Abstract
The innate immune system elicits the first wave of immune responses against pathogen infection. Its operational modes are complex and have yet to be defined. Here, we report the identification of an innate immune regulator termed TAPE (TBK1-associated protein in endolysosomes), previously known as CC2D1A/Freud-1/Aki-1, which modulates the TLR3 and TLR4 pathways. We found that TAPE activated the TBK1, NF-κB, and ERK pathways leading to IFN-β and inflammatory cytokine induction. TAPE was shown to colocalize with endosomal marker Rab5 and lysosomal marker LAMP1 in mammalian cells, suggesting that TAPE resided in endolysosomes. Knockdown of TAPE selectively impaired the TLR3 and endocytic TLR4 pathways to IFN-β induction. Furthermore, TAPE interacted and synergized with Trif to activate IFN-β. TAPE knockdown failed to block Trif-mediated IFN-β induction, whereas Trif knockdown impaired the TLR3 and TAPE cooperation on IFN-β induction, suggesting that TAPE acts upstream of Trif. Together, our data demonstrate a central role for TAPE in linking TLR3 and TLR4 to innate immune defenses at an early step.
Keywords: Innate Immunity, Interferon, MAPKs, NF-kappaB transcription Factor, Toll-like receptors (TLR), TRIF, TAPE, TBK1
Introduction
The mammalian innate immune system serves the first line of host defense against pathogen infection meanwhile links to the adaptive immune system for inducing the full spectrum of immune responses. Innate immune recognition of invading pathogens is mediated by host pattern-recognition receptors (PRRs),2 which detect conserved microbial components known as pathogen-associated molecular patterns. Intracellular innate immune regulators help translate the PRR signals into the innate immune responses. Four major families of PRRs have been identified as follows: Toll-like receptors (TLRs), RIG-I-like receptors, NOD-like receptors, and C-type lectin receptors (1). Recently, several cytosolic nucleic acid sensors have also been found, including DAI (2), AIM-2 (3–6), RNA polymerase III (7, 8), and LRRFIP1 (9). PRRs are located in discrete subcellular compartments, like the cell surface, endolysosome, and cytoplasm, to detect invading pathogens by different routes. Upon engaging with pathogen-associated molecular patterns, PRRs trigger major downstream pathways, including NF-κB, MAPK, and/or IRF3/7, to induce the production of inflammatory cytokines and/or type I interferons (IFNs) thereby leading to antimicrobial immune responses.
TLRs represent the prototypical family of PRR and consist of two subgroups, cell surface TLRs and endosomal TLRs (1). Cell surface TLRs (e.g. TLR1, TLR2, TLR4, TLR5, and TLR6) are mainly responsible for detecting microbial lipids, lipopeptides, and peptidoglycans from extracellular pathogens. Endosomal TLRs (e.g. TLR3, TLR7/8, and TLR9) are located in the endolysosomal compartments to detect microbial nucleic acids from the endocytic, phagocytic, and autophagic pathways. Among cell surface TLRs, TLR4 mainly cooperates with cell surface CD14 and MD2 to detect lipopolysaccharide (LPS) from Gram-negative bacteria, which is a component causing sepsis. A recent study showed that TLR4 complexes with TLR6 and CD36 to recognize sterile ligands such as amyloid-β and oxidized low density lipoprotein (LDL), which are components implicated in Alzheimer disease and atherosclerosis, respectively (10). Among endosomal TLRs, TLR3 is involved in sensing viral double-stranded RNA and poly(I-C) (polyinosinic-polycytidylic acid, a synthetic double-stranded RNA analog). Studies showed that TLR3 might play a protective or an adverse role in defending viral infection in a virus-dependent manner (11–13). TLR7 and TLR8 detect single-stranded RNA from RNA viruses or bacteria, whereas TLR9 detects DNA-bearing CpG motifs from DNA viruses or bacteria.
Several TIR domain-containing adaptors play critical roles in linking TLRs to downstream signaling pathways (14). MyD88 is a TIR domain-containing adaptor involved in all TLR and IL-1R pathways except TLR3, whereas Trif (also called Ticam-1) is another TIR domain-containing adaptor critical for the endosomal TLR3 pathway and the endocytic TLR4 pathway (14, 15). Interestingly, TLR4 is the only TLR to use both the MyD88-dependent and Trif-dependent pathways to achieve optimal innate immune responses (16, 17). Two IKK-related kinases, TBK1 (also known as NAK) and IKKϵ (also known as IKKi), play key roles in linking the endosomal TLR (TLR3 and TLR4) and cytosolic RIG-I-like receptor signals to the activation of transcriptional factors IRF3 and IRF7 for type I IFN production (18, 19). In addition, TBK1 is essential for bridging innate and adaptive immunity in response to DNA vaccines (20). IKKϵ is also shown to phosphorylate STAT1 to regulate expression of a set of IFN-β inducible genes (21). Several adaptors are shown to regulate the activity of TBK1 and IKKϵ, including TANK, NAP1, SINTBAD, and SIKE (22–25).
TAPE, previously known as CC2D1A (coiled-coil and C2 domain-containing 1A)/Freud-1, was first reported to act as a transcriptional repressor of the serotonin-1A (5-HT1A) receptor gene (26, 27). CC2D1A/Freud-1 contains four DM14 domains in the N-terminal region, a helix-loop-helix domain, and a C2 domain in the distal C terminus. A deletion in this gene has been linked to nonsyndromic mental retardation (28). Another study reports that CC2D1A/Freud-1, also named Aki1 (Akt kinase-interacting protein 1), acts as a scaffold to regulate the PDK1/Akt pathway during EGF receptor signaling (29). To our knowledge, the role of CC2D1A/Freud-1 in the immune system has not yet been rigorously examined except for a large scale analysis indicating a potential role for TAPE as a NF-κB activator (30). In light of a novel role for CC2D1A/Freud-1 in innate immunity demonstrated below, we here propose an alternative name, designated TAPE (TBK1-associated protein in endolysosomes). Expression of TAPE led to the phosphorylation of ERK and the activation of the IFN-β, interferon-stimulated response element (ISRE), and NF-κB promoters. TAPE interacted and cooperated with innate immune regulators, including TBK1, IKKϵ, and Trif, to induce synergistic IFN-β activation. TAPE was shown to colocalize with an early endosomal protein Rab5 and a lysosomal protein Lamp1 in mammalian cells, whereas its deletion mutants disturbed the distributions of Rab5 and Lamp1, suggesting a regulatory role for TAPE in endolysosomes. By overexpression and knockdown analyses, TAPE was shown to modulate the TLR3 and endocytic TLR4 pathways. Interestingly, TAPE acted upstream of Trif to regulate the TLR3 pathway. Therefore, our results suggest that TAPE serves as an innate immune regulator to link TLR3 and TLR4 to the Trif-dependent signaling pathways.
EXPERIMENTAL PROCEDURES
Cell, Viruses, and Reagents
HEK293 and HEK293T cells were grown in DMEM with 7% cosmic calf serum (Hyclone). Vero cells were grown in DMEM with 10% fatal bovine serum (FBS, Hyclone). Vesicular stomatitis virus and herpes simplex virus-1 (HSV-1 strain KOS) were described previously (31, 32). Flagellin (number tlrl-bsfla), poly(I-C) (number tlrl-pic), FSL-1(number tlrl-fsl), and Pam3CSK4(number tlrl-pms) were purchased from Invivogen. LPS (number L3024) was purchased from Sigma. Antibodies of anti-TRIF (number 4596), anti-TBK1 (number 3013), anti-MAPK (number 9926), and anti-phospho-MAPK (number 9910) were purchased from Cell Signaling. Anti-Myc (clone 4A6, number 05-724) and anti-actin (MAB1501) antibodies were purchased from Millipore. Anti-HA (clone HA-7, H9658) and anti-FLAG (clone M2, number F1804) antibodies were purchased from Sigma. Anti-TAPE antibody was generated against a synthetic peptide flanking the human TAPE residues 920–937.
Plasmids
The full-length human TAPE cDNA (IMAGE clone 6585236 from Invitrogen) was amplified by PCR and then cloned into pEGFP-N1 (Clontech) to generate an expression construct hTAPE-EGFP with a C-terminal EGFP tag. Similarly, the hTAPE-N-EGFP (amino acids 1–660) and hTAPE-C-EGFP (amino acids 763–951) deletion mutants were generated by PCR cloning. The hTAPE-ΔC2-EGFP mutant was generated by sequential cloning of hTAPE-N and hTAPE-C into pEGFP-N1, resulting in the replacement of the C2 domain (amino acids 661–762) with KLM residues. To generate the N-terminal HA-tagged human TAPE expression constructs, including HA-hTAPE (amino acids 1–951), HA-hTAPE-N (amino acids 1–660), and HA-hTAPE-C (amino acids 763–951), the full-length human TAPE cDNA (IMAGE clone) was amplified by PCR using 5′ forward primers containing an HA tag sequence and then cloned into pcDNA6.0/Myc-His (Invitrogen). Similarly, hTAPE-ΔC2-EGFP was used as a template for PCR cloning of HA-hTAPE-ΔC2. Mouse TBK1 was amplified by PCR and then cloned into pcDNA3.0-HA. Mouse TAPE (mTAPE, accession BC027028) was purchased from Open Biosystems. The following plasmids were obtained from Addgene and their identification numbers are included: pcDNA3-CD14 (Addgene plasmid 13645), pFLAG-CMV1-hMD2 (Addgene plasmid 13028), pcDNA3-TLR1-YFP (Addgene plasmid 13014), pcDNA3-TLR2-YFP (Addgene plasmid 13016), pcDNA3-TLR6-CFP (Addgene plasmid 13021), and pGL3-ELAM-luc (Addgene plasmid 13029); the plasmids were developed by D. Golenbock (University of Massachusetts Medical School). MyD88-FLAG (Addgene plasmid 13093), hTLR3-FLAG (Addgene plasmid 13084), mTLR4-FLAG (Addgene plasmid 13087) and hTLR5-FLAG (Addgene plasmid 13088) were developed by R. Medzhitov (Yale University). IKKβ-K44M (Addgene plasmid 11104) was developed by A. Rao (Harvard University). DsRed-Rab5 WT (Addgene plasmid 13050) was developed by R. Pagano (Mayo Clinic). Lamp1-RFP (Addgene plasmid 1817) was developed by W. Mothes (Yale University). Other plasmids were kindly provided as follows: TBK1-Myc, TBK1-K38A-Myc, IKKϵ-Myc, and IKKϵ-K38A-Myc by U. Siebenlist (National Institutes of Health), FLAG-TRIF and IFN-β-Luc by K. Fitzgerald (University of Massachusetts Medical School), and ISRE-Luc by R. Lin (McGill University).
Primers
The following primers were used for cloning the indicated genes: hTAPE-EGFP using forward 5′-GCTAGCATGCACAAGAGGAAAGGA-3′ and reverse 5′-ACCGGTAACCTGCGGAGCCGCTG-3′; hTAPE-ΔC2-EGFP using two pairs of primers, 1st forward 5′-GCTAGCATGCACAAGAGGAAAGGA-3′ and 1st reverse 5′-AAGCTTGAAGAGGAGCATGTCGTT-3′ (for the TAPE-N fragment); 2nd forward 5′-AAGCTTATGATAGCATGTGAGGTC-3′ and 2nd reverse 5′-ACCGGTAACCTGCGGAGCCGCTG-3′ (for the TAPE-C fragment); hTAPE-C-EGFP using forward 5′-AAGCTTATGATAGCATGTGAGGTC-3′ and reverse 5′-ACCGGTAACCTGCGGAGCCGCTG-3′; hTAPE-N-EGFP using forward 5′-GCTAGCATGCACAAG AGGAAAGGA-3′ and reverse 5′-AAGCTTGAAGAGGAGCATGTCGTT-3′. For cloning N-terminally HA-tagged hTAPE full-length (FL), hTAPE-N, hTAPE-C, and hTAPE-ΔC2, primers were used as follows: hTAPE-FL, hTAPE-N, and hTAPE-ΔC2 forward (containing HA tag sequence) 5′-GGATCCATGTACCCATACGATGTTCCAGATTACGCTCACAAGAGGAAAGGA-3′; hTAPE-FL, hTAPE-ΔC2, and TAPE-C reverse 5′-GAATTCCTATCACCTGCGGAGCCG-3′; hTAPE-N reverse 5′-GAATTCTCAGAAGAGGAGCATGTC-3′ and hTAPE-C forward (containing HA tag sequence) 5′-GGATCCATGTACCCATACGATGTTCCAGATTACGCTATAGCATGTGAGGTC-3′; mTBK1-HA, mTbk1 sense 5′-GGATCCCAGAGCACCTCCAAC-3′ and mTBK1 reverse 5′-GAATTCCTAAAGACAGTCCACATTGCG-3′.
RNA Interference
All siRNA oligonucleotides were synthesized by Invitrogen. The sequences of the siRNA oligonucleotides are as follows (only the sense strands are shown): control (GFP), 5′-GCAGAAGAACGGCAUCAAG-3′; hTAPE-1, 5′-GGCGCUCUAUCAGACAGCAAUUGAA-3′; hTAPE-2, 5′-CGCAUCGUCAAGCAAUACCAA-3′; TBK1, 5′-GACAGAAGUUGUGAUCACA-3′; IKKβ-1, 5′-GUACAGCGAGCAAACCGAG-3′; IKKβ-2, 5′-CUUAGAUACCUUCAUGAAA-3′; hIPS1, 5′-CCACCUUGAUGCCUGUGAA-3′; TRIF, 5′-GACCAGACGCCACUCCAAC-3′; hMyD88, 5′-CUGGAACAGACAAACUAUC-3′. mMyD88 siRNAs (sc-35987) were purchased from Santa Cruz Biotechnology. Two types of TAPE-1 siRNAs were used as follows: one was regular siRNA, and the other called Stealth (st) siRNA was chemically modified and used for most of knockdown experiments. HEK293 cells seeded onto 24-well plates (5 × 104/well) were transfected with the indicated siRNAs (20 nm) using RNAiMAX (Invitrogen). 48 h after transfection, cells were used for reporter assays or plaque assays as described.
Plaque Assay
For overexpression experiments, HEK293 cells (in the 24-well plate) transfected with HA-TAPE or a control vector (pcDNA6.0/Myc-His) were infected with the indicated viruses. Virus titers in culture supernatants were collected and measured. Vero cells were infected with serial dilutions of viruses and were overlaid with DMEM containing 2% FBS and 1% methylcellulose. Plaques were counted after 48–72 h post-infection. For knockdown experiments, HEK293 cells were transfected with the indicated siRNAs for 48 h, and then were transfected with the indicated receptors. The following procedures of virus infection were similar to the overexpression experiments described above.
Enzyme-linked Immunosorbent Assay (ELISA)
HEK293 cells transiently transfected with HA-TAPE or a control vector (pcDNA6.0/Myc-His) were cultured 24 h. Chemokine IL-8 in the supernatants was measured by ELISA according to the manufacturer's instructions (R&D Systems). HEK293 cells treated with the indicated siRNAs were transfected with TLR3 and then stimulated with poly(I-C) (50 μg/ml). IFN-β and regulated on activation normal T cell expressed and secreted (RANTES) in the supernatants were measured by ELISA according to the manufacturer's instructions (R&D Systems).
RT-PCR
HEK293 cells treated with the indicated siRNAs were transfected with TLR3 and then stimulated with poly(I-C) (50 μg/ml). Total RNA was isolated from cells using TRIzol reagent (Invitrogen). After DNase (Promega) treatment, RNA was reversely transcribed into cDNA using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. cDNA was amplified by PCR using specific primers (Gapdh sense 5′-TGATGACATCAAGAAGGTGGTGAAG-3′; Gapdh antisense 5′-TCCTTGGAGGCCATGTGGGCCAT-3′; Ifnb sense 5′-CTCCTCCAAATTGCTCTCCT-3′; Ifnb antisense 5′-CCTTGGCCTTCAGGTAATGC-3′).
Confocal Microscopy
HEK293 cells in the 6-well plate were transfected with TAPE-EGFP together with Rab5-DsRed (an early endosomal marker) or Lamp1-RFP (a lysosomal marker). 24 h later, cells were passed into a chamber slide (Nunc Lab-TekTM II) and incubated at 37 °C for 24 h. Cells were then fixed and mounted for confocal microscopic examination (Leica TCS SP2).
Reporter Assay
HEK293 cells seeded on 24-well plates were transiently transfected with a luciferase reporter plasmid (e.g. IFN-β-Luc, ELAM-Luc, or ISRE-Luc) together with an internal control reporter plasmid (pRL-TK or β-actin-β-gal) and indicated expression constructs by Lipofectamine 2000 (Invitrogen). An empty vector (pcDNA3.0 or pcDNA6.0/Myc-His) was used to equalize the total amount of plasmids. 24 h after transfection or indicated stimulation, the luciferase activity in lysates was measured by the Dual-Luciferase assay kit (Promega). In the case of actin-β-gal as an internal control, the activity of β-galactosidase was measured by the Galacto-Light PlusTM system (Applied Biosystems), and then relative luciferase activity was determined.
Coimmunoprecipitation and Western Blot Analysis
HEK293T cells were transfected with the indicated plasmids. 24 h after transfection, cells were lysed in 1% Triton X-100 lysis buffer (1% Triton X-100, 50 mm Tris (pH 8.0), 150 mm NaCl, 2 mm EDTA) with protease and phosphatase inhibitors. Lysates were incubated with the indicated antibodies for immunoprecipitation. Immunoprecipitates were subjected to Western blot analyses with the indicated antibodies.
RESULTS
TAPE Activates the TBK1-IFN-β Pathway
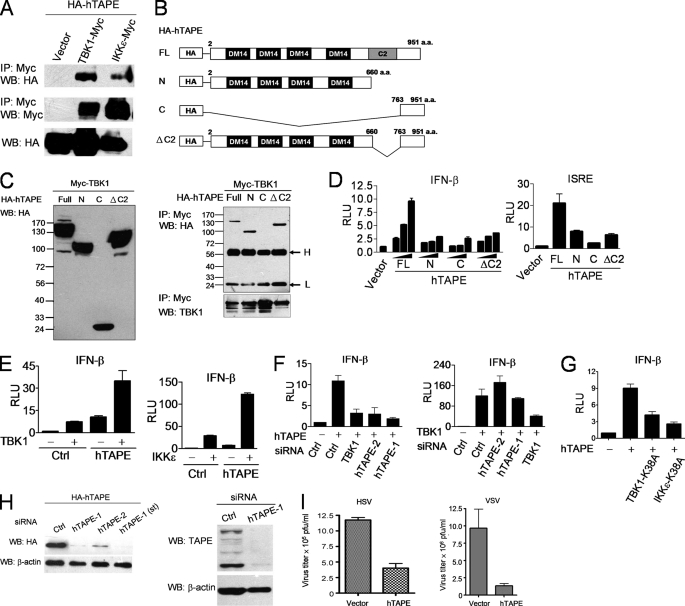
A previous study showed that TBK1 and TAPE are present in an immunocomplex targeting the nuclear hormone receptor coactivator SRC-2 (33). To explore the potential interplay between TBK1 and TAPE in innate immune defenses, we first confirmed whether these molecules are associated in mammalian cells. Our results from coimmunoprecipitation showed that HA-tagged human TAPE (HA-hTAPE) was associated with TBK1-Myc or IKKϵ-Myc in HEK293T cells (Fig. 1A). To gain the mechanistic insight into the TAPE-TBK1 interaction, we generated three N-terminal HA-tagged deletion mutants of hTAPE, designated hTAPE-N containing the N-terminal region, hTAPE-C containing the C-terminal region, and hTAPE-ΔC2 lacking the C2 domain, to dissect the TAPE region for binding TBK1 (Fig. 1B). The protein expression levels of hTAPE and its deletion mutants were examined (Fig. 1C, left panel). We found that the TAPE full-length and deletion mutants containing the N-terminal region were associated with TBK1 (Fig. 1C, right panel), suggesting that the N-terminal region of TAPE is essential for interacting with TBK1. A similar result was also found using the C-terminally enhanced GFP-tagged TAPE full-length and deletion mutants for experiments (supplemental Fig. S1). In light of this interaction, we assessed if TAPE regulates the TBK1-IFN-β pathway. Results from reporter assays showed that overexpression of hTAPE induced the highest IFN-β promoter activity in a dose-dependent manner compared with its deletion mutants (Fig. 1D, left panel). Likewise, hTAPE induced the activation of the interferon-stimulated response element (ISRE) promoter, which is regulated by type I IFNs (Fig. 1D, right panel). Furthermore, hTAPE displayed the ability to synergize with TBK1 or IKKϵ to activate IFN-β (Fig. 1E). We then determined the position of TAPE in the TBK1-IFN-β pathway. Using the small interfering RNA (siRNA) approach, we demonstrated that TBK1 knockdown impaired hTAPE-induced IFN-β activation, whereas hTAPE knockdown failed to block TBK1-induced IFN-β activation (Fig. 1F). These results suggest that hTAPE functions upstream of TBK1 to activate IFN-β. In line with this finding, we showed that the kinase-defective TBK1 and IKKϵ mutants blocked hTAPE-induced IFN-β activation (Fig. 1G). The knockdown effect of siRNAs on the endogenous and ectopic expression of hTAPE in HEK293 cells was confirmed by immunoblotting (Fig. 1H). Next, we investigated the effect of TAPE on defending against virus infection. Expression of hTAPE in HEK293 cells led to lower viral titers of herpes simplex virus-1 (HSV-1, a DNA virus) and vesicular stomatitis virus (vesicular stomatitis virus, an RNA virus) (Fig. 1I). Together, these results indicate that TAPE acts as an innate immune regulator to trigger the TBK1-IFN-β pathway and antiviral responses.
FIGURE 1.
TAPE activates the TBK1-IFN-β pathway. A, 293T cells were transfected with HA-tagged human TAPE (HA-hTAPE) together with a control vector, TBK1-Myc or IKKϵ-Myc. Cell lysates were subjected to the immunoprecipitation (IP)-Western blot (WB) analysis using the indicated antibodies. B, schematic structure of the full-length (FL) HA-hTAPE and its deletion mutants, HA-TAPE-N, HA-TAPE-C, and HA-TAPE-ΔC2. a.a., amino acids. C, 293T cells were transfected with TBK1-Myc together with HA-hTAPE or its deletion mutants. Expression of hTAPE and its deletion mutants were examined by Western blot (left panel). TBK1 interaction with hTAPE and its deletion mutants was examined by the immunoprecipitation-Western blot analysis (right panel). Labeled H and L represent immunoglobulin heavy and light chain, respectively. D, 293 cells were transfected with an IFN-β-Luc reporter plasmid together with increasing amounts of HA-hTAPE or its deletion mutants and then analyzed for the IFN-β promoter activity (left panel). A similar approach was employed to assess the effect of hTAPE and its deletion mutants on the interferon-stimulated response element promoter activity (right panel). Luciferase reporter activity was reported by relative light units (RLU). E, 293 cells were transfected with IFN-β-Luc together with HA-hTAPE plus TBK1-Myc or IKKϵ-Myc to analyze the synergistic effect on the IFN-β promoter activity. F, 293 cells treated with indicated siRNAs were transfected with IFN-β-Luc together with hTAPE (left) or TBK1 (right) and then analyzed for the IFN-β promoter activity. G, TBK1 and IKKϵ kinase-defective mutants block TAPE-induced IFN-β activation. 293 cells were transfected with IFN-β-Luc, together with hTAPE plus a control vector, the kinase-defective mutant TBK1-K38A or IKKϵ-K38A for analyzing the IFN-β promoter activity. H, RNA interference resulted in decreased expression of TAPE as determined by Western blotting. Western blot analysis of transfected HA-hTAPE (left panel, anti-HA antibody) or endogenous TAPE (right panel, anti-TAPE antibody) in 293 cells treated with TAPE siRNAs or control siRNA. I, 293 cells were transfected with HA-hTAPE or a control vector, and then infected with HSV-1 (multiplicity of infection of 1, left panel) or vesicular stomatitis virus (VSV) (multiplicity of infection of 0.001, right panel) for viral plaque assays. Values represent the mean ± S.E. of duplicate (F and G) or triplicate (D, E, and I) samples. Data are from one experiment representative of two or three.
TAPE Activates the NF-κB and ERK Pathways
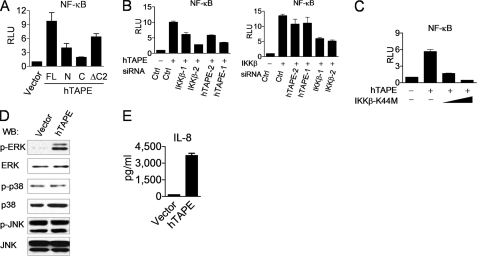
Next we explored whether TAPE activates other innate immune pathways, like the NF-κB and MAPK pathways, which are essential for the production of inflammatory cytokines. Our results from reporter assays showed that expression of hTAPE induced the highest NF-κB promoter activity compared with its deletion mutants (Fig. 2A). Furthermore, hTAPE-induced NF-κB activation was impaired by IKKβ knockdown or a kinase-defective IKKβ mutant (IKKβ-K44M) (Fig. 2, B, left panel, and C). Conversely, hTAPE knockdown showed no significant blocking effect on IKKβ-induced NF-κB activation (Fig. 2B, right panel). These data suggest that TAPE functions upstream of IKKβ to activate the NF-κB pathway. The effect of TAPE on the MAPK pathway was measured by examining the phosphorylation levels of three major MAPK members, including ERK, JNK, and p38. We observed that overexpression of hTAPE induced the phosphorylation of ERK but not JNK or p38, indicating a functional role for TAPE in regulating the ERK pathway (Fig. 2D). In addition, hTAPE expression in HEK293 cells led to the increased interleukin-8 (IL-8) production (Fig. 2E), whose promoter contains the binding sites for transcriptional factors NF-κB and AP-1. These data together indicate that TAPE possesses the ability to activate two other innate immune pathways for inflammatory cytokine production.
FIGURE 2.
TAPE activates the NF-κB and ERK pathways. A, 293 cells were transfected with an NF-κB reporter plasmid (pELAM-Luc) together with HA-hTAPE or its deletion mutants and then analyzed for the NF-κB promoter activity. RLU, relative light units; FL, full length. B, 293 cells treated with indicated siRNAs were transfected with pELAM-Luc together hTAPE (left) or IKKβ (right) and then analyzed for the NF-κB promoter activity. C, 293 cells were transfected with pELAM-Luc, together with hTAPE and increasing amounts of kinase-defective mutant IKKβ-K44M, for analyzing the NF-κB promoter activity. D and E, 293 cells were transfected with a control vector or HA-hTAPE. Cell lysates (D) were analyzed for Western blot (WB) with indicated MAPK and phospho-MAPK antibodies. Supernatants (E) were measured by ELISA for IL-8 production. Values represent the mean ± S.E. of duplicate (B and E) or triplicate (A and C) samples. Data are from one experiment representative of two or three.
Localization of TAPE in Endolysosomes
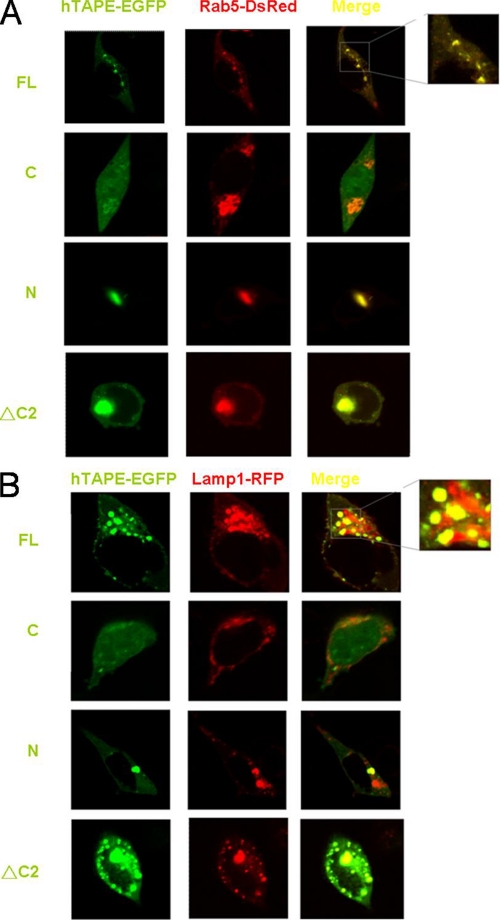
Several lines of evidence prompted us to examine the subcellular localization of TAPE in mammalian cells. First, innate immune regulators, like IPS-1/MAVS, STING, and Unc93b, reside and function in distinct vesicular compartments such as endolysosomes, mitochondria, and endoplasmic reticulum (34–36). Second, the C2 domain is one of the phospholipid-binding domains for proteins to target lipid membranes (37). Third, a Drosophila ortholog of TAPE called Lgd (lethal giant discs) regulates endosomal trafficking (38–40). The C2 domain of Lgd is responsible for binding phospholipids (38). Given these facts, we used confocal microscopy to examine the subcellular distribution of TAPE. The full-length and deletion mutants of hTAPE with a C-terminally enhanced GFP tag were used for this purpose. Expression of hTAPE-EGFP displayed a punctate pattern in HEK293 cells (Fig. 3A, top row). Furthermore, hTAPE-EGFP was found to highly overlap with an endosomal protein Rab5 with a DsRed tag (Rab5-DsRed) in HEK293 cells (Fig. 3A, top row). Similarly, hTAPE-EGFP showed the substantial overlap with a lysosomal protein Lamp1 with a red fluorescent protein tag (Lamp1-RFP) (Fig. 3B, top row). These data suggest that TAPE is localized in endolysosomal compartments in mammalian cells. In contrast, we found that expression of hTAPE mutants disturbed the punctate patterns of Rab5 and Lamp1 distribution in HEK293 cells (Fig. 3, A and B). Expression of hTAPE-Ν-EGFP led to the formation of a large merged aggregate with Rab5-DsRed (Fig. 3A, 3rd row). A similar event was observed in the case of coexpression of hTAPE-ΔC2-EGFP with Rab5-DsRed (Fig. 3A, bottom row). To a lesser extent, these hTAPE mutants formed a merged aggregate with Lamp1-RFP and some merged speckles in the case of hTAPE-ΔC2-EGFP (Fig. 3B, 3rd and bottom rows). The hTAPE-C-EGFP mutant showed a diffuse distribution in HEK293 cells but caused the formation of large Rab5-DsRed aggregates without overlap with them (Fig. 3A, 2nd row). Expression of hTAPE-C-EGFP with Lamp1-RFP showed distinct dispersed distributions without evident overlap (Fig. 3B). Together, our results suggest that TAPE, like its Drosophila ortholog Lgd, resides in endolysosomes and may regulate the distribution of these vesicular compartments through its multiple domains.
FIGURE 3.
TAPE is localized in endolysosomes. A and B, 293 cells were transfected with EGFP-tagged hTAPE and its deletion mutants, together with an endosomal protein Rab5-DsRed (A) or a lysosomal protein Lamp1-RFP (B). Transfected cells were then passed into a chamber slide for fixation 24 h after passing. Fixed cells were examined by confocal microscopy. The yellow signals in merged panels indicate colocalization of two molecules. FL, full length.
TAPE Functions Upstream of Trif to Regulate the TLR3 Pathway
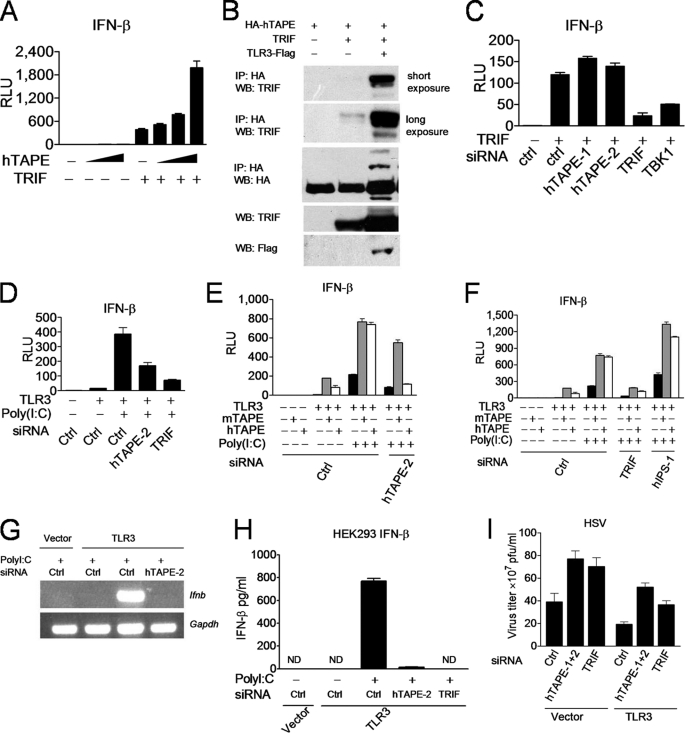
In light of TAPE localization in endolysosomes and its role in the TBK1-IFN-β pathway, we determined whether TAPE is involved in the endosomal TLR3 and TLR4 pathways. We first examined the interplay between TAPE and Trif, a key adaptor linking the TLR3 and TLR4 signals to the TBK1-IFN-β pathway. Expression of hTAPE synergized with Trif to activate IFN-β in an hTAPE dose-dependent manner (Fig. 4A). TAPE also formed a complex with Trif in mammalian cells, and this interaction was further enhanced in the presence of TLR3, implying a regulatory role for TLR3 in TAPE-Trif interaction (Fig. 4B). Notably, knockdown of hTAPE, unlike TBK1, failed to impair Trif-induced IFN-β activation (Fig. 4C). These results suggest that TAPE functions either upstream of or in parallel with Trif during TLR3 signaling. Subsequently, we assessed the involvement of TAPE in the TLR3 pathway. Knockdown of hTAPE, like Trif, impaired TLR3-induced IFN-β activation by poly(I-C) stimulation (Fig. 4D). The off-target silencing effect is a common concern raised by the siRNA approach (41). To rule out this possibility, we used a mouse TAPE (mTAPE) expression construct to rescue the blocking effect caused by hTAPE knockdown. Our results from rescuing experiments demonstrated that in the presence of a control siRNA, expression of mTAPE or hTAPE synergized with TLR3 for IFN-β activation, and the poly(I-C) treatment further enhanced this activation (Fig. 4E). However, only expression of mTAPE but not hTAPE rescued the impairment of TLR3-mediated IFN-β activation caused by hTAPE knockdown and further induced synergistic activation (Fig. 4E). Knockdown of Trif but not IPS-1 blocked synergistic IFN-β activation by TLR3 cooperation with hTAPE or mTAPE (Fig. 4F).
FIGURE 4.
TAPE interacts with and acts upstream of Trif in the TLR3 pathway. A, 293 cells were transfected with IFN-β-Luc together with Trif and increasing amounts of HA-hTAPE for analyzing the synergistic effect. RLU, relative light units. B, 293T cells were cotransfected with HA-TAPE, Trif, or FLAG-TLR3 at indicated combinations. Cell lysates were subjected to the immunoprecipitation-Western blot analysis (IP-WB) analysis. C, 293 cells treated with indicated siRNAs were transfected with IFN-β-Luc together with Trif for analyzing the IFN-β promoter activity. D, 293 cells treated with indicated siRNAs were transfected with IFN-β-Luc (left panel) together with TLR3, and then stimulated with poly(I-C) (20 μg/ml) for analyzing the IFN-β promoter activity. E and F, 293 cells treated with indicated siRNA were transfected with IFN-β-Luc together with TLR3, mouse TAPE (mTAPE), or hTAPE as indicated combinations, and then stimulated with poly(I-C) (20 μg/ml) for analyzing the IFN-β promoter activity. Results were separately displayed as E and F. G, 293 cells treated with indicated siRNAs were transfected with TLR3 and then stimulated with poly(I-C) (50 μg/ml) for analyzing the IFN-β (Ifnb) or Gapdh (loading control) by RT-PCR. H, 293 cells treated with indicated siRNAs were transfected with TLR3 and then stimulated with poly(I-C) (50 μg/ml). Supernatants were measured by ELISA for IFN-β production. Values represent the mean ± S.E. of triplicate samples. ND means not detectable. I, 293 cells treated with indicated siRNAs were transfected with TLR3 or a control vector and then infected with HSV-1 (multiplicity of infection of 1) for viral plaque assay. Values represent the mean ± S.E. of duplicate (A, C, D, E, and F) or triplicate (I) samples. Data are from one experiment representative of two or three.
We further confirmed the effect of hTAPE knockdown on the endogenous IFN-β induction. Knockdown of hTAPE impaired induction of endogenous IFN-β from the RNA level to the cytokine production (Fig. 4, G and H). A similar blocking effect of hTAPE knockdown on regulated on activation normal T cell expressed and secreted (RANTES) production was also observed (supplemental Fig. S2). TLR3 is shown to implicate in host defenses against HSV-1 infection (11). Results from plaque assays also showed that hTAPE is critical for inducing optimal TLR3-mediated antiviral responses against HSV-1 (Fig. 4I). Collectively, these results suggest that TAPE is a novel regulator in the TLR3-Trif pathway.
TAPE Is Critical for Linking the TLR4-Trif Branch Pathway to IFN-β Activation
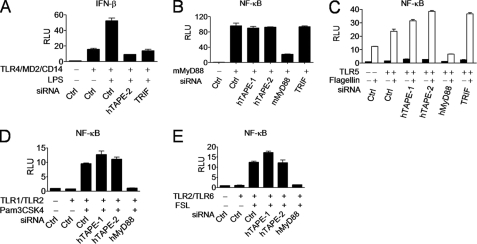
It is established that in addition to TLR3, Trif acts downstream of the endocytic TLR4-TRAM complex to activate IFN-β and further enhance NF-κB activation. Thus, we determined whether TAPE is involved in the endocytic TLR4-Trif pathway. Our results showed that knockdown of hTAPE, like Trif, impaired TLR4-induced IFN-β activation by lipopolysaccharide stimulation (Fig. 5A). We also assessed the relationship between TAPE and MyD88, which is a key adaptor involved in all TLRs except TLR3. Knockdown of hTAPE failed to block MyD88-induced NF-κB activation (Fig. 5B), suggesting that TAPE is not involved in the MyD88 downstream pathway. We further determined whether TAPE is involved in the cell surface TLR5 pathway, which activates the NF-κB promoter via MyD88 in case TAPE functions upstream of MyD88 to mediate the TLR5 pathway. Unlike MyD88, hTAPE knockdown failed to block TLR5-induced NF-κB activation by flagellin stimulation (Fig. 5C). Similarly, we found no blocking effect of hTAPE knockdown on other surface TLR pathways, such as the TLR1/TLR2 (Fig. 5D) and TLR2/TLR6 combinations (Fig. 5E). Together, these data demonstrate a selective role for TAPE in the endocytic TLR4-Trif pathway but not other cell surface TLR-MyD88 pathways.
FIGURE 5.
TAPE is implicated in the TLR4-Trif branch pathway. A, 293 cells treated with the indicated siRNAs were transfected with IFN-β-Luc together with TLR4, MD2, and CD14 and then stimulated with LPS (100 ng/ml) for analyzing the IFN-β promoter activity. B–E, 293 cells treated with indicated siRNAs were transfected with pELAM-Luc, together with MyD88 (B), TLR5 plus flagellin (4 μg/ml) stimulation (C), TLR1 and TLR2 plus Pam3CSK4 (0.5 μg/ml) stimulation (D), or TLR2 and TLR6 plus FSL-1 (0.5 μg/ml) stimulation (E), for analyzing the NF-κB promoter activity. Values represent the mean ± S.E. of duplicate samples (A–E). Data are from one experiment representative of two or three. RLU, relative light units; Ctrl, control.
DISCUSSION
Much progress has been made in decoding action modes of the innate immune system by identification and characterization of PRRs and innate immune regulators. However, signaling networks underlying the innate immune system remains to be further investigated to appreciate the complexity of innate immune defenses. Through this work we have revealed TAPE/CC2D1A to be a novel regulator of innate immune defenses. TAPE/CC2D1A showed the ability to activate the TBK1, NF-κB, and ERK pathways for the production of IFN-β and inflammatory cytokines. A recent work also demonstrated that TAPE/CC2D1A activates the NF-κB pathway via Traf2, TAK1, and IKKs (42).
Similar to its Drosophila ortholog Lgd, TAPE/CC2D1A predominantly resided in endolysosomes, and its deletion mutants disturbed the distribution of endolysosomes. In support of this notion, a recent study using siRNA screening revealed that TAPE/CC2D1A is one of the genes implicated in endosomal trafficking in mammalian cells (43). The C2 domain of Drosophila Lgd is responsible for binding phospholipids to anchor on the lipid membranes (38). In line with this evidence, we also found that TAPE/CC2D1A bound phospholipids (data not shown). In addition, the N- and distal C-terminal regions of TAPE/CC2D1A may also play critical roles in regulating the distribution of endolysosomes. TAPE/CC2D1A deletion mutants, which were shown to disturb the endolysosomal distribution, were also impaired in their ability to activate innate immune pathways. These findings together suggest that TAPE/CC2D1A is a regulator of endolysosomes. It is plausible that the endolysosomal localization of TAPE/CC2D1A is associated with its functions in innate immune signaling. Nonetheless, it seems that TAPE/CC2D1A localizes in other cellular compartments to exert different functions. For example, a previous study showed that TAPE/CC2D1A shuttles between the cytoplasm and the nucleus, but in the nucleus it acts as a transcriptional repressor (26). Another study showed that TAPE/Aki1 localizes in centrosomes to regulate centriole cohesion (44). Further analyses are needed to explore the subcellular localizations and functions of TAPE/CC2D1A.
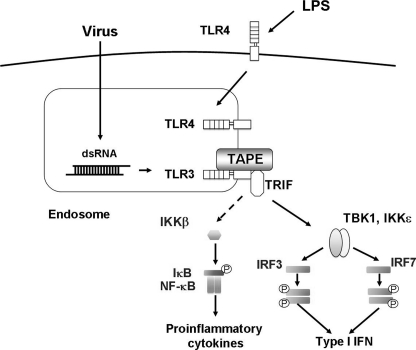
TAPE/CC2D1A was shown to link the endosomal TLR3 and TLR4 to downstream pathways but was not implicated in modulating the cell surface TLR-MyD88 pathways, including TLR1, TLR2, TLR5, and TLR6. In particular, TAPE/CC2D1A was critical for the optimal IFN-β production during TLR3 and TLR4 signaling. Our results indicated that TAPE/CC2D1A knockdown had no significant blocking effect on IL-8 production upon TLR3 stimulation (data not shown). This suggests that TAPE/CC2D1A is not essential for linking TLR3 to the NF-κB pathway. Because no enzymatic activity or domain was found in TAPE/CC2D1A, it is likely that TAPE/CC2D1A functions as an adaptor or a scaffold coupling TLR3 to the Trif-mediated downstream pathways (Fig. 6). We speculate a similar scenario for TAPE/CC2D1A in modulating the endocytic TLR4-Trif pathway. Emerging evidence indicates that endolysosomes function as type I IFN-inducing organelles for endosomal TLRs (45). Our findings that an endolysosomal adaptor TAPE/CC2D1A coupled TLR3 and TLR4 to the Trif-mediated downstream pathways further support this notion.
FIGURE 6.
Model for TAPE in endosomal TLR3 and TLR4 signaling. TAPE acts as an adaptor or a scaffold to couple endosomal TLR3 and TLR4 to the Trif-mediated downstream pathways.
Given the importance of TAPE/CC2D1A in regulating the TLR3 and TLR4 pathways described above, it is rational to speculate that TAPE/CC2D1A contributes to innate immune defenses against pathogen infections such as viruses and Gram-negative bacteria. Generation of TAPE/CC2D1A-deficient mice is warranted to assess the in vivo functions of TAPE/CC2D1A in innate immune defenses against pathogen infections. On the other hand, pathogens like viruses and bacteria have developed strategies to evade host innate immune detection for their survival through targeting PRRs or innate immune regulators (46). Thus, it is of interest to determine whether microbial components may target TAPE/CC2D1A to disrupt host innate immune defenses.
Supplementary Material
Acknowledgments
We thank K. Fitzgerald (University of Massachusetts Medical School), R. Lin (McGill University), U. Siebenlist (National Institutes of Health), and other scientists mentioned under the “Experimental Procedures” for plasmids. We thank Yen-Jen Cheng, Lin-Fang Chen, Chia-Kai Wu, and Hong-Yu Tseng for technical assistance.
This work was support by National Science Council in Taiwan Grants NSC 97-2320-B-006-014-MY3 and NSC 98-2321-B-006-008 and by National Cheng-Kung University Intramural Grant C023.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- PRR
- pattern-recognition receptor
- TAPE
- TBK1-associated protein in endolysosomes
- TLR
- Toll-like receptor
- IKK
- Iκ kinase
- EGFP
- enhanced GFP.
REFERENCES
- 1. Takeuchi O., Akira S. (2010) Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 2. Takaoka A., Wang Z., Choi M. K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., Ohba Y., Taniguchi T. (2007) Nature 448, 501–505 [DOI] [PubMed] [Google Scholar]
- 3. Roberts T. L., Idris A., Dunn J. A., Kelly G. M., Burnton C. M., Hodgson S., Hardy L. L., Garceau V., Sweet M. J., Ross I. L., Hume D. A., Stacey K. J. (2009) Science 323, 1057–1060 [DOI] [PubMed] [Google Scholar]
- 4. Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K. L., Superti-Furga G. (2009) Nat. Immunol. 10, 266–272 [DOI] [PubMed] [Google Scholar]
- 5. Fernandes-Alnemri T., Yu J. W., Datta P., Wu J., Alnemri E. S. (2009) Nature 458, 509–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiu Y. H., Macmillan J. B., Chen Z. J. (2009) Cell 138, 576–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ablasser A., Bauernfeind F., Hartmann G., Latz E., Fitzgerald K. A., Hornung V. (2009) Nat. Immunol. 10, 1065–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang P., An H., Liu X., Wen M., Zheng Y., Rui Y., Cao X. (2010) Nat. Immunol. 11, 487–494 [DOI] [PubMed] [Google Scholar]
- 10. Stewart C. R., Stuart L. M., Wilkinson K., van Gils J. M., Deng J., Halle A., Rayner K. J., Boyer L., Zhong R., Frazier W. A., Lacy-Hulbert A., El Khoury J., Golenbock D. T., Moore K. J. (2010) Nat. Immunol. 11, 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang S. Y., Jouanguy E., Ugolini S., Smahi A., Elain G., Romero P., Segal D., Sancho-Shimizu V., Lorenzo L., Puel A., Picard C., Chapgier A., Plancoulaine S., Titeux M., Cognet C., von Bernuth H., Ku C. L., Casrouge A., Zhang X. X., Barreiro L., Leonard J., Hamilton C., Lebon P., Héron B., Vallée L., Quintana-Murci L., Hovnanian A., Rozenberg F., Vivier E., Geissmann F., Tardieu M., Abel L., Casanova J. L. (2007) Science 317, 1522–1527 [DOI] [PubMed] [Google Scholar]
- 12. Wang T., Town T., Alexopoulou L., Anderson J. F., Fikrig E., Flavell R. A. (2004) Nat. Med. 10, 1366–1373 [DOI] [PubMed] [Google Scholar]
- 13. Le Goffic R., Balloy V., Lagranderie M., Alexopoulou L., Escriou N., Flavell R., Chignard M., Si-Tahar M. (2006) PLoS Pathog. 2, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. O'Neill L. A., Bowie A. G. (2007) Nat. Rev. Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 15. Seya T., Matsumoto M., Ebihara T., Oshiumi H. (2009) Immunol. Rev. 227, 44–53 [DOI] [PubMed] [Google Scholar]
- 16. Kawai T., Akira S. (2010) Nat. Immunol. 11, 373–384 [DOI] [PubMed] [Google Scholar]
- 17. Kagan J. C., Su T., Horng T., Chow A., Akira S., Medzhitov R. (2008) Nat. Immunol. 9, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. (2003) Nat. Immunol. 4, 491–496 [DOI] [PubMed] [Google Scholar]
- 19. Sharma S., tenOever B. R., Grandvaux N., Zhou G. P., Lin R., Hiscott J. (2003) Science 300, 1148–1151 [DOI] [PubMed] [Google Scholar]
- 20. Ishii K. J., Kawagoe T., Koyama S., Matsui K., Kumar H., Kawai T., Uematsu S., Takeuchi O., Takeshita F., Coban C., Akira S. (2008) Nature 451, 725–729 [DOI] [PubMed] [Google Scholar]
- 21. Tenoever B. R., Ng S. L., Chua M. A., McWhirter S. M., García-Sastre A., Maniatis T. (2007) Science 315, 1274–1278 [DOI] [PubMed] [Google Scholar]
- 22. Huang J., Liu T., Xu L. G., Chen D., Zhai Z., Shu H. B. (2005) EMBO J. 24, 4018–4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sasai M., Shingai M., Funami K., Yoneyama M., Fujita T., Matsumoto M., Seya T. (2006) J. Immunol. 177, 8676–8683 [DOI] [PubMed] [Google Scholar]
- 24. Kawagoe T., Takeuchi O., Takabatake Y., Kato H., Isaka Y., Tsujimura T., Akira S. (2009) Nat. Immunol. 10, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryzhakov G., Randow F. (2007) EMBO J. 26, 3180–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rogaeva A., Albert P. R. (2007) Eur. J. Neurosci. 26, 965–974 [DOI] [PubMed] [Google Scholar]
- 27. Ou X. M., Lemonde S., Jafar-Nejad H., Bown C. D., Goto A., Rogaeva A., Albert P. R. (2003) J. Neurosci. 23, 7415–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogaeva A., Galaraga K., Albert P. R. (2007) J. Neurosci. Res. 85, 2833–2838 [DOI] [PubMed] [Google Scholar]
- 29. Nakamura A., Naito M., Tsuruo T., Fujita N. (2008) Mol. Cell. Biol. 28, 5996–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuda A., Suzuki Y., Honda G., Muramatsu S., Matsuzaki O., Nagano Y., Doi T., Shimotohno K., Harada T., Nishida E., Hayashi H., Sugano S. (2003) Oncogene 22, 3307–3318 [DOI] [PubMed] [Google Scholar]
- 31. Chen S. H., Oakes J. E., Lausch R. N. (1993) Antiviral Res. 22, 15–29 [DOI] [PubMed] [Google Scholar]
- 32. Chen S. H., Yao H. W., Chen I. T., Shieh B., Li C., Chen S. H. (2008) J. Clin. Invest. 118, 3470–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung S. Y., Malovannaya A., Wei J., O'Malley B. W., Qin J. (2005) Mol. Endocrinol. 19, 2451–2465 [DOI] [PubMed] [Google Scholar]
- 34. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 35. Ishikawa H., Barber G. N. (2008) Nature 455, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tabeta K., Hoebe K., Janssen E. M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., Shamel L., Herskovits A. A., Portnoy D. A., Cooke M., Tarantino L. M., Wiltshire T., Steinberg B. E., Grinstein S., Beutler B. (2006) Nat. Immunol. 7, 156–164 [DOI] [PubMed] [Google Scholar]
- 37. Lemmon M. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 38. Gallagher C. M., Knoblich J. A. (2006) Dev. Cell 11, 641–653 [DOI] [PubMed] [Google Scholar]
- 39. Jaekel R., Klein T. (2006) Dev. Cell 11, 655–669 [DOI] [PubMed] [Google Scholar]
- 40. Childress J. L., Acar M., Tao C., Halder G. (2006) Curr. Biol. 16, 2228–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jackson A. L., Linsley P. S. (2010) Nat. Rev. Drug Discov. 9, 57–67 [DOI] [PubMed] [Google Scholar]
- 42. Zhao M., Li X. D., Chen Z. (2010) J. Biol. Chem. 285, 24372–24380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Collinet C., Stöter M., Bradshaw C. R., Samusik N., Rink J. C., Kenski D., Habermann B., Buchholz F., Henschel R., Mueller M. S., Nagel W. E., Fava E., Kalaidzidis Y., Zerial M. (2010) Nature 464, 243–249 [DOI] [PubMed] [Google Scholar]
- 44. Nakamura A., Arai H., Fujita N. (2009) J. Cell Biol. 187, 607–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Barton G. M., Kagan J. C. (2009) Nat. Rev. Immunol. 9, 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bowie A. G., Unterholzner L. (2008) Nat. Rev. Immunol. 8, 911–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.