitro translated proteins on the same blot. Based on data in B and D, combined levels of PER1 and PER2 peak at 24 h after serum shock. 1× in vitro translated proteins is roughly equal to 1 fmol. D, relative abundance of endogenous clock proteins at 24 h in MEFs. Results are shown as means ± S.E. of three experiments. CLK, CLOCK; BM, BMAL1.
Overexpression of CLOCK-BMAL1 Reduces Amplitudes but Does Not Substantially Disturb Circadian Bioluminescence Rhythms
Although previous studies demonstrated that the interacting feedback loops controlling CLOCK and BMAL1 oscillations are dispensable for circadian rhythm generation and that constitutive expression of exogenous BMAL1 can rescue arrhythmicity in the Bmal1 mutant mouse and cells derived from the mouse (19, 23, 24), it has not been shown how overexpression of both CLOCK and BMAL1 would affect the clock and circadian rhythms in vivo. Based on our quantitative findings in MEFs, overexpression of either one would not effectively increase the levels of the CLOCK-BMAL1 complex; CLOCK and BMAL1 levels are similar in MEFs (unlike in liver), so either component could be limiting for complex formation. Furthermore, in reporter assays in cultured cells, expression of CLOCK or BMAL1 alone does not increase E-box-mediated transcription of the reporter gene (16, 45). Strong transcriptional activation can be observed only when CLOCK and BMAL1 are coexpressed, indicating that heterodimer formation is a prerequisite for E-box-mediated transcriptional activation of the clock and clock-controlled genes. Because in vitro reporter assays showed that transcriptional activation can be increased by CLOCK-BMAL1 in a dose-responsive manner (22), it was expected that co-overexpression of CLOCK and BMAL1 could compromise the molecular clock by exceeding the capacity of the negative complex to inhibit transcription.
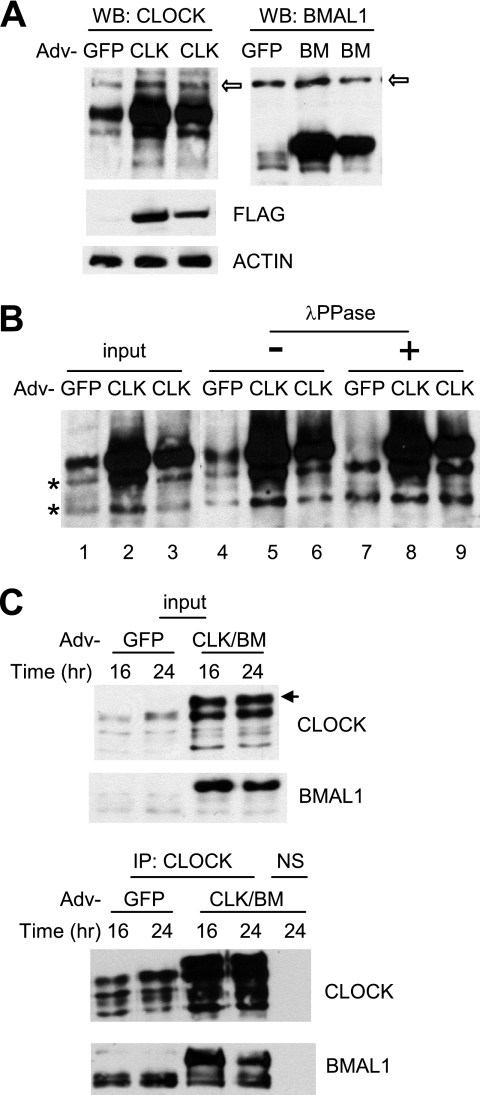
To effectively express CLOCK and BMAL1 in MEFs, we generated adenoviral constructs as described previously (15). When these constructs were tested in a reporter assay, adenoviral coexpression of CLOCK and BMAL1 activated transcription from a Per1-luciferase reporter gene to a level comparable with that produced by transfection-mediated expression of CLOCK and BMAL1 (supplemental Fig. S2A). The adenoviral expression of CLOCK and BMAL1 also activated transcription of the luciferase reporter from a Per2 promoter and could be inhibited by CRY1 (supplemental Fig. S2B), demonstrating that the virus-expressed CLOCK and BMAL1 are functionally normal in terms of activation and inhibition. When compared with endogenous proteins, the levels of exogenous CLOCK and BMAL1 were 3–5- and ∼10-fold higher in MEFs, respectively (Fig. 2A). The size of the exogenous BMAL1 was distinctively larger than that of the endogenous BMAL1 (the smear of the endogenous band is due mainly to phosphorylation as in in vivo tissues (supplemental Fig. S3)), but the exogenous CLOCK co-migrated with the larger hyperphosphorylated species of endogenous CLOCK (Fig. 2A) (14). Phosphatase treatment of immunoprecipitated CLOCK from control cells resolved the four species of in vivo CLOCK into two non-phosphorylated species (Fig. 2B, compare lanes 4 and 7), as reported previously in liver (14). However, the main exogenous CLOCK band was not affected by the treatment (Fig. 2B, compare lanes 5, 6, 8, and 9), suggesting that CLOCK is not phosphorylated when expressed alone, as has been demonstrated previously (22). Exogenous CLOCK was efficiently phosphorylated (similar to in vivo CLOCK) when BMAL1 was coexpressed (indicated by the arrow in Fig. 2C, top panel) (22). In addition, immunoprecipitation revealed that exogenous CLOCK and BMAL1 were involved in formation of the positive complex (Fig. 2C, bottom panels). In summary, the exogenous CLOCK and BMAL1 are potent in activating transcription, can form a heterodimer, and are post-translationally regulated like their in vivo counterparts.
FIGURE 2.
Adenoviral CLOCK and BMAL1 are functional in cultured cells. A, adenoviral constructs overexpress exogenous CLOCK and BMAL1 proteins in MEFs. MEFs were infected with GFP, CLOCK (CLK), or BMAL1 (BM) adenovirus (Adv) and harvested 24 h later. 1× (center lane) and 0.5× (last lane of each blot) titers were used for CLOCK and BMAL1. The white arrows indicate nonspecific bands. Note that exogenous CLOCK co-migrated with a hyperphosphorylated endogenous CLOCK isoform (see B). Expression of FLAG-CLOCK was confirmed by FLAG immunoblot (center panel). B, resolution of exogenous versus endogenous CLOCK protein. MEFs were infected with GFP or CLOCK adenovirus and harvested as described above. MEF extracts (input) were immunoprecipitated and treated without (−) or with (+) λ-protein phosphatase (λPPase). Note that phosphatase treatment resulted in two non-phosphorylated CLOCK isoforms in GFP MEFs (indicated by two asterisks) but three isoforms in CLOCK MEFs, indicating that the top band is exogenous CLOCK. C, exogenous CLOCK and BMAL1 form a heterodimer. MEFs were infected with GFP or CLOCK/BMAL1 adenovirus and harvested at 16 and 24 h. Bottom panels are immunoprecipitated (IP) samples from the top panels, which are input samples for immunoprecipitation. Note an extra band in CLOCK/BMAL1 (CLK/BM) MEFs in the top panel. This is a hyperphosphorylated exogenous CLOCK isoform that can be seen only when co-infected with BMAL1 adenovirus. NS, nonspecific immunoprecipitation.
To determine how increased levels of the positive components in the feedback loop affect circadian rhythms, CLOCK and BMAL1 were overexpressed in Per2Luc MEFs (Fig. 3, A and B). When CLOCK and BMAL1 were individually expressed, overexpression of CLOCK and BMAL1 significantly shortened and lengthened the circadian period, respectively. It is not clear why the circadian period is differentially regulated by overexpressed CLOCK and BMAL1. It could be due to different expression levels or functional antagonism of the overexpressed CLOCK and BMAL1 in the negative feedback loop. In any case, intact robust circadian rhythms in CLOCK- or BMAL1-overexpressing cells are consistent with our stoichiometry data showing that expression of either protein would not effectively increase levels of the positive complex because CLOCK and BMAL1 in MEFs are almost equimolar. Thus, both CLOCK and BMAL1 were coexpressed in MEFs to efficiently increase the levels of the CLOCK-BMAL1 complex (Fig. 3C). The bioluminescence was still rhythmic in the cells and was not substantially disrupted compared with control cells, as assessed by the ClockLab software. In contrast to the transient reporter assays, basal levels of bioluminescence were only slightly elevated. However, amplitudes were reduced by the increased PER2-LUC basal levels, which is attributable to a higher ratio of the CLOCK-BMAL1 complex to the PER-CRY complex as compared with control cells and thus less robust feedback inhibition. This was most notable in the 2nd and 3rd days after serum shock, before amplitudes of bioluminescence started to damp quickly even in control cells. We could not observe significant changes in the period between CLOCK/BMAL1- and GFP-expressing cells (for GFP, n = 11 samples; for CLOCK/BMAL1, n = 10 samples; p = 0.36), suggesting that the differential regulation of the period by overexpression of the individual activators may be due to functional antagonism of CLOCK and BMAL1 overexpression on the circadian clock.
FIGURE 3.
PER2-LUC rhythms are relatively intact in CLOCK/BMAL1-overexpressing MEFs. A and B, overexpression of CLOCK or BMAL1 alters the circadian period. Period quantification is shown as means ± S.E. (for GFP, n = 13 samples; for CLOCK, n = 10; and for BMAL1, n = 8) in B. *, p < 0.05; **, p < 0.01. C, coexpression of CLOCK and BMAL1 reduces rhythm amplitude but does not severely disrupt circadian rhythms. Note that basal lines were elevated around the second and third peaks in CLOCK/BMAL1 (CLK/BM) MEFs. D, a mutant BMAL1 lacking the DNA-binding domain significantly disrupts circadian rhythms. More traces from a different experiment are shown in supplemental Fig. S4. bHLH, basic helix-loop-helix; PAC, PAS-associated C-terminal domain. E, expression levels of mutant BMAL1 (mutBM) are similar to those of adenovirus (Adv)-expressed wild-type BMAL1 in MEFs. MEFs were harvested 24 h after adenoviral infection.
Although in vitro characterization of our adenoviral constructs indicated that they express active transcription factors in cultured cells, the mild phenotype in MEFs suggests that the adenoviral proteins may not regulate in vivo target clock genes as efficiently as the in vitro data suggested. To validate that adenoviral expression did not grossly impair protein function, we tested if an exogenous dominant-negative BMAL1 mutant could disrupt the endogenous CLOCK-BMAL1 complex and thus compromise PER2-LUC rhythms. We generated a BMAL1 mutant lacking the DNA-binding basic region (Fig. 3D) (46). The BMAL1 mutant could still interact with CLOCK through helix-loop-helix and PAS domains, but the resulting complex would not bind E-box motifs, and thus the BMAL1 mutant would act as a dominant-negative mutant (47, 48). Overexpression of the BMAL1 mutant effectively disrupted circadian rhythms and lowered basal levels of bioluminescence, suggesting that transactivation of the endogenous Per2-luc gene had been compromised, as expected (Fig. 3, D and E). Taken together, the data show that the mild phenotype despite the coexpression of CLOCK and BMAL1 is not due to ineffectiveness of the exogenous CLOCK and BMAL1 expressed by the adenoviral vector.
Differential Regulation of CLOCK-BMAL1 Target Genes by Overexpressed CLOCK/BMAL1
We investigated clock and clock-controlled genes for their expression levels in CLOCK/BMAL1-overexpressing cells to determine how transcription of the target genes is affected by overexpressed CLOCK-BMAL1. Consistent with the bioluminescence data, mRNA levels of Per2-luc were only slightly elevated, and the same was true of Per1 (Fig. 4A). Immunoblot data for PER2-LUC were also in accordance with the mRNA data (Fig. 4B). However, mRNA levels for the clock-controlled genes Rev-erbα and Dbp were increased dramatically (Fig. 4A), demonstrating that the exogenous CLOCK/BMAL1 can indeed form an active transcriptional heterodimer and up-regulate at least some of their in vivo target genes, as observed in the transient reporter assays. There was no significant difference in Cry1 mRNA levels between CLOCK/BMAL1- and GFP-overexpressing cells. To test if the differential transcriptional regulation may result from differences in CLOCK-BMAL1 binding to promoters of the genes (i.e. if elevated CLOCK-BMAL1 levels lead to increased binding of CLOCK-BMAL1 in the promoters of those genes with enhanced transcriptional activation), we measured CLOCK-BMAL1 binding to E-box motifs in Per1, Per2, and Dbp genes using ChIP (Fig. 4C). CLOCK-BMAL1 binding to the promoter of Dbp was increased in proportion to the levels of total CLOCK/BMAL1, suggesting that enhanced binding of CLOCK-BMAL1 is indeed responsible for the high levels of Dbp mRNA (Fig. 4C). However, CLOCK-BMAL1 binding to the promoters of Per1 and Per2 was not significantly changed in CLOCK/BMAL1-overexpressing cells (Fig. 4C), suggesting that CLOCK-BMAL1 binding to E-box motifs in Per genes is already saturated with endogenous levels of CLOCK and BMAL1, and the detailed mechanism for activation and inhibition in Per genes may be different from those for other target genes, as suggested previously (e.g. Refs. 14, 39, and 49). For Per1 and Per2 genes, transcriptional activation and inhibition may occur with constitutive binding (but rhythmic activity) of CLOCK-BMAL1 to the promoters, whereas in other target genes such as Dbp, rhythmic transcriptional activation is paralleled by rhythmic CLOCK-BMAL1 binding to the promoter (14, 39, 49).
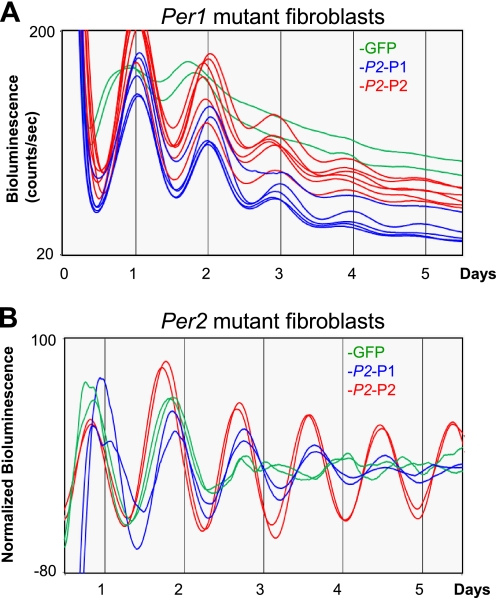
Increased PER Levels in Per1 or Per2 Mutant Fibroblasts Rescue Arrhythmicity in the Cells
Our quantitative data on clock protein expression in fibroblasts suggest an intuitive explanation at the biochemical level for why cultured fibroblasts derived from Per1, Per2, and Cry1 mutant mice are arrhythmic (7): the lower levels of PER and CRY2 expression in the fibroblasts (Fig. 1D) render the cells more sensitive to loss of further components. Note that this explanation assumes that PER1 and PER2, and CRY1 and CRY2, are redundant in the feedback loop. Thus, PER1 levels in Per2 mutant cells, PER2 levels in Per1 mutant cells, or CRY2 levels in Cry1 mutant cells are not adequately expressed to allow formation of functionally sufficient negative complexes to counterbalance the CLOCK-BMAL1 complexes. If the arrhythmicity in Per mutant fibroblasts is indeed caused by insufficient levels of PER and therefore insufficient levels of PER-CRY complex relative to CLOCK-BMAL1, rather than by functional differences between PER1 and PER2, then arrhythmicity should be rescued by increasing either PER1 or PER2 levels in the Per mutant cells. To test this possibility, we exogenously expressed PER1 or PER2 via the Per2 promoter in the mutant cells (Fig. 5). Consistent with our prediction of functional redundancy between PER1 and PER2, exogenous expression of PER1 and PER2 in Per1−/−/Per2Luc mutant cells rescued the arrhythmicity equally well (Fig. 5A). The robustness of rhythms in exogenous PER-expressing Per1 mutant cells was apparently achieved by deeper troughs in PER2-LUC expression (as opposed to higher peaks), consistent with intensified negative feedback. Because the Per2 mutant cells do not have any endogenous reporter that can be monitored in real time, a luciferase reporter under the control of a Per2 promoter (exogenous Per2 promoter-luciferase reporter) was introduced into the cells, as has been done previously (15). As in Per1 mutant cells, both exogenous Per2 promoter-driven PER1 and PER2 rescued the arrhythmicity of Per2 mutant fibroblasts, but PER2-expressing Per2 mutant cells exhibited more robust rhythms than PER1-expressing mutant cells, suggesting that Per1 and Per2 may not be equally functional and redundant in the mutant cells. Because it has been shown that Per1 (and most likely Per2, too) is required for cell-autonomous rhythm generation in individual fibroblasts (7), our data strongly suggest that modulation of the relative ratio between the positive and negative complexes affects the cell-autonomous oscillators, rather than reinforcing synchronization among fibroblasts for a longer period (50, 51). Overall, our data strongly support our hypothesis that the major negative feedback loop in the clock mechanism is driven by stoichiometric interaction between the positive and negative complexes, and cell-autonomous robustness can be modulated by regulating the relative abundance between them.
FIGURE 5.
Exogenous expression of PER1 or PER2 via the Per2 promoter can rescue arrhythmicity in Per1 and Per2 mutant fibroblasts. A, rescued rhythmicity in Per1 mutant fibroblasts by exogenous expression of PER. Results are representative of at least five experiments. B, rescued rhythmicity in Per2 mutant fibroblasts by exogenous expression of PER. Note that bioluminescence rhythms are presented after the base-line luminescence is subtracted because of increased base line of wild-type luciferase. Two traces of each sample are shown, representative of three experiments. Rhythms were not detectible after the second peak in GFP-overexpressing Per2 mutant fibroblasts.
Overexpression of PER2 or PER1 under the Control of a Per2 Promoter Enhances Robustness of Circadian Rhythms in Wild-type MEFs
We also tested the circadian system by increasing the levels of the negative complex PER-CRY by overexpressing the limiting components PER1 and PER2 using the Per2 promoter in WT MEFs. In a previous study, we showed that circadian rhythms and the molecular clock are completely compromised when exogenous PER2 is constitutively overexpressed or expressed in antiphase to endogenous Per2, whereas circadian rhythms are maintained if exogenous Per2 is expressed in phase with endogenous Per2 (15). During the course of that study, we noticed that bioluminescence rhythms are more robust and last longer in MEFs expressing PER2 via the exogenous Per2 promoter as compared with GFP-expressing MEFs. On the basis of our hypothesis and the above results, we speculate that the negative feedback loop in the clock may have been reinforced by improved stoichiometry between the positive and negative complexes in the MEFs. Endogenous PER2 levels are approximately one-third to one-fourth of those of CLOCK-BMAL1 (Fig. 1D); the overexpression of PER2 was ∼4-fold higher than that of endogenous PER2 (15). Thus, the total levels of PER1/2-CRY1/2 would have been close to or a little more than those of CLOCK-BMAL1 in the Per2-PER2-overexpressing MEFs. PER-CRY levels are also similar to CLOCK-BMAL1 levels in mouse liver, which has very high amplitude clock protein and mRNA rhythms compared with cultured fibroblasts.
To determine quantitatively how increased levels of the negative complex PER-CRY affect circadian rhythms, we exogenously expressed the limiting components of the negative complex, PER2 and PER1, via the Per2 promoter in WT MEFs (Fig. 6). Consistent with our hypothesis and previous data, the circadian period was significantly shortened, and robust circadian bioluminescence rhythms were maintained much longer in Per2 promoter-PER2 MEFs than in GFP MEFs (Fig. 6, A and C). Enhanced robustness was most dramatic when amplitudes were quantitatively compared between the groups of cells using later days of the bioluminescence record (Fig. 6C). We believe that the enhanced robustness by exogenous PER2 expression was achieved in a cell-autonomous manner: increased PER would have reinforced the negative feedback loop as it has done in Per mutant cells, thus making the feedback loop more resistant to intrinsic damping processes in individual cells. However, we cannot rule out that PER overexpression may have reduced the variability in the intrinsic circadian periods of the cells; the resulting improvement in synchronization may have also contributed to the robustness of ensemble rhythms (50, 51). In any case, our data demonstrate that robustness of circadian rhythms can be modulated at the population level by changing the stoichiometric relationship between clock components in individual oscillators. Similar effects were observed with expression of exogenous PER1 via the Per2 promoter (Fig. 6, B and C), further supporting that both PER1 and PER2 are rate-limiting and functionally redundant in the circadian negative feedback loop.
DISCUSSION
Numerous genetic and biochemical studies have revealed essential clock genes and posit a convincing model that these gene products constitute a self-sustaining negative feedback loop via physical interactions. This model implies that stoichiometry of clock components is crucial for rhythm generation, and yet this has not been intensively explored. Thus, by studying the stoichiometric relationship between the activator and inhibitor complexes, we tested the viability of the current model of how the clock works and refined our understanding of one of its most fundamental aspects.
It has been observed that clock protein and mRNA rhythms are much less robust in cultured fibroblasts, even immediately after synchronization, than in mouse tissues in vivo. The lower amplitude of rhythms in cultured fibroblasts may be explained by the relative abundance of CLOCK-BMAL1 over PER-CRY (Fig. 1D), as opposed to the situation in mouse liver, where CLOCK-BMAL1 levels are similar to those of PER-CRY (14). This hypothesis is strongly supported by our findings that robustness of circadian rhythms in WT and Per mutant cells can be dramatically enhanced by increasing levels of PER-CRY close to those of CLOCK-BMAL1.
Using our model that the stoichiometry between CLOCK-BMAL1 and PER-CRY is a key determinant of rhythm amplitude, we could explain why Cry1 mutant fibroblasts are almost arrhythmic, whereas Cry2 mutant fibroblasts are robustly rhythmic (7). Because in vitro reporter assays suggested that CRY1 and CRY2 are almost equally potent as inhibitors of CLOCK-BMAL1-mediated transcription (16, 17), our data imply that different expression levels of CRY1 and CRY2 account for the circadian phenotypes in Cry mutant fibroblasts. In the absence of CRY1 in the MEFs, CRY2 would be the limiting factor in forming the PER-CRY complex, and the low levels of endogenous CRY2 expression (Fig. 1D) may result in insufficient levels of the PER-CRY complex to close the negative feedback loop and generate sustained circadian rhythms.
To rigorously test the importance of stoichiometry, we used adenoviral vectors to alter the ratio of CLOCK-BMAL1 to PER-CRY. In good agreement with studies in Drosophila (52–55), overexpression of PER under the control of a Per2 promoter, to be in phase with endogenous PER, significantly shortened the circadian period (Fig. 6) (15). This is probably due to more rapid accumulation of the negative complex PER-CRY, leading to advanced repression as suggested by Kadener et al. (52). We also found that exogenous PER1 expression in Per2 mutant cells and exogenous PER2 expression in Per1 mutant cells could rescue the defective circadian clock function; the fact that restoring the balance of PER-CRY complexes to CLOCK-BMAL1 is sufficient for rhythmicity strongly supports our conclusion that proper stoichiometry is essential for the robustness of circadian rhythms and confirms the redundancy of PER1 and PER2 in the feedback mechanism.
In our ChIP experiments, CLOCK-BMAL1 overexpression increased the CLOCK binding to E-box motifs in the Dbp gene, suggesting that increased mRNA levels of Dbp are due to increased transcription through CLOCK-BMAL1-mediated activation and that the E-box motifs are not saturated with endogenous levels of CLOCK-BMAL1. However, in Per genes in the same cells, CLOCK binding to E-box motifs did not significantly increase, despite the >3-fold increase in CLOCK/BMAL1 levels compared with endogenous levels. The modest increase in Per transcription in CLOCK-BMAL1-overexpressing cells may be explained by saturation of E-box motifs with endogenous levels of the transcription factors. Per expression would still oscillate in the cells because CLOCK-BMAL1 bound to E-box motifs would be rhythmically inhibited by oscillating PER-CRY. The slight weakening of inhibition may have occurred because increased amounts of CLOCK/BMAL1 would titrate out PER/CRY to some degree before they disrupt the CLOCK-BMAL1 activity on the E-box motifs. Consistent with this prediction, we observed increased basal levels in bioluminescence rhythms. The persistence of rhythms despite a large excess of positive elements may also suggest that a catalytic mechanism for PER-CRY-mediated inhibition of CLOCK-BMAL1 plays a role in the mammalian clockwork, as has been described for Neurospora and Drosophila.
Resilience of Per transcription has been reported in other genetically altered mice. Per mRNA levels are much more mildly modulated compared with other direct target genes such as Dbp and Rev-erbα in Clock- or Bmal1-null mutant mice (24, 57). In addition, circadian gene expression is maintained even when global transcriptional rates in fibroblasts are dramatically reduced by treatment with transcription inhibitor drugs (58). Because it has been shown that Per oscillations are most critical for circadian rhythm generation (15), resilience of circadian transcription may have been sustained because of reduced yet rhythmic Per expression in the drug-treated cells. The resilience of the molecular clock to wide ranges of clock protein levels may be a conserved feature of eukaryotic clock systems because it has also been shown in Neurospora (56).
In conclusion, our findings on circadian clock protein stoichiometry provide new insights into mechanisms underlying rhythm amplitude variation and into the effects of manipulating clock gene expression. Our data suggest that modulation of Per expression may be an attractive target for pharmacological intervention to enhance or restore circadian rhythms in humans suffering from certain circadian disorders or from age-related circadian disturbances (59). Also, because the transacting element (CLOCK-BMAL1) is the same for Per and other target genes, it is tempting to speculate that resilience of Per transcription and the circadian clock is at least partially encoded in Per loci.
Supplementary Material
Acknowledgments
We thank Jiangqin Lai for excellent technical assistance during the project and Dennis Chang for assistance with manuscript revisions.
This work was supported, in whole or in part, by National Institutes of Health Grant NS-053616 (to C. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Allada R., Emery P., Takahashi J. S., Rosbash M. (2001) Annu. Rev. Neurosci. 24, 1091–1119 [DOI] [PubMed] [Google Scholar]
- 2. Green C. B., Takahashi J. S., Bass J. (2008) Cell 134, 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lowrey P. L., Takahashi J. S. (2004) Annu. Rev. Genomics Hum. Genet. 5, 407–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reppert S. M., Weaver D. R. (2002) Nature 418, 935–941 [DOI] [PubMed] [Google Scholar]
- 5. Schibler U. (2005) EMBO Rep. 6, S9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yagita K., Tamanini F., van Der Horst G. T., Okamura H. (2001) Science 292, 278–281 [DOI] [PubMed] [Google Scholar]
- 7. Liu A. C., Welsh D. K., Ko C. H., Tran H. G., Zhang E. E., Priest A. A., Buhr E. D., Singer O., Meeker K., Verma I. M., Doyle F. J., 3rd, Takahashi J. S., Kay S. A. (2007) Cell 129, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dibner C., Schibler U., Albrecht U. (2010) Annu. Rev. Physiol. 72, 517–549 [DOI] [PubMed] [Google Scholar]
- 9. Gallego M., Virshup D. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 139–148 [DOI] [PubMed] [Google Scholar]
- 10. Liu Y., Bell-Pedersen D. (2006) Eukaryotic Cell 5, 1184–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Panda S., Hogenesch J. B., Kay S. A. (2002) Nature 417, 329–335 [DOI] [PubMed] [Google Scholar]
- 12. Loros J. J., Dunlap J. C. (2001) Annu. Rev. Physiol. 63, 757–794 [DOI] [PubMed] [Google Scholar]
- 13. Cuninkova L., Brown S. A. (2008) Ann. N.Y. Acad. Sci. 1129, 358–370 [DOI] [PubMed] [Google Scholar]
- 14. Lee C., Etchegaray J. P., Cagampang F. R., Loudon A. S., Reppert S. M. (2001) Cell 107, 855–867 [DOI] [PubMed] [Google Scholar]
- 15. Chen R., Schirmer A., Lee Y., Lee H., Kumar V., Yoo S. H., Takahashi J. S., Lee C. (2009) Mol. Cell 36, 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999) Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 17. Griffin E. A., Jr., Staknis D., Weitz C. J. (1999) Science 286, 768–771 [DOI] [PubMed] [Google Scholar]
- 18. Sato T. K., Yamada R. G., Ukai H., Baggs J. E., Miraglia L. J., Kobayashi T. J., Welsh D. K., Kay S. A., Ueda H. R., Hogenesch J. B. (2006) Nat. Genet. 38, 312–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. (2002) Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 20. Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P. (2007) Nature 450, 1086–1090 [DOI] [PubMed] [Google Scholar]
- 21. Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J. J., Sassone-Corsi P. (2005) Science 309, 1390–1394 [DOI] [PubMed] [Google Scholar]
- 22. Kondratov R. V., Chernov M. V., Kondratova A. A., Gorbacheva V. Y., Gudkov A. V., Antoch M. P. (2003) Genes Dev. 17, 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDearmon E. L., Patel K. N., Ko C. H., Walisser J. A., Schook A. C., Chong J. L., Wilsbacher L. D., Song E. J., Hong H. K., Bradfield C. A., Takahashi J. S. (2006) Science 314, 1304–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K., Kay S. A. (2008) PLoS Genet. 4, e1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Menet J. S., Abruzzi K. C., Desrochers J., Rodriguez J., Rosbash M. (2010) Genes Dev. 24, 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denault D. L., Loros J. J., Dunlap J. C. (2001) EMBO J. 20, 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schafmeier T., Haase A., Káldi K., Scholz J., Fuchs M., Brunner M. (2005) Cell 122, 235–246 [DOI] [PubMed] [Google Scholar]
- 28. He Q., Shu H., Cheng P., Chen S., Wang L., Liu Y. (2005) J. Biol. Chem. 280, 17526–17532 [DOI] [PubMed] [Google Scholar]
- 29. He Q., Liu Y. (2005) Genes Dev. 19, 2888–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reppert S. M., Weaver D. R. (2001) Annu. Rev. Physiol. 63, 647–676 [DOI] [PubMed] [Google Scholar]
- 31. Yoo S. H., Yamazaki S., Lowrey P. L., Shimomura K., Ko C. H., Buhr E. D., Siepka S. M., Hong H. K., Oh W. J., Yoo O. J., Menaker M., Takahashi J. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Etchegaray J. P., Machida K. K., Noton E., Constance C. M., Dallmann R., Di Napoli M. N., DeBruyne J. P., Lambert C. M., Yu E. A., Reppert S. M., Weaver D. R. (2009) Mol. Cell. Biol. 29, 3853–3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DeBruyne J. P., Weaver D. R., Reppert S. M. (2007) Nat. Neurosci. 10, 543–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee C., Weaver D. R., Reppert S. M. (2004) Mol. Cell. Biol. 24, 584–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Balsalobre A., Damiola F., Schibler U. (1998) Cell 93, 929–937 [DOI] [PubMed] [Google Scholar]
- 36. Shearman L. P., Jin X., Lee C., Reppert S. M., Weaver D. R. (2000) Mol. Cell. Biol. 20, 6269–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ripperger J. A., Schibler U. (2006) Nat. Genet. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 40. Hida A., Koike N., Hirose M., Hattori M., Sakaki Y., Tei H. (2000) Genomics 65, 224–233 [DOI] [PubMed] [Google Scholar]
- 41. Lee H., Chen R., Lee Y., Yoo S., Lee C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 21359–21364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., Schibler U. (2008) Cell 134, 317–328 [DOI] [PubMed] [Google Scholar]
- 43. Kim E. Y., Edery I. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6178–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu W., Zheng H., Houl J. H., Dauwalder B., Hardin P. E. (2006) Genes Dev. 20, 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 46. Pabo C. O., Sauer R. T. (1992) Annu. Rev. Biochem. 61, 1053–1095 [DOI] [PubMed] [Google Scholar]
- 47. Duffield G. E., Watson N. P., Mantani A., Peirson S. N., Robles-Murguia M., Loros J. J., Israel M. A., Dunlap J. C. (2009) Curr. Biol. 19, 297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benezra R., Davis R. L., Lockshon D., Turner D. L., Weintraub H. (1990) Cell 61, 49–59 [DOI] [PubMed] [Google Scholar]
- 49. Etchegaray J. P., Lee C., Wade P. A., Reppert S. M. (2003) Nature 421, 177–182 [DOI] [PubMed] [Google Scholar]
- 50. Welsh D. K., Yoo S. H., Liu A. C., Takahashi J. S., Kay S. A. (2004) Curr. Biol. 14, 2289–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 52. Kadener S., Menet J. S., Schoer R., Rosbash M. (2008) PLoS Biol. 6, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rutila J. E., Edery I., Hall J. C., Rosbash M. (1992) J. Neurogenet. 8, 101–113 [DOI] [PubMed] [Google Scholar]
- 54. Baylies M. K., Bargiello T. A., Jackson F. R., Young M. W. (1987) Nature 326, 390–392 [DOI] [PubMed] [Google Scholar]
- 55. Hao H., Glossop N. R., Lyons L., Qiu J., Morrish B., Cheng Y., Helfrich-Förster C., Hardin P. (1999) J. Neurosci. 19, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng P., Yang Y., Liu Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Debruyne J. P., Noton E., Lambert C. M., Maywood E. S., Weaver D. R., Reppert S. M. (2006) Neuron 50, 465–477 [DOI] [PubMed] [Google Scholar]
- 58. Dibner C., Sage D., Unser M., Bauer C., d'Eysmond T., Naef F., Schibler U. (2009) EMBO J. 28, 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reddy A. B., O'Neill J. S. (2010) Trends Cell Biol. 20, 36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.