Abstract
Hypoxia modulates gene expression and affects multiple aspects of endothelial cell biology. Fibulin-5 (FBLN5) is an extracellular matrix protein essential for elastic fiber assembly and vasculogenesis that participates in vascular remodeling and controls endothelial cell adhesion, motility, and proliferation. In this context, we aimed to analyze FBLN5 regulation by hypoxia in endothelial cells. Hypoxia (1% O2) increased FBLN5 mRNA levels in endothelial cells in a time-dependent manner. Maximal induction (∼2.5-fold) was achieved after 24 h of hypoxia. This effect paralleled an increase in both intracellular and extracellular FBLN5 protein levels. The increase in FBLN5 mRNA levels observed in hypoxic cells was blocked by inhibitors of the PI3K/Akt/mTOR pathway (LY294002 and rapamycin) and mimicked by dimethyl oxal glycine, which prevents proline hydroxylase-mediated degradation of HIF-1α. Silencing of HIF-1α completely prevented hypoxia-induced FBLN5 up-regulation. Accordingly, both hypoxia and HIF-1α overexpression increased FBLN5 transcriptional activity. Serial promoter deletion and mutagenesis studies revealed the involvement of a putative hypoxia response element (HRE) located at −78 bp. In fact, EMSA and ChIP assays demonstrated increased HIF-1 binding to this site in hypoxic cells. Interestingly, the rate of endothelial cells undergoing apoptosis in cultures exposed to hypoxia increased in FBLN5 knockdown cells, suggesting that hypoxia-induced FBLN5 expression contributes to preserve cell survival. These results provide evidence that HIF-1 signaling underlies the increase of FBLN5 expression elicited by hypoxia in endothelial cells and suggest that FBLN5 induction could be involved in the adaptive survival response of endothelial cells to hypoxia.
Keywords: Apoptosis, Endothelium, Extracellular Matrix, Gene Regulation, Hypoxia, Endothelial Cells, Fibulin 5
Introduction
Endothelial cells are primary sensors to the hypoxic stress characteristic of some physiological processes and diseases such as cancer, ischemic disorders, chronic inflammation, and atherosclerosis. The adaptive response of endothelial cells to hypoxia involves a complex coordinated response affecting multiple aspects of endothelial cell biology (1). Hypoxia is the major driving force for neovascularization through the modulation of a cascade of genes and proteins that promote endothelial cell sprouting and proliferation (2). Furthermore, hypoxia triggers cell survival, activates DNA repair processes, enhances glucose uptake and metabolism, alters vascular tone and coagulant function, modulates endothelial permeability and cell adhesive properties, and promotes extracellular matrix (ECM)4 remodeling (1, 3, 4).
The hypoxia-inducible factor-1 (HIF-1) is a critical mediator of cellular responses to hypoxia (2). HIF-1, the prototype of this family, is a heterodimeric basic helix-loop-helix transcription factor composed of HIF-1β (constitutive subunit) and HIF-1α (oxygen-sensitive subunit). In normoxic conditions, HIF-1α is degraded by a mechanism involving hydroxylation of two prolyl residues by specific prolyl hydroxylases, ubiquitylation, and proteasomal degradation through a von Hippel-Lindau-dependent pathway. Conversely, under hypoxic conditions, HIF-1α hydroxylation is inhibited leading to increased HIF-1α levels that, in turn, results in an enhancement of HIF-1-dependent transcriptional responses (5–7). HIF-1 controls multiple aspects of endothelial behavior including cell proliferation, chemotaxis, ECM penetration, and wound healing (8).
Fibulin 5 (FBLN5) is a widely expressed ECM glycoprotein that colocalizes with elastic fibers and is essential for proper elastic fiber assembly and vasculogenesis (9–13). Altered FBLN5 expression has been linked to several pathological processes, including tumor progression (14). In the vascular wall, the up-regulation of FBLN5 observed in atherosclerotic plaques and in neointimal thickening induced by balloon injury suggests its role in vascular remodeling (10, 15, 16). In fact, FBLN5 binds to αvβ3, αvβ5, and α9β1 integrins and promotes endothelial cell adhesion via its Arg-Gly-Asp (RGD) motif (10, 11, 15). Furthermore, it has been reported that FBLN5 inhibits endothelial and vascular smooth muscle cell proliferation and migration (16–18) and displays anti-angiogenic properties both in vivo and in vitro (19, 20).
FBLN5 has been associated with different vascular processes involving ECM remodeling; however, little is known about the mechanisms underlying its regulation in vascular cells. This study identifies FBLN5 as a hypoxia-responsive gene in endothelial cells, dissects the molecular mechanisms underlying this regulation, and delineates its biological significance.
EXPERIMENTAL PROCEDURES
Cell Culture
Bovine aortic endothelial cells (BAEC; Clonetics) were cultured in RPMI 1640 (Invitrogen), supplemented with 10% FCS (Biological Industries), antibiotics (0.1 mg/ml streptomycin, 100 units/ml penicillin G), and 2 mm l-glutamine (Invitrogen) as described previously (21). Human umbilical vein endothelial cells (HUVEC) were obtained by collagenase digestion and cultured in medium M199 (Biological Industries) supplemented with 20 mm HEPES, pH 7.4 (Invitrogen), 30 μg/ml endothelial cell growth supplement (Millipore), 100 μg/ml heparin (Sigma), 20% FCS, and antibiotics (22). Mouse lung endothelial cells were isolated from lungs of C57BL/6 mice by collagenase A (Roche Applied Science) digestion followed by selection with intercellular adhesion molecule 2-coated magnetic beads (Invitrogen) as described previously (23). Endothelial cells were used between the third and fifth passage. The human epithelioid carcinoma HeLa cell line was cultured in DMEM (Invitrogen) supplemented with 10% FCS and antibiotics. Cells were maintained in standard culture conditions (21% O2, 5% CO2, 95% humidity) until subconfluence. Cells were exposed to hypoxia (1% O2, 5% CO2, balanced with N2) in a Forma Series II hypoxic incubator (model 3141; Thermo Electron Corp.). The mammalian target of rapamycin (mTOR) inhibitor rapamycin (100 nm; Sigma) and PI3K inhibitor LY294002 (10 μm; Calbiochem) were added 1 h prior the hypoxic stimulus. No cytotoxicity, analyzed by the trypan blue exclusion test and the XTT-based assay for cell viability (Roche Applied Science), was observed after the treatment with either of these compounds.
Real-time PCR
Total RNA was isolated using UltraspecTM (Biotecx) following manufacturer's instructions. RNA (1 μg) was reverse transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) in the presence of random hexamers. Quantification of mRNA levels was performed by real-time PCR using an ABI PRISM 7900HT sequence detection system (Applied Biosystems) and specific primers and probes provided by the Assay-on-Demand system for FBLN2 (Hs00157482_m1), FBLN3 (Hs00244575_m1), FBLN5 (Hs00197064_m1), HIF-1α (Hs00153153_m1), HIF-2α (Hs01026149_m1), and vascular endothelial growth factor (VEGF)-A (Hs00173626_m1). TATA-binding protein was used as an endogenous control (Hs99999910_m1) (24). Each sample was amplified in duplicate.
Confocal Microscopy
HUVEC were plated at a density of 2 × 104 cells onto 12-mm diameter glass-bottomed dishes (Willco Wells B.V.) coated with gelatin. Cells were maintained in normoxia or exposed to hypoxia as described above. Afterward, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Tween® 20. The permeabilization step was avoided in those experiments aimed to demonstrate the extracellular localization of FBLN5. Then, cells were incubated with a mouse monoclonal antibody against FBLN5 (Abcam (ab36611); 1:2000) for 1 h, washed, and incubated for 1 h with a fluorescence-conjugated secondary antibody (donkey anti-mouse Alexa Fluor 488; Molecular Probes). Nuclei and actin fibers were stained with Hoechst 33342 and Alexa Fluor 633-phalloidin, respectively (Molecular Probes). Fluorescence images were acquired with a Leica DMIRE2 confocal microscope using Leica Confocal Software.
FBLN5 Promoter Constructs
A 1667-bp fragment of the human FBLN5 promoter (−1650 to +17 related to the start codon) was amplified by PCR from genomic DNA and cloned into the pGL3 basic luciferase reporter vector (Promega) using KpnI-XhoI restriction sites (pFBLN5–1650). Two serial deletion constructs (−635 to +17 and −329 to +17) were generated by PCR amplification from pFBLN5–1650 (pFBLN5–635 and pFBLN5–329, respectively). Primers used were: pFBLN5–1650 (upper primer), 5′-TTAAGGTACCCCTTTGGTTGCCCCTTACTTTATT-3′; pFBLN5–635 (upper primer), 5′-GAGTTCGGTACCCTTTCCTTCCGGAGGCGA-3′; pFBLN5–329 (upper primer), 5′-TGTAATCGGTACCAGCTGTGTCCAGACTG-3′; and the reverse primer, 5′-TAACTCAGGTCCAAGACGCGCGAGGAGGAGATGCGAA-3′. (The KpnI sites in upper primers and the XhoI site in the reverse primer are underlined.) All constructs were confirmed by DNA sequencing.
Transient Transfection Assays
Transient transfections of BAEC were performed with LipofectinTM (Invitrogen) and the FBLN5 constructs described above together with pSVβ-galactosidase as an internal control (Promega) (25). In co-transfection experiments, performed in cells maintained under normoxic conditions, a HIF-1α expression vector (pHIF-1α), kindly provided by Dr. L. E. Huang (Laboratory of Human Carcinogenesis, NCI, National Institutes of Health, Bethesda, MD), or the corresponding empty vector (pCDNA3) were used (26). Transient transfection assays of subconfluent BAEC were performed in six-well plates with 1 μg/well of the indicated construct, 0.3 μg/well of pSVβ- galactosidase, and 3 μl of LipofectinTM. The complexes DNA/liposome were added to the cells for 7 h. After 24 h, transfected cells were exposed to hypoxia for 18 h. Luciferase activity was measured in cell lysates using the Luciferase assay kit (Promega) and a luminometer (Orion I, Berthold Detection Systems) according to the manufacturer. Results were normalized by β-galactosidase activity using the Enzyme Assay SystemTM (Promega). In some EMSA experiments, HeLa cells grown in 100-mm2 plates were transfected with 8 μg/plate of the HIF-1α expression vector (pHIF-1α) or its corresponding empty vector (pcDNA3) and 32 μl of LipofectamineTM (Invitrogen). Cells were incubated for 3 h with the DNA-liposome complex. Plates were processed to obtain nuclear extracts 40 h after transfection.
Site-directed Mutagenesis
The FBLN5 HRE site located at position −78 was mutated using the QuikChange II Site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions (27). The HRE mutation was introduced with primers: 5′-CAGAGGAGGCCGtttTGCCCGAGCTCCTCC-3′ and 5′-GGAGGAGCTCGGGCAaaaCGGCCTCCTCTG-3′ (putative site is underlined, and changes are indicated in lowercase letters) using the pFBLN5–329 vector as a template. Mutation was confirmed by DNA sequencing.
siRNA Transfection
Silencer predesigned siRNA targeting HIF-1α (42840, Ambion), HIF-2α (s4699, Ambion), FBLN5 (L-017621-00; Dharmacon) or the corresponding control siRNAs, SilencerTM Negative Control no. 1 (Ambion) and ON-TARGETplus siCONTROL (catalog no. D-001810-10; Dharmacon) were transfected in HUVEC using an Amaxa NucleofectorTM and the HUVEC Nucleofector kit according to the manufacturer's instructions (Amaxa) as described previously (28). Briefly, electroporation was carried out with 1 × 106 cells and 1 μg of siRNA with the U-001 program. After electroporation, cells were resuspended in 1.5 ml of prewarmed cell culture medium and seeded in six-well plates (350,000 cells/well). After 24 h, cells were subjected to hypoxic or normoxic conditions. Gene knockdown was verified by real time-PCR and/or Western-blot as indicated.
Nuclear Extract Preparation
HeLa cells were grown in 100-mm dishes under normoxic or hypoxic conditions for 24 h. Alternatively, HeLa were transfected with the HIF-1α expression vector (pHIF-1α) or its corresponding empty vector (pcDNA3) as described above. Cells were collected in ice-cold PBS and nuclear extracts were obtained using the NucBuster Protein Extraction Kit (Novagen) according to the manufacturer's recommendations. Proteins were quantified by the BCA protein assayTM (Pierce), and nuclear extracts were aliquoted and stored at −80 °C until used. The increase in nuclear levels of HIF-1α elicited by either hypoxia or HIF-1α overexpression was verified by Western blot.
EMSA
Two double-stranded DNA probes, one containing the putative wild-type HRE element present in the FBLN5 promoter (positions from −78 to −74) and the other mutated at three bases of this HRE, were generated from the annealing of single-stranded complementary oligonucleotides as follows: FBLN5-HRE, 5′-GAGGAGGCCGACGTGCCCGAGCTCC-3′ and its complement; and FBLN5mut-HRE, 5′-GAGGAGGCCGTTTTGCCCGAGCTCC-3′ and its complement. (Wild-type and mutated HRE sites are underlined.) Purified probes were end-labeled with [γ-32P]ATP (PerkinElmer Life Science) and T4 polynucleotide kinase (Roche Applied Science) (29). EMSA assays were performed using the Novagen's EMSA accessory kit. Briefly, nuclear proteins (4 μg) were incubated for 15 min on ice with 0.01 units poly(dI-dC)·poly(dI-dC) and 500 ng of sonicated salmon sperm DNA, in 20 mm HEPES, pH 8, 0.2 mm EDTA, 100 mm KCl, 20% glycerol, and 0.5 mm DTT. Approximately 40,000 cpm of the appropriate 32P end-labeled probe were then added to the reaction mixture and incubated for 30 min on ice in a final volume of 20 μl. For the supershift assay, 2 μg of an anti-HIF-1α antibody (ab16066; Abcam) were added to the reactions. Protein-DNA complexes were resolved by electrophoresis at 4 °C on 5% polyacrylamide, 5% glycerol gels in 0.5× TBE. The gels were dried and subjected to autoradiography using a Storage Phosphor Screen (GE Healthcare). Shifted bands were detected using a Typhoon 9400 scanner (Amersham Biosciences).
ChIP Assay
ChIP assays were performed using the HIF-1α ExactaChIPTM kit according to the manufacturer's protocol (R&D Systems). Briefly, HUVEC cultured under normoxic and hypoxic conditions were cross-linked with 1% formaldehyde for 15 min. The cross-linked reaction was stopped by adding glycine to a final concentration of 125 mm. Then, cells were washed, harvested by scraping, lysed, and sonicated to shear chromatin to DNA fragments of 0.5–1 kb. Lysates were centrifuged, and an aliquot of supernatant (20 μl) was saved as input DNA. Supernatants were then immunoprecipitated with an anti-HIF-1α antibody (5 μg) or an IgG as a control for nonspecific binding of DNA. Immunoprecipitates were recovered by addition of streptavidin-agarose beads (Sigma). After washing, 100 μl of chelating resin solution (R&D Systems) were added to the beads and boiled for 10 min. Finally, DNA was purified and concentrated using the QIAquick PCR purification kit (Qiagen). Purified DNA was analyzed by conventional and real-time PCR with primers designed to amplify the human FBLN5 gene from −245 to +59 (sequence positions relative to the ATG): upper primer, 5′-GCTAAGCAAAACCAGGTGCT-3′ and lower primer, 5′-GTGCGAAGGCGAGAAGAAA-3′. As a positive control, a conventional PCR analysis using VEGF-specific oligonucleotides provided by the kit was also carried out. Real-time PCR was performed in samples from two individual ChIP assays by triplicate with the QuantifastTM SYBR® Green PCR kit (Qiagen). The relative abundance of specific sequences in immunoprecipitated DNA was determined using the ΔΔCt method. Quantifications were corrected to account for 4% inputs (4).
Western Blot Analysis
Whole-cell extracts were obtained from endothelial cell cultures as described (30). Briefly, cells were washed twice with PBS and lysed with a lysis buffer containing 50 mm HEPES, pH 7.4, 150 mm NaCl, 100 mm NaF, 10 mm sodium pyrophosphate, 10 mm EDTA, 2 mm Na3VO4, 1 mm PMSF, 5 μm leupeptin, and 0.5% SDS. Nuclear and whole-cell extracts were resolved by SDS-PAGE and transferred to 0.45-μm nitrocellulose filters (Bio-Rad). In some experiments, supernatants from HeLa cultured under normoxia or exposed to hypoxia and concentrated using Amicon Ultra 10K filter units (Millipore) were used. Blots were incubated with antibodies directed against HIF-1α (NB100–449A, Novus; 1:500), HIF-2α (ab199, Abcam; 1:300), FBLN5 (sc30170; Santa Cruz Biotechnology; 1:300); Akt (9272, Cell Signaling); phosphorylated-Akt (Ser473) (antibody no. 9271, Cell Signaling), ERK1/2 (antibody no. 9102, Cell Signaling) and phospho-ERK1/2 (Thr202/Tyr204) (antibody no. 9106, Cell Signaling). Bound antibodies were detected after incubation with a HRP-conjugated goat anti-rabbit IgG and using the SuperSignal West Dura Extended Duration Substrate (Pierce). Equal loading of protein in each lane was verified by Ponceau staining and by β-actin (for whole-cell extracts) or nucleolin (for nuclear extracts) signal (31, 32).
Annexin V Binding by Flow Cytometry
Annexin V binding was used as an index of cell apoptosis. HUVEC were transfected with a siRNA against FBLN5 or with a SilencerTM Negative Control no. 1 as described above. Then, cells were arrested and exposed to hypoxia for 16 h. Finally, cells were trypsinized, resuspended in 1× annexin V binding buffer (BD Pharmingen) and incubated with annexin V conjugated with FITC (annexin V-FITC) (BD Pharmingen) and propidium iodide (PI) (BD Pharmingen). Annexin V-FITC and PI binding was analyzed by FACS in an EPICS® XLTM Flow Cytometer. Results were expressed as percentage of total cell population. Annexin V-FITC fluorescence (abscissa) was plotted versus PI uptake (ordinate). Data were gated for damaged cells (annexin V− and PI+), necrotic cells (annexin V+ and PI+), viable cells (annexin V− and PI−), and apoptotic cells (annexin V+ and PI−).
Statistical Analysis
Data are expressed as mean ± S.E. Means were compared by one-factor analysis of variance followed by Fisher protected least significant difference to assess specific group differences. Differences were considered significant at p < 0.05.
RESULTS
Hypoxia Induces FBLN5 Expression in Endothelial Cells
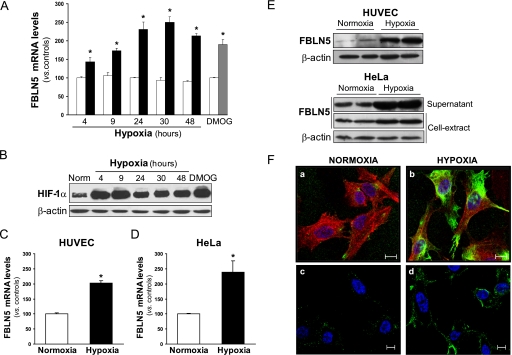
It has been previously demonstrated that FBLN5 regulates endothelial cell function (16–20). Here, we tested whether hypoxia could modulate FBLN5 expression in endothelial cells. We found that FBLN5 expression was enhanced in a time-dependent manner in BAEC exposed to hypoxia (1% O2). FBLN5 mRNA levels progressively increased from 4 h to reach a plateau after 24–48 h of hypoxia (2.5-fold) (Fig. 1A). Similarly, dimethyl oxal glycine, which prevents proline hydroxylase-mediated proteasomal degradation of HIF-1α mimicking hypoxia (33), markedly up-regulated FBLN5 expression in endothelial cells (Fig. 1A). Dimethyl oxal glycine and hypoxia strongly raised HIF-1α protein levels, with an increase observed at 4 h and even after 48 h of hypoxia exposure (Fig. 1B). In these conditions, mRNA levels of VEGF, a well known hypoxia-regulated gene were also up-regulated (data not shown). The induction of FBLN5 expression under hypoxic conditions was also observed in other primary endothelial cells such as mouse lung endothelial cells (data not shown) and HUVEC (Fig. 1C) and in the human epithelioid carcinoma HeLa cell line (Fig. 1D), indicating that such regulation is not restricted to endothelial cells. FBLN5 protein levels evaluated by Western blot in whole-cell lysates revealed a marked enhancement of FBLN5 expression in both HUVEC and HeLa exposed to hypoxia (Fig. 1E). In agreement, immunocytochemistry analysis demonstrated that hypoxia markedly increased intracellular FBLN5 levels. FBLN5 staining was mainly localized in the perinuclear region and in the cell periphery (Fig. 1F, panels a and b). Hypoxia also augmented the extracellular presence of FBLN5 as we showed by confocal microscopy studies performed in nonpermeabilized cells (Fig. 1F, panels c and d) and by immunoblot analysis of concentrated supernatants from HeLa cultures (Fig. 1E, lower panel).
FIGURE 1.
Hypoxia increases FBLN5 expression. BAEC were maintained under normoxic conditions (white bars), exposed to hypoxia (1% O2; black bars) during different times or treated with dimethyl oxal glycine (0.5 mm, 24 h; gray bar). A, FBLN5 mRNA levels were evaluated by real-time PCR and were normalized by TATA-binding protein expression levels. Data are expressed as mean ± S.E. (n = 9; *, p < 0.05 versus control cells). B, HIF-1α protein levels were evaluated in cell lysates by Western blot. Unchanged levels of β-actin are shown as a loading control (Norm, normoxia). Autoradiograms are representative of two independent experiments performed in duplicate. C and D, FBLN5 mRNA levels were analyzed in HUVEC (C) and HeLa (D) exposed to hypoxia for 24 h. Data are expressed as mean ± S.E. (n = 9; *, p < 0.05 versus control cells). E, HUVEC and HeLa were maintained under normoxia or exposed to hypoxia for 48 h. FBLN5 protein levels were analyzed by Western blot from whole-cell extracts or supernatant from cell cultures. β-Actin levels were used as a loading control. A representative autoradiogram of three independent experiments performed by duplicate is shown. F, FBLN5 localization was analyzed by immunocytochemistry in HUVEC maintained in normoxia (a and c) or exposed to hypoxia (b and d). In permeabilized cells, intracellular FBLN5 staining (green) was detected in the perinuclear region and in cell periphery (a and b). Extracellular localization was analyzed in nonpermeabilized cells (c and d). Cells were counterstained with Hoechst to highlight nuclei (blue) and with Alexa Fluor 633-phalloidin to visualize F-actin (red). Bar, 10 μm.
PI3K/Akt/mTOR Axis Is Involved in FBLN5 Induction by Hypoxia
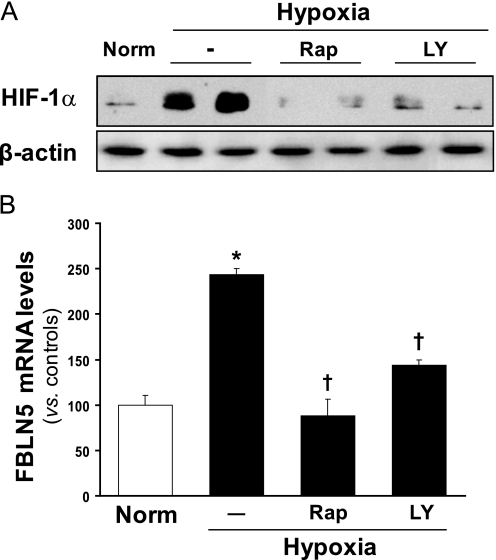
The PI3K/Akt/mTOR pathway has been implicated in the regulation of HIF-1 protein levels (6). To define the role of this signaling pathway in the induction of FBLN5 by hypoxia, we examined the effect of LY294002 and rapamycin, compounds that inhibit PI3K and mTOR, respectively. Western blot analysis confirmed that both inhibitors prevented the induction of HIF-1α protein levels caused by hypoxia (Fig. 2A). Accordingly, the increase in FBLN5 mRNA levels observed in hypoxic cells was completely blocked by pretreatment with either LY294002 or rapamycin (Fig. 2B). These compounds neither alter HIF-1α protein levels nor FBLN5 expression under basal conditions (normoxia; data not shown). These results evidence the involvement of the PI3K/Akt/mTOR pathway on FBLN5 up-regulation by hypoxia.
FIGURE 2.
The increase in FBLN5 expression elicited by hypoxia is mediated by the PI3K/Akt/mTOR pathway. BAEC were preincubated with rapamycin (Rap; 100 nm) or LY294002 (LY; 10 μm) and exposed to normoxia (Norm, controls) or hypoxia (1% O2, 24 h). A, HIF-1α protein levels were analyzed by Western blot. β-Actin levels were used as a loading control. A representative autoradiogram of three independent experiments performed by duplicate is shown. B, FBLN5 mRNA levels were evaluated by real-time PCR. Data are expressed as mean ± S.E. (n = 9; * and †, p < 0.05 versus control cells and versus hypoxic cells, respectively).
HIF-1α Inhibition Prevents FBLN5 Induction by Hypoxia
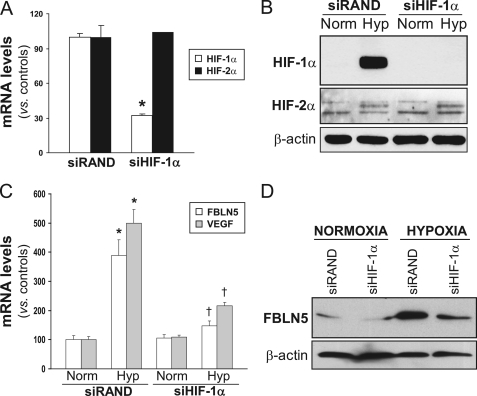
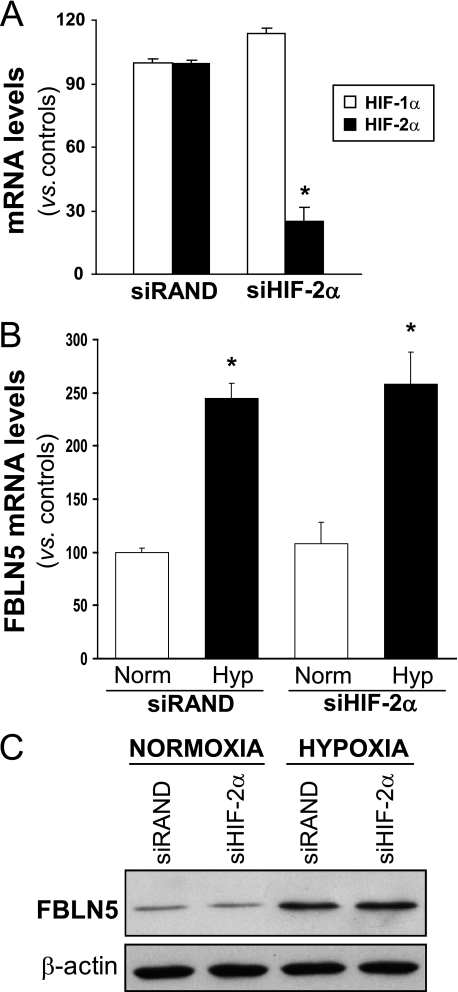
To further evaluate the contribution of HIF-1α in FBLN5 regulation, knockdown experiments using a specific siRNA against HIF-1α (siHIF-1α) were performed. siHIF-1α strongly reduced HIF-1α mRNA levels and prevented the increase of HIF-1α protein levels elicited by hypoxia (Fig. 3, A and B). In these conditions HIF-1α knockdown counteracted the up-regulation of FBLN5 mRNA and protein levels triggered by hypoxia (Fig. 3, C and D). As expected, hypoxia-induced up-regulation of VEGF, a well established HIF-1α target gene, was abolished by siHIF-1α (Fig. 3C). HIF-1α knockdown neither affected HIF-2α mRNA levels (Fig. 3A) nor prevented the hypoxia-mediated induction of HIF-2α protein levels in HUVEC (Fig. 3B), thus confirming the specificity of our approach. It should be noted that, as described previously (34), the up-regulation of HIF-2α expression by hypoxia in these cells is markedly lower than that of HIF-1α (Fig. 3B), suggesting a major role of HIF-1α on hypoxia-induced FBLN5 up-regulation. In agreement, the specific silencing of HIF-2α (Fig. 4A) was not able to prevent the up-regulation of FBLN5 observed under hypoxic conditions (Fig. 4, B and C).
FIGURE 3.
HIF-1α silencing abrogates hypoxia-induced FBLN5 up-regulation. A, HUVEC were transfected with a HIF-1α specific siRNA (siHIF-1α) or a control siRNA (siRAND), and HIF-1α and HIF-2α mRNA levels were determined by real-time PCR. Data are expressed as mean ± S.E. (n = 6; *, p < 0.05 versus siRAND cells). B, Western blot analysis confirms the blockade of HIF-1α protein levels by siHIF-1α under hypoxic conditions, whereas HIF-2α expression remained unchanged (Norm, normoxia; Hyp, hypoxia). C, HIF-1α silencing prevents the hypoxia-mediated up-regulation of FBLN5 and VEGF mRNA levels evaluated by real-time PCR. Results are expressed as mean ± S.E. (n = 9; * and †, p < 0.05 versus normoxic cells transfected with a siRAND and versus hypoxic cells transfected with a siRAND, respectively). D, FBLN5 protein levels were analyzed by Western blot in HUVEC transfected with a siHIF-1α or a control siRNA (siRAND) under normoxic or hypoxic conditions. A representative autoradiogram of two independent experiments is shown.
FIGURE 4.
HIF-2α silencing does not alter hypoxia-induced FBLN5 up-regulation. A, HIF-1α and HIF-2α mRNA levels, determined by real-time PCR, were analyzed in HUVEC transfected with a HIF-2α-specific siRNA (siHIF-2α) or a control siRNA (siRAND). Data are expressed as mean ± S.E. (n = 6; *, p < 0.05 versus siRAND cells). HIF-2α knockdown did not affect the up-regulation of FBLN5 mRNA (B) and protein levels (C) triggered by hypoxia (Norm, normoxia; Hyp, hypoxia). Results are expressed as mean ± S.E. (n = 6; *, p < 0.05 versus normoxic cells transfected with a siRAND).
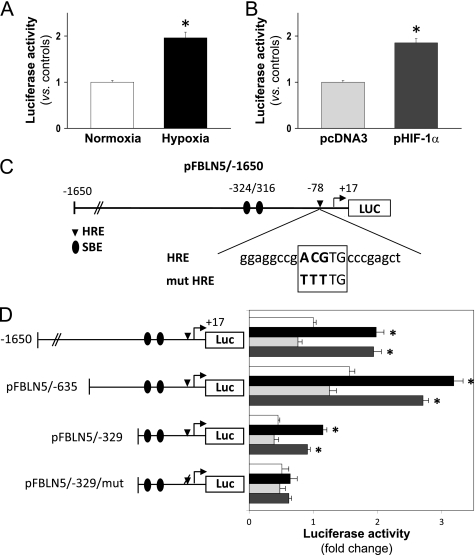
Hypoxia Increases FBLN5 Transcriptional Activity through an HRE
Data described above suggest that under hypoxic conditions HIF-1α could increase FBLN5 transcriptional activity. To examine this hypothesis, a pGL3 luciferase reporter construct harboring 1650 bp of the FBLN5 promoter (upstream ATG) was generated and used in transient transfection studies. As shown in Fig. 5A, hypoxia induced FBLN5 transcriptional activity ∼2-fold. Similarly, cotransfection with a HIF-1α expression vector (pHIF-1α) increased FBLN5 promoter activity as compared with empty vector-transduced cells (Fig. 5B). A serial promoter deletion analysis determined that the proximal 329-bp region retains hypoxia and HIF-1α responsiveness (Fig. 5D). Of note, further deletions completely abolished transcriptional activity (data not shown) in agreement with a previous report that describes the requirement of sequences between −392 and −315 for basal promoter activity (35). In silico studies using MatInspector software for the analysis of transcription binding sites identified a putative HRE located at −78 bp (Fig. 5C). To test the functionality of this motif, we used a site-directed mutagenesis approach. Fig. 5D shows that mutation of this HRE sequence completely abrogated the induction of FBLN5 promoter activity elicited by both hypoxia and HIF-1α overexpression. Thus, these data suggest that the HRE site identified in our analysis confers FBLN5 hypoxia responsiveness.
FIGURE 5.
Hypoxia up-regulates FBLN5 transcriptional activity through an HRE. A, BAEC transfected with the pFBLN5–1650 luciferase (Luc) construct were exposed to normoxia (white bars) or hypoxia (black bars) for 18 h. B, alternatively, cells maintained under normoxic conditions were co-transfected with an HIF-1α expression vector (pHIF-1α; dark gray bars) or the corresponding empty vector (pcDNA3; light gray bars). Luciferase and β-galactosidase activities were determined as described under “Experimental Procedures.” Results are expressed as mean ± S.E. (n = 9). C, scheme corresponding to the FBLN5 promoter region analyzed in transient transfection studies. The location of two Smad binding sites (SBE) and the sequence of the putative HRE motif are shown. Changes introduced by mutagenesis are indicated in boldface. D, a promoter serial deletion study was performed using the wild-type pFBLN5-1650, pFBLN5-635, and pFBLN5-329 luciferase constructs or the HRE-mutated vector pFBLN5-329/mut. Position of the wild-type HRE motif (indicated as a diamond) or its mutated form (indicated as a deleted diamond) are shown. Cells were cultured under normoxia (white bars) or hypoxia (black bars), co-transfected with a HIF-1α expression vector (dark gray bars) or the corresponding empty vector (pcDNA3; light gray bars). Luciferase activity is expressed as fold-change using pFBLN5-1650 (in normoxia) as a reference value. Results are expressed as mean ± S.E. (n = 9; *, p < 0.05 versus normoxic or pcDNA3-transfected cells).
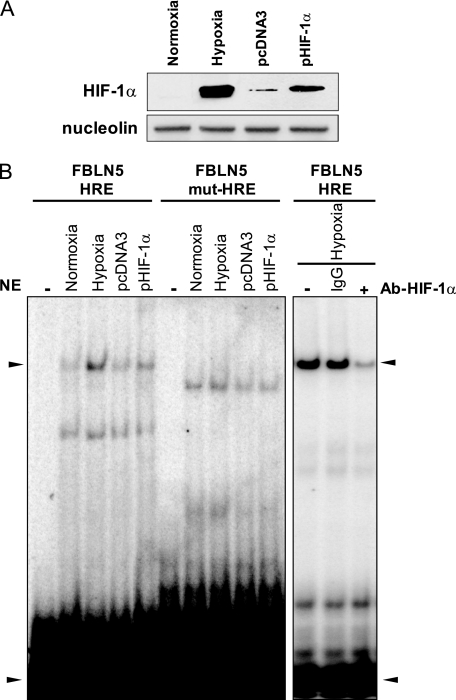
HIF-1α Binds to FBLN5 HRE Motif
To characterize the functionality of the putative HRE element described above, we performed EMSA assays. HeLa cells, rather than HUVEC, were used in these experiments because of their higher transfection efficiency by the liposome system used. HeLa cells were either subjected to hypoxia or transfected with a HIF-1α expression vector (pHIF-1α) or the corresponding empty vector (pcDNA3), and nuclear extracts were obtained. These extracts were used in binding reactions with 32P end-labeled probes corresponding to the wild-type HRE (FBLN5-HRE) or its mutated form (FBLN5mut-HRE). Western blot assays verified that both hypoxia and HIF-1α overexpression increased nuclear HIF-1α protein levels (Fig. 6A). As shown in Fig. 6B, EMSA assays conducted with the wild-type HRE probe evidenced a pattern of two main retarded bands. Both hypoxia and HIF-1α overexpression enhanced the intensity of the upper-shifted band. In fact, the intensity of the upper retarded band in cells exposed to hypoxia or transduced with pHIF-1α correlated with levels of HIF-1α detected in these cells by Western blot (Fig. 6A). Conversely, mutation of the putative HRE element changed the pattern of shifted bands and abrogated the regulation caused by either hypoxia or HIF-1α overexpression The altered pattern of bands when mutated probes are used has been described previously with both HRE and non-HRE probes (36, 37) and could be derived from the partial and unspecific recognition of these sequences by nuclear proteins. Finally, an antibody against HIF-1α strongly reduced the intensity of the upper shifted band suggesting the interaction of HIF-1α with this sequence. These data indicate that hypoxia promotes the binding of HIF-1 to the HRE sequence present in the FBLN5 promoter.
FIGURE 6.
Hypoxia induces the binding of HIF-1 to an HRE in the FBLN5 promoter. HeLa cells were subjected to hypoxic or normoxic conditions or co-transfected with a HIF-1α expression vector (pHIF-1α) or the empty vector (pcDNA3) and nuclear extracts were obtained. A, nuclear levels of HIF-1α were determined by Western blot. Nucleolin levels were used as a loading control. B, the DNA binding activity of HIF-1α was evaluated by EMSA using an oligonucleotide probe corresponding to the putative HRE binding site in the FBLN5 promoter (FBLN5-HRE). The binding to this motif was prevented when the HRE site was mutated (FBLN5mut-HRE). Upon addition of a specific antibody against HIF-1α (Ab-HIF-1α), the intensity of the shifted band was strongly reduced. A nonspecific IgG was used as a control. The arrowheads indicate the position of the specific retarded band and the excess of free probe. A representative autoradiogram of two independent experiments is shown. NE, nuclear extracts.
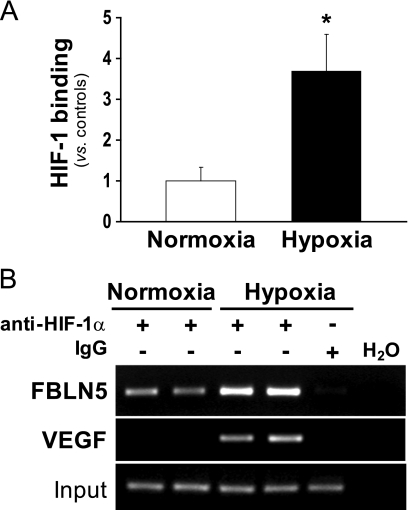
HIF-1α Binds to FBLN5 Promoter
To confirm that hypoxia promotes HIF-1 recruitment to the FBLN5 promoter, ChIP assays were carried out. HUVEC were subjected to normoxic or hypoxic conditions, and chromatin immunoprecipitations were performed with a specific HIF-1α antibody. Consistent with data from EMSA studies, results of ChIP analysis using primers flanking the FBLN5 HRE site (−245/+59) revealed that hypoxia induced the recruitment of HIF-1α to the FBLN5 proximal promoter (Fig. 7A). The specificity of chromatin immunoprecipitation was confirmed by the absence of signal in the IgG control. Furthermore, preimmunoprecipitation samples evidenced equivalent DNA input (Fig. 7B). As a positive control, an increase in HIF-1α recruitment to the VEGF promoter, a well known HIF-1α target gene, was also observed in cells under hypoxic conditions (Fig. 7B). Overall, these results provide convincing evidence that the −78 sequence within the FBLN5 proximal promoter functions as a HRE and is capable of conferring hypoxia-mediated regulation of FBLN5 expression.
FIGURE 7.
HIF-1α binds to the FBLN5 promoter in hypoxic cells. The relative in vivo association of HIF-1α with the human FBLN5 promoter was analyzed by ChIP in HUVEC incubated under normoxic or hypoxic conditions. Chromatin was sheared by sonication, and immunoprecipitations were performed with an anti-HIF-1α antibody or a nonspecific goat IgG. A, the enrichment of HIF-1α was quantified by real-time PCR using FBLN5 promoter-specific primers. PCR reactions using total input DNA saved before immunoprecipitation and H2O were also carried out. Data were normalized to the total input DNA and represented as mean ± S.E. of two independent experiments performed in duplicate (*, p < 0.05 versus control cells). B, agarose gel electrophoresis of PCR products amplified using specific primers for both FBLN5 and VEGF promoters. Equal input DNA and control IgG immunoprecipitations are shown.
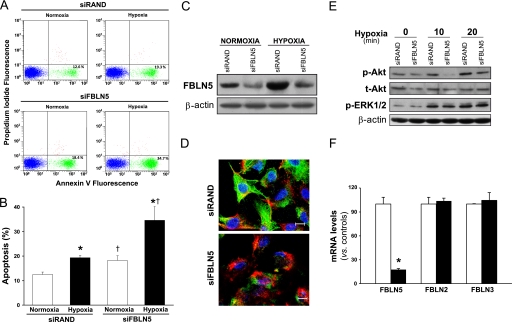
FBLN5 Knockdown Potentiates Hypoxia-induced Apoptosis in Endothelial Cells
We aimed to establish whether hypoxia-induced FBLN5 up-regulation could be related to the survival response triggered by hypoxia in endothelial cells. HUVEC transfected with a siRNA against FBLN5 (siFBLN5) or with a control siRNA (siRAND) were maintained under normoxic or hypoxic conditions and cells were stained with annexin V-FITC and PI to identify apoptotic cells. As we and others (4, 38) had described previously, HUVEC subjected to hypoxia show an increased apoptotic rate (evaluated by annexin V+ and PI staining) compared with normoxic cells. FACS analysis showed that the rate of endothelial cells undergoing apoptosis in cultures exposed to hypoxia increased in FBLN5 knockdown cells (from 19.33 ± 0.86% to 34.63 ± 5.68%) (Fig. 8, A and B). A slight increase in apoptosis in cells maintained under normoxia was also observed. siFBLN5 efficiently inhibited FBLN5 expression as confirmed by Western blot (Fig. 8C), immunofluorescence (Fig. 8D), and real-time PCR (Fig. 8F) without affecting FBLN2 or FBLN3 expression (Fig. 8F). The reduced survival response observed after FBLN5 knockdown was associated to a decrease in basal levels of both total and phosphorylated Akt and to a lesser activation of Akt under hypoxia. In these conditions, ERK1/2 activation remained unchanged (Fig. 8E). Altogether, our results indicate that hypoxia-induced FBLN5 expression seems to contribute to preserve endothelial cell survival.
FIGURE 8.
FBLN5 knockdown potentiates apoptosis under hypoxia. HUVEC were transfected with a FBLN5-specific siRNA (siFBLN5) or a control siRNA (siRAND) and exposed to normoxia or hypoxia during 24 h. Apoptosis was assessed by annexin V/propidium iodide staining and FACS analysis. A, representative FACS analyses corresponding to one experiment are shown. B, graphical representation showing the percentage of apoptotic cells under each experimental condition. Data are reported as mean ± S.E. (n = 9; *, p < 0.05 versus cells transfected with the same siRNA and maintained in normoxia; †, versus cells transfected with siRAND and maintained under the same experimental condition). C, Western blot analysis demonstrate the efficiency of FBLN5 knockdown in both cells maintained under normoxia or exposed to hypoxia. D, representative confocal images showing FBLN5 staining (in green) from cells transfected with either siRAND or siFBLN5 and exposed to hypoxia. Nuclei were stained with Hoechst (blue), and F-actin is visualized in red. Bar, 10 μm. E, HUVEC were transfected with siRAND or siFBLN5 for 48 h and were exposed to hypoxia for the indicated times. The effect of FBLN5 silencing on total Akt (t-Akt) and on the activation of Akt (p-Akt) and ERK1/2 (p-ERK1/2) was evaluated by Western blot. F, mRNA levels corresponding to FBLN2, FBLN3, and FBLN5 in HUVEC transfected with siRAND (white bars) or siFBLN5 (black bars) were analyzed by real-time PCR (n = 9; *, p < 0.05 versus cells transfected with siRAND.).
DISCUSSION
FBLN5 is an ECM protein crucial for elastic fiber assembly. It serves as an organizer of elastogenesis that scaffolds elastic fiber components in the proximity of cell surface (11, 13, 39). This protein plays a key role in physiological processes such as tissue development, remodeling, and repair (10, 16, 40–42). Conversely, aberrant FBLN5 expression has been associated with several types of tumors (14) and pelvic organ prolapse (43). In addition, mutations in the FBLN5 gene are related with macular degeneration and severe forms of cutis laxa (44, 45). Interestingly, FBLN5 acts as a multifunctional protein involved in cell-cell and matrix-cell signaling pathways key for vascular cell proliferation, adhesion, and migration (11, 15, 17, 46–48), as well as for vascular redox state (49, 50) and synthesis of ECM proteases (51, 52). Our current work demonstrates that hypoxic stress induces FBLN5 expression in vascular endothelial cells (human, bovine, or mouse endothelial cells), an effect that is extensive to other cell types. Furthermore, our results point to HIF-1 as responsible for FBLN5 regulation by hypoxia and identify FBLN5 as a novel HIF-1 target gene. Interestingly, FBLN5 up-regulation seems to be part of the adaptive response of endothelial cells to survive to hypoxia.
The transcriptional response to hypoxia is primarily mediated by HIF transcription factors. Accordingly, our data show that inhibition of HIF prolyl hydroxylases (by dimethyl oxal glycine) and the consequent HIF-1α stabilization (33), recapitulates the increase in FBLN5 expression elicited by hypoxia in endothelial cells. The role of HIF-1 on FBLN5 regulation is further supported by data from HIF-1α knockdown experiments and by the induction of FBLN5 transcriptional activity elicited by HIF-1α overexpression, which mimics that achieved by hypoxia. In addition, the inhibition of the PI3K/Akt/mTOR pathway, which is involved in the regulation of HIF-1 (6), prevented the induction of FBLN5 by hypoxia. Although HIF-2α also mediates the adaptive response to hypoxia, its marginal induction in HUVEC exposed to hypoxia (see Fig. 3B) (34) and the fact that HIF-2α silencing did not affect the hypoxia-mediated increase of FBLN5 expression support a major role of HIF-1α on FBLN5 regulation.
Our serial deletion studies delimited hypoxia responsiveness to a proximal promoter region of 329 bp. In silico analysis of this sequence evidenced the presence of two Smad response elements reported previously (35) and, more interestingly, a putative HRE located at −78. Although Smad proteins have been involved in the cellular response to hypoxia (53), mutation of the HRE site totally abrogated hypoxic FBLN5 induction. Accordingly, EMSA and ChIP data further support that this HRE motif located in the proximal FBLN5 promoter confers hypoxia sensitivity. As few studies have been focused on the characterization of the FBLN5 promoter, the HRE motif identified in our study is one of the few FBLN5 regulatory elements described so far.
FBLN5 is able to regulate processes in a tissue- and cell type-specific manner (47). Indeed, in vascular cells, FBLN5 differentially regulates cell proliferation and motility and ECM protease activity compared with other cells such as fibroblasts or fibrosarcoma cells (51, 52). The mechanism underlying these disparate effects is unknown, but it could be related with differences in the ECM environment or in the binding of FBLN5 to integrins. It should be noted that, although FBLN5 supports VSMC adhesion through integrins (15), it fails to activate integrin signaling (46). In fact, it has been recently proposed that FBLN5 could act as an “integrin antagonist,” a concept that opens up new expectations for FBLN5 as a critical regulator of cell homeostasis.
As described above, FBLN5 favors endothelial cell attachment to the ECM, contributing to preserve structural and functional characteristics of the endothelial cell monolayer (48). Furthermore, FBLN5 cooperates with FBLN2 to maintain the integrity of the adult vessel wall after injury and to avoid abnormal remodeling (54). Although hypoxia is a well established proangiogenic stimulus, FBLN5 exhibits antiangiogenic activities both in vitro and in vivo (18–20, 52). However, hypoxia should not be considered as a mere regulator of angiogenesis. Preliminary data suggest that FBLN5 could contribute to the hypoxia-induced increase in endothelial cell adhesion (data not shown) in agreement with previous reports (17). More interestingly, our data indicate that the up-regulation of FBLN5 expression in ischemic conditions is associated with the survival response of endothelial cells to hypoxia. Accordingly, fibroblasts harboring a defective FBLN5 form reported in recessive cutis laxa disease showed increased apoptosis compared with cells that express the wild-type FBLN5 form (55). The observation that prolonged blockade of FBLN5 results in a reduction of both total and hypoxia-induced phosphorylated Akt is consistent with a role of FBLN5 in cell survival, as activation of Akt is critical for cell survival to hypoxia (4, 56). The mechanisms underlying the prosurvival properties of FBLN5 remain complex and not well understood. Recently, it has been described that FBLN5 regulates reactive oxygen species production in endothelial cells (50). Furthermore, FBLN5 is able to activate integrin signaling, involved in cell survival (57, 58), although, as we mentioned above, in some scenarios, it has been postulated as an integrin antagonist. The reported antiapoptotic effect of FBLN5 could involve specific cell-type mechanisms, and the alteration of integrin expression and ECM composition elicited by hypoxia adds further complexity to ECM-cell signaling (59–61). As an integrin-binding ECM component, it is commonly accepted that the biological activity of FBLN5, is derived from its extracellular location. Zhou et al. (62) have recently described that FBLN5 binds and colocalizes with Nogo-B in both the cytosol and the cell surface of HeLa, thereby facilitating FBLN5 secretion; however, they neither analyzed whether FBLN5 could affect Nogo-B function nor attributed an “intracellular function” for FBLN5. Therefore, there is no evidence supporting a functional role for intracellular FBLN5. However, taking into account that the molecular mechanisms underlying the biology of FBLN5 remain to be fully elucidated and that hypoxia increases both intracellular and extracellular FBLN5 levels, we cannot rule out a role for intracellular FBLN5 in cell survival. Therefore, further studies will be necessary to clarify the mechanisms by which FBLN5 regulates endothelial cell survival. In summary, our study identifies FBLN5 as a new HIF-1 target gene involved in the orchestrated adaptive response of endothelial cells to hypoxic stress.
Acknowledgments
We are grateful to Dr. L. E. Huang for providing the HIF-1α expression vector. We also thank Silvia Aguiló and Esther Peña for technical assistance.
This work was supported in part by Grants PS09/01797 and SAF2009-11949 and Red Temática de Investigación Cardiovascular (RD06/0014/0027) from the Ministerio de Ciencia e Innovación-Instituto de Salud Carlos III (Fondo de Investigaciones Sanitarias).
- ECM
- extracellular matrix
- FBLN
- fibulin
- HIF
- hypoxia-inducible factor
- BAEC
- bovine aortic endothelial cells
- HUVEC
- human umbilical vein endothelial cells
- mTOR
- mammalian target of rapamycin
- VEGF
- vascular endothelial growth factor
- HRE
- hypoxia response element
- PI
- propidium iodide.
REFERENCES
- 1. Semenza G. L. (2000) J. Clin. Invest. 106, 809–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pugh C. W., Ratcliffe P. J. (2003) Nat. Med. 9, 677–684 [DOI] [PubMed] [Google Scholar]
- 3. Economopoulou M., Langer H. F., Celeste A., Orlova V. V., Choi E. Y., Ma M., Vassilopoulos A., Callen E., Deng C., Bassing C. H., Boehm M., Nussenzweig A., Chavakis T. (2009) Nat. Med. 15, 553–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martorell L., Gentile M., Rius J., Rodríguez C., Crespo J., Badimon L., Martínez-González J. (2009) Mol. Cell. Biol. 29, 5828–5842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Safran M., Kaelin W. G., Jr. (2003) J. Clin. Invest. 111, 779–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Semenza G. L. (2009) Physiology 24, 97–106 [DOI] [PubMed] [Google Scholar]
- 7. del Peso L., Castellanos M. C., Temes E., Martin-Puig S., Cuevas Y., Olmos G., Landazuri M. O. (2003) J. Biol. Chem. 278, 48690–48695 [DOI] [PubMed] [Google Scholar]
- 8. Tang N., Wang L., Esko J., Giordano F. J., Huang Y., Gerber H. P., Ferrara N., Johnson R. S. (2004) Cancer Cell 6, 485–495 [DOI] [PubMed] [Google Scholar]
- 9. Choi J., Bergdahl A., Zheng Q., Starcher B., Yanagisawa H., Davis E. C. (2009) Matrix Biol. 28, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kowal R. C., Richardson J. A., Miano J. M., Olson E. N. (1999) Circ. Res. 84, 1166–1176 [DOI] [PubMed] [Google Scholar]
- 11. Nakamura T., Lozano P. R., Ikeda Y., Iwanaga Y., Hinek A., Minamisawa S., Cheng C. F., Kobuke K., Dalton N., Takada Y., Tashiro K., Ross J., Jr., Honjo T., Chien K. R. (2002) Nature 415, 171–175 [DOI] [PubMed] [Google Scholar]
- 12. Nonaka R., Onoue S., Wachi H., Sato F., Urban Z., Starcher B. C., Seyama Y. (2009) Clin. Biochem. 42, 713–721 [DOI] [PubMed] [Google Scholar]
- 13. Yanagisawa H., Davis E. C., Starcher B. C., Ouchi T., Yanagisawa M., Richardson J. A., Olson E. N. (2002) Nature 415, 168–171 [DOI] [PubMed] [Google Scholar]
- 14. Albig A. R., Schiemann W. P. (2005) Future Oncol. 1, 23–35 [DOI] [PubMed] [Google Scholar]
- 15. Nakamura T., Ruiz-Lozano P., Lindner V., Yabe D., Taniwaki M., Furukawa Y., Kobuke K., Tashiro K., Lu Z., Andon N. L., Schaub R., Matsumori A., Sasayama S., Chien K. R., Honjo T. (1999) J. Biol. Chem. 274, 22476–22483 [DOI] [PubMed] [Google Scholar]
- 16. Spencer J. A., Hacker S. L., Davis E. C., Mecham R. P., Knutsen R. H., Li D. Y., Gerard R. D., Richardson J. A., Olson E. N., Yanagisawa H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2946–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Preis M., Cohen T., Sarnatzki Y., Ben Yosef Y., Schneiderman J., Gluzman Z., Koren B., Lewis B. S., Shaul Y., Flugelman M. Y. (2006) Biochem. Biophys. Res. Commun. 348, 1024–1033 [DOI] [PubMed] [Google Scholar]
- 18. Xie L., Palmsten K., MacDonald B., Kieran M. W., Potenta S., Vong S., Kalluri R. (2008) Exp. Biol. Med. 233, 155–162 [DOI] [PubMed] [Google Scholar]
- 19. Albig A. R., Schiemann W. P. (2004) DNA Cell Biol. 23, 367–379 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan K. M., Bissonnette R., Yanagisawa H., Hussain S. N., Davis E. C. (2007) Lab. Invest. 87, 818–827 [DOI] [PubMed] [Google Scholar]
- 21. Rodríguez C., Alcudia J. F., Martínez-González J., Guadall A., Raposo B., Sánchez-Gómez S., Badimon L. (2009) Cardiovasc. Res. 83, 595–603 [DOI] [PubMed] [Google Scholar]
- 22. Rodríguez C., Alcudia J. F., Martínez-González J., Raposo B., Navarro M. A., Badimon L. (2008) Atherosclerosis 196, 558–564 [DOI] [PubMed] [Google Scholar]
- 23. Orbe J., Rodríguez J. A., Calvayrac O., Rodríguez-Calvo R., Rodríguez C., Roncal C., Martínez de Lizarrondo S., Barrenetxe J., Reverter J. C., Martínez-González J., Páramo J. A. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 2109–2116 [DOI] [PubMed] [Google Scholar]
- 24. Martorell L., Martínez-González J., Crespo J., Calvayrac O., Badimon L. (2007) J. Thromb. Haemost. 5, 1766–1773 [DOI] [PubMed] [Google Scholar]
- 25. Raposo B., Rodríguez C., Martínez-González J., Badimon L. (2004) Atherosclerosis 177, 1–8 [DOI] [PubMed] [Google Scholar]
- 26. Huang L. E., Gu J., Schau M., Bunn H. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7987–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rius J., Martínez-González J., Crespo J., Badimon L. (2006) Atherosclerosis 184, 276–282 [DOI] [PubMed] [Google Scholar]
- 28. González-Díez M., Rodríguez C., Badimon L., Martínez-González J. (2008) Thromb. Haemost. 100, 119–126 [DOI] [PubMed] [Google Scholar]
- 29. Rodriguez C., Raposo B., Martínez-González J., Llorente-Cortés V., Vilahur G., Badimon L. (2003) Cardiovasc. Res. 58, 178–185 [DOI] [PubMed] [Google Scholar]
- 30. Martínez-González J., Rodríguez-Rodríguez R., González-Díez M., Rodríguez C., Herrera M. D., Ruiz-Gutierrez V., Badimon L. (2008) J. Nutr. 138, 443–448 [DOI] [PubMed] [Google Scholar]
- 31. Martínez-González J., Escudero I., Badimon L. (2004) Atherosclerosis 174, 305–313 [DOI] [PubMed] [Google Scholar]
- 32. Abedin S. A., Thorne J. L., Battaglia S., Maguire O., Hornung L. B., Doherty A. P., Mills I. G., Campbell M. J. (2009) Carcinogenesis 30, 449–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av., Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 34. Hu C. J., Wang L. Y., Chodosh L. A., Keith B., Simon M. C. (2003) Mol. Cell. Biol. 23, 9361–9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuang P. P., Joyce-Brady M., Zhang X. H., Jean J. C., Goldstein R. H. (2006) Am. J. Physiol. Cell. Physiol. 291, C1412–1421 [DOI] [PubMed] [Google Scholar]
- 36. Luo Y., Jiang C., Belanger A. J., Akita G. Y., Wadsworth S. C., Gregory R. J., Vincent K. A. (2006) Mol. Pharmacol. 69, 1953–1962 [DOI] [PubMed] [Google Scholar]
- 37. Wolf D., Cammas F., Losson R., Goff S. P. (2008) J. Virol. 82, 4675–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Madge L. A., Pober J. S. (2000) J. Biol. Chem. 275, 15458–15465 [DOI] [PubMed] [Google Scholar]
- 39. Hirai M., Ohbayashi T., Horiguchi M., Okawa K., Hagiwara A., Chien K. R., Kita T., Nakamura T. (2007) J. Cell Biol. 176, 1061–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jean J. C., Eruchalu I., Cao Y. X., Joyce-Brady M. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 282, L75–82 [DOI] [PubMed] [Google Scholar]
- 41. Kuang P. P., Goldstein R. H., Liu Y., Rishikof D. C., Jean J. C., Joyce-Brady M. (2003) Am. J. Physiol. Lung Cell. Mol. Physiol. 285, L1147–1152 [DOI] [PubMed] [Google Scholar]
- 42. Lee M. J., Roy N. K., Mogford J. E., Schiemann W. P., Mustoe T. A. (2004) J. Am. Coll. Surg. 199, 403–410 [DOI] [PubMed] [Google Scholar]
- 43. Söderberg M. W., Byström B., Kalamajski S., Malmström A., Ekman-Ordeberg G. (2009) Mol. Hum. Reprod. 15, 251–257 [DOI] [PubMed] [Google Scholar]
- 44. Loeys B., Van Maldergem L., Mortier G., Coucke P., Gerniers S., Naeyaert J. M., De Paepe A. (2002) Hum. Mol. Genet. 11, 2113–2118 [DOI] [PubMed] [Google Scholar]
- 45. Stone E. M., Braun T. A., Russell S. R., Kuehn M. H., Lotery A. J., Moore P. A., Eastman C. G., Casavant T. L., Sheffield V. C. (2004) N. Engl. J. Med. 351, 346–353 [DOI] [PubMed] [Google Scholar]
- 46. Lomas A. C., Mellody K. T., Freeman L. J., Bax D. V., Shuttleworth C. A., Kielty C. M. (2007) Biochem. J. 405, 417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schiemann W. P., Blobe G. C., Kalume D. E., Pandey A., Lodish H. F. (2002) J. Biol. Chem. 277, 27367–27377 [DOI] [PubMed] [Google Scholar]
- 48. Williamson M. R., Shuttleworth A., Canfield A. E., Black R. A., Kielty C. M. (2007) Biomaterials 28, 5307–5318 [DOI] [PubMed] [Google Scholar]
- 49. Nguyen A. D., Itoh S., Jeney V., Yanagisawa H., Fujimoto M., Ushio-Fukai M., Fukai T. (2004) Circ. Res. 95, 1067–1074 [DOI] [PubMed] [Google Scholar]
- 50. Schluterman M. K., Chapman S. L., Korpanty G., Ozumi K., Fukai T., Yanagisawa H., Brekken R. A. (2010) Dis. Model Mech. 3, 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee Y. H., Albig A. R., Regner M., Schiemann B. J., Schiemann W. P. (2008) Carcinogenesis 29, 2243–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Albig A. R., Neil J. R., Schiemann W. P. (2006) Cancer Res. 66, 2621–2629 [DOI] [PubMed] [Google Scholar]
- 53. Zhang H., Akman H. O., Smith E. L., Zhao J., Murphy-Ullrich J. E., Batuman O. A. (2003) Blood 101, 2253–2260 [DOI] [PubMed] [Google Scholar]
- 54. Chapman S. L., Sicot F. X., Davis E. C., Huang J., Sasaki T., Chu M. L., Yanagisawa H. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hu Q., Loeys B. L., Coucke P. J., De Paepe A., Mecham R. P., Choi J., Davis E. C., Urban Z. (2006) Hum. Mol. Genet. 15, 3379–3386 [DOI] [PubMed] [Google Scholar]
- 56. Alvarez-Tejado M., Naranjo-Suarez S., Jiménez C., Carrera A. C., Landázuri M. O., del Peso L. (2001) J. Biol. Chem. 276, 22368–22374 [DOI] [PubMed] [Google Scholar]
- 57. Yue W., Sun Q., Landreneau R., Wu C., Siegfried J. M., Yu J., Zhang L. (2009) Cancer Res. 69, 6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Avraamides C. J., Garmy-Susini B., Varner J. A. (2008) Nat. Rev. Cancer 8, 604–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rohwer N., Welzel M., Daskalow K., Pfander D., Wiedenmann B., Detjen K., Cramer T. (2008) Cancer Res. 68, 10113–10120 [DOI] [PubMed] [Google Scholar]
- 60. Ryu M. H., Park H. M., Chung J., Lee C. H., Park H. R. (2010) Biochem. Biophys. Res. Commun. 393, 11–15 [DOI] [PubMed] [Google Scholar]
- 61. Walton H. L., Corjay M. H., Mohamed S. N., Mousa S. A., Santomenna L. D., Reilly T. M. (2000) J. Cell. Biochem. 78, 674–680 [DOI] [PubMed] [Google Scholar]
- 62. Zhou S., Xiao W., Wan Q., Yi C., Xiao F., Liu Y., Qi Y. (2010) Biochem. Biophys. Res. Commun. 398, 247–253 [DOI] [PubMed] [Google Scholar]