Abstract
Post-translational histone modifications play important roles in regulating gene expression programs, which in turn determine cell fate and lineage commitment during development. One such modification is histone ubiquitination, which primarily targets histone H2A and H2B. Although ubiquitination of H2A and H2B has been generally linked to gene silencing and gene activation, respectively, the functions of histone ubiquitination during eukaryote development are not well understood. Here, we identified USP12 and USP46 as histone H2A and H2B deubiquitinases that regulate Xenopus development. USP12 and USP46 prefer nucleosomal substrates and deubiquitinate both histone H2A and H2B in vitro and in vivo. WDR48, a WD40 repeat-containing protein, interacts with USP12 and USP46 and is required for the histone deubiquitination activity. Overexpression of either gene leads to gastrulation defects without affecting mesodermal cell fate, whereas knockdown of USP12 in Xenopus embryos results in reduction of a subset of mesodermal genes at gastrula stages. Immunohistochemical staining and chromatin immunoprecipitation assays revealed that USP12 regulates histone deubiquitination in the mesoderm and at specific gene promoters during Xenopus development. Taken together, this study identifies USP12 and USP46 as histone deubiquitinases for H2A and H2B and reveals that USP12 regulates Xenopus development during gastrula stages.
Keywords: Chromatin Histone Modification, Chromatin Regulation, Development, Gene Regulation, Histone Modification, Xenopus
Introduction
Eukaryotic development requires precise control of gene expression patterns that are essential for cellular identity and differentiation (1, 2). Genomic DNA in eukaryotic cells is organized into a chromatin structure by association with histone and non-histone proteins (3, 4), and the structure of chromatin is believed to play a critical role in regulating chromatin-templated nuclear processes such as transcription (5, 6). Post-translational modifications of histones represent a major mechanism by which cells control the structure and function of chromatin. An increasing list of histone-modifying enzymes and histone modifications has been shown to be critical for normal development and to play causal roles in the pathogenesis of certain human diseases (7–9).
Of the vast variety of histone modifications, histone ubiquitination is unique, in which a 76-amino acid bulky protein is attached primarily to histone H2A and H2B (10, 11). The recent characterization of ubiquitin ligase hPRC1L and deubiquitinase Ubp-M (USP16) for histone H2A revealed critical functions for this modification in gene silencing, X inactivation, cell cycle progression, and DNA damage repair (12–15). In addition to Ubp-M, 2A-DUB (MYSM1) and USP21 were also identified as H2A-specific deubiquitinases (16, 17). These enzymes might function in different cellular processes, for example, 2A-DUB in androgen receptor-mediated gene activation and USP21 in liver regeneration (16, 17). Recently, the Drosophila PcG gene calypso was found to encode a ubiquitin C-terminal hydrolase BAP1, which specifically deubiquitinates histone H2A and regulates Hox gene repression (18). It will be interesting to determine the relationship between BAP1 and USP16 in H2A deubiquitination and Hox gene expression in different organisms. H2B ubiquitination is conserved, and enzymes catalyzing this modification were first identified in Saccharomyces cerevisiae as Rad6 and Bre1 (19–21). Their mammalian counterparts Rad6A/B and RNF20/40 were also shown to mediate H2B ubiquitination (22–24). H2B ubiquitination can be reversed in S. cerevisiae by Ubp-8 and Ubp-10 (25–28). Ubp-8 orthologs were identified in Drosophila as Nonstop and in human as USP22 (29–31). In addition, USP7 has also been implicated in H2B deubiquitination (32), and USP3 has been shown to deubiquitinate both H2A and H2B (33). Reducing USP3 levels results in replication defects that cause activation of the ataxia telangiectasia mutated/ataxia telangiectasia and Rad3-related DNA damage response and delayed progression through cell cycle S phase (33). Recent studies revealed that H2B ubiquitination has pleiotropic effects, with positive and negative influences on transcription, depending on particular genes, the given chromatin contexts, and specific regions of the genes (34, 35). Intriguingly, active transcription proves to be essential for H2B ubiquitination (35). Despite these advances, how histone ubiquitination regulates higher eukaryotic development is less understood.
By following the histone H2A deubiquitination activity coupled with conventional chromatography, we previously reported the purification and functional characterization of a histone H2A-specific deubiquitinase Ubp-M (14). During the purification, we noticed that there is a weak H2A deubiquitination activity that is independent of Ubp-M. We report here the purification of this activity as USP12 and USP46. Our studies further reveal that USP12 and USP46 interact with a WD40 repeat-containing protein WDR48, which is required for the histone deubiquitination activity (36). USP12 and USP46 prefer nucleosomal substrates in vitro and significantly deubiquitinate both histone H2A and H2B in vitro and in vivo. Our studies further reveal that USP12 regulates cell fate determination and the gastrulation process during Xenopus embryonic development.
EXPERIMENTAL PROCEDURES
Substrate Preparation and in Vitro Histone Deubiquitination Assay
Preparation of ubH2A-containing mononucleosomes and in vitro histone deubiquitination assays was performed as described previously (14). To purify mononucleosomes containing human ubiquitinated H2B, the yeast FLAG-tagged H2B in plasmid pZS144 (HTA1-FLAG-HTB1 (CEN, TRP1)) (37) was replaced with a fragment containing the human FLAG-H2B. The resulting plasmid pZS200 (HTA1-FLAG-hH2B (CEN, TRP1)) was introduced into a UBP8 and UBP10 double deletion strain YZS608 (HTA1-HTB1 (CEN, URA3), ubp8Δ::KanMX4, ubp10Δ::NatMX; derived from Y131) (38) to obtain the strain YZS609 (HTA1-HTB1 (CEN, URA3), HTA1-FLAG-hH2B, (CEN, TRP1), ubp8Δ::KanMX4, ubp10Δ::NatMX), in which both human FLAG-H2B and yeast H2B are expressed. Human ubH2B-containing mononucleosomes were purified by anti-FLAG immunoprecipitation as described previously (39). Core histones containing ubH2A and ubH2B were prepared from mononucleosomes with small scale hydroxyapatite columns as described previously (14).
Purification and Identification of the Ubp-M-independent H2A Deubiquitination Activity
HeLa nuclear proteins were separated into nuclear extracts and nuclear pellets as described previously (40). Nuclear pellet (8.86 g) was loaded onto a customer-packaged 1,450-ml DE52 column equilibrated with buffer D (20 mm Tris-HCl, pH 7.9, 0.1 mm EDTA, 10% glycerol, 1 mm DTT, 0.1 mm PMSF, 0.025% Nonidet P-40) containing 20 mm (NH4)2SO4 (BD20). Proteins bound to the column were step-eluted with BD350 and BD500. The BD350 fraction was dialyzed against buffer D containing 100 mm KCl (BC100) before loading onto a customer-packaged 500-ml P11 column. Bound proteins were step-eluted with BC300, BC500, and BC1000. The BC500 fraction was dialyzed against BD20 and loaded onto a 45-ml DEAE5PW column (TOSOH Bioscience). Bound proteins were eluted with an 8-column volume linear gradient from BD20 to BD500. Fractions containing H2A deubiquitination activity were pooled and adjusted to BD1000 with saturated (NH4)2SO4 before loading onto a 22-ml FPLC phenyl-Sepharose column (GE Healthcare). Bound proteins were eluted with a 10-column volume linear gradient from BD1000 to BD0. The Ubp-M independent H2A deubiquitination activity was eluted between BD500 and BD420. These fractions were then dialyzed against buffer P (5 mm HEPES-KOH, pH 7.5, 0.04 m KCl, 0.01 mm CaCl2, 10% glycerol, 1 mm DTT, 0.1 mm PMSF) containing 10 mm potassium phosphate (BP10) and loaded onto a 5-ml hydroxyapatite column (Bio-Rad). Bound proteins were eluted with a 20-column volume linear gradient from BP10 to BP600. Fractions containing the H2A deubiquitination activity were dialyzed against BC50 before loading onto a 1-ml Mono Q column (GE Healthcare). Proteins bound to the column were eluted with a 20-column volume linear gradient from BC50 to BC500 (pH 6, 20 mm MES-KOH instead of Tris-HCl). Active fractions were pooled and further separated in a 1-ml Mono S column (GE Healthcare) in a 20-column volume linear gradient from BC50 to BC500. Active fractions from the Mono S column were pooled and loaded onto a Superose 6 column (GE Healthcare) equilibrated with BC500. To identify the protein that co-elutes with the H2A deubiquitination activity, fraction 64 of the Superose 6 column was concentrated and resolved on an SDS-polyacrylamide gel. The candidate bands that correlated to the activity were excised. To determine other deubiquitinases in fraction 64, the entire gel lane was divided into 22 pieces and also subjected to mass spectrometry analysis with a high resolution linear quadrupole ion trap Fourier transform ion cyclotron resonance mass spectrometer (LTQ3 FT-ICR MS, ICR, Thermo Fisher Scientific).
Briefly, individual gel slices were destained with a 5:8 ratio of potassium ferricyanide/sodium thiosulfate, reduced with 10 mm DTT at 37 °C for 45 min, alkylated with 50 mm iodoacetamide at 37 °C for 45 min, and digested with trypsin overnight at 37 °C. Peptides were extracted from the gel using 50% acetonitrile and concentrated in a speed vacuum. Tryptic digests were loaded onto a 100-μm diameter, 11-cm pulled tip packed column with Jupiter 5-μm C18 reversed-phase beads (Phenomenex) using a Micro AS autosampler and LC nanopump (Eksigent). An acetonitrile gradient in 0.1% formic acid was run from 5 to 40% over 50 min at a flow rate of 650 nl/min. The eluting peptides were analyzed by collision-induced dissociation fragmentation on a LTQ FT-ICR. The LTQ FT-ICR parameters were set as described previously (41). Fully tryptic human peptides were identified using TurboSequest version 27 (revision 12, Thermo Fisher Scientific) (42), MASCOT 2.2 (Matrix Biosciences) (43), and Protein Prospector version 5.2.2 (University of California, San Francisco) (44) algorithms with a parent ion mass accuracy of 10.0 ppm from the UniRef100 data base (06/2009). SEQUEST and MASCOT results were further refined through the trans-proteomic pipeline using the Peptide Prophet (45) and Protein Prophet (46) models.
Purification and Identification of the USP12 and USP46 Complex
To identify proteins that interact with USP12 and USP46, stable cell lines expressing FLAG-HA-USP12 and FLAG-HA-USP46 were established as described previously (47). Briefly, USP12 and USP46 cDNAs with an N-terminal FLAG and HA tag (FH-USP12 and FH-USP46) were cloned into the retroviral pMIGR1 vector. pMIGR1-FH-USP12 (or pMIGR1-FH-USP46) vectors and ϕampho plasmids were used to co-transfect 293T cells to produce recombinant retrovirus. The retrovirus in the supernatant was filtered with a 0.45-μm pore size filter and was used to infect HeLa S3 cells by spinoculation. Because GFP and FLAG-HA-USP12 (or FLAG-HA-USP46) were produced in the same bicistronic transcript, cell sorting for GFP expression was done twice to enrich GFP-positive cells for over 90% of cells that express GFP and USP12 (or USP46).
To purify the USP12 and USP46 complexes, the nuclear pellet (1.5 g) from HeLa S3 stable cell lines expressing FLAG-HA-USP12 or FLAG-HA-USP46 were loaded onto a 220-ml DE52 column and step-eluted with BD350 and BD500. The BD350 fraction was then loaded onto a 120-ml P11 column and step-eluted with BC100, BC300, BC500, and BC1000. The BC500 fraction, which contains USP12 and USP46, was loaded on a 45-ml DEAE5PW column (TOSOH Bioscience), and bound proteins were eluted with an 8-column volume linear gradient from BD20 to BD500. Fractions that contain USP12 and USP46 were pooled and were used as input for anti-FLAG immunoprecipitation. The eluate from anti-FLAG immunoprecipitation was further purified on a Superose 6 column (GE Healthcare). Protein identification was performed as described above.
USP12 and USP46 Knockdown, Reverse Transcription-Polymerase Chain Reaction (RT-PCR), and Chromatin Immunoprecipitation (ChIP) Assays
siRNA oligonucleotides against USP12 and USP46 were purchased from Invitrogen in a purified, annealed duplex form and transfected into cells with Lipofectamine 2000. Semi-quantitative RT-PCR and ChIP were performed as described previously (14, 48). Primers used are as follows: USP12, 5′-AGACCCAACGTGGGTTGATGAGAT-3′ and 5′-GAACCACAACAGCAACAAGGTCGT-3′; USP46, 5′-AAACCAGAACTCACCTGGGTCCAT-3′ and 5′-ATCTGTGCAGCTGCTCCATGTACTTGAACC-3′; HOXA9, 5′-ACGTGGACTCGTTCCTGCTG-3′ and 5′-AGGTTTAATGCCATAAGGCCG-3′; HOXB1, 5′-TCAGGCGGTTGACAGCTATG-3′ and 5′-ATGCTGCGGAGGATATGGC-3′; HOXC5, 5′-TGTGGGAACTATGGATCGGC-3′ and 5′-ACGGGTAAATCTGTGGCGG-3′; and HOXD10, 5′-GATTCCTTGATCAGTGCCTGC-3′ and 5′-GCCGAAATGAGTTTGTTGCG-3′.
Xenopus Manipulation
Embryos were obtained, maintained, and microinjected with mRNAs or MOs as described previously (49). RNAs were synthesized using mMessage mMachine in vitro transcription kit (Ambion). Standard control MO (Gene Tools Inc.), USP12/46-specific MOs (total 40 ng/embryo), and mRNAs (0.5–2 ng/embryo) were injected into the animal pole or dorsal marginal zone region of 2- or 4-cell stage embryos. The injected embryos were observed at tadpole stages for their morphology or processed at gastrula stages for marker expression analysis by RT-PCR or in situ hybridization (14). Immunofluorescent staining of embryonic sections was performed as follows. Briefly, after dewaxing and hydration, antigen was retrieved by boiling sections in 10 mm sodium citrate (pH 6). Blocking was done using 10% normal goat serum. Anti-ubH2A (1:50) antibody, biotinylated anti-mouse antibody (1:1,000) and streptavidin Cy3 (1:1,000) was used.
Constructs, Antibodies, and Recombinant Baculovirus
cDNAs for USP12 and USP46 were cloned from a HeLa cDNA library and verified by DNA sequencing. To generate antibodies against USP12 and USP46, full-length USP12 and USP46 were cloned into pGEX-KG, and purified recombinant proteins were used to immunize rabbits. For recombinant baculovirus production, USP12 and USP46 cDNAs were cloned into the pFastBacHTb vector (Invitrogen) with a FLAG tag at the N terminus. Recombinant baculoviruses expressing FLAG-USP12 and FLAG-USP46 were generated and amplified following the manufacturer's protocol. Anti-ubH2A (E6C5; 1:500) was purchased from Millipore (catalog no. 05-678) and anti-ubH2B (1:500) was provided by Dr. Moshe Oren. ubH2A and ubH2B signals were detected using the LI-COR Odyssey Infrared Imaging System (Lincoln, NE) and IR dye-conjugated secondary antibodies (1:5,000) as described by the manufacturer. FLAG-USP22 plasmid was kindly provided by Dr. Steven McMahon, and FLAG-USP22 was purified as described previously (29, 50).
RESULTS
Purification of the Ubp-M-independent H2A Deubiquitination Activity
In an effort to determine the functional significance of histone deubiquitination in higher eukaryotes, we set out to identify the enzyme(s) that are responsible for this process. By following the histone deubiquitination activity coupled with conventional chromatography, we previously reported the purification and functional characterization of a histone H2A-specific deubiquitinase Ubp-M (USP16) (14). During these studies, however, we noticed that HeLa cells contain a histone H2A deubiquitination activity that is independent of Ubp-M (Fig. 1A, compare lanes 4 and 5 with 9). To identify the enzyme(s) that are responsible for this activity, we purified this activity through an eight-column purification scheme (Fig. 1B). As shown in Fig. 1C, in the Superose 6 column, the H2A deubiquitination activity was eluted between fractions 61 and 67, with an apparent molecular mass between 220 and 443 kDa (top three panels). Of the three fractions, fraction 64 has the strongest activity, and fraction 61 has a stronger activity than fraction 67. Silver staining of an SDS-PAGE containing the same fractions revealed that two minor bands, which migrated between 37 and 50 kDa, co-eluted with the histone deubiquitination activity (Fig. 1C, top panel; marked with dots in an enlarged image in Fig. 1D). To determine the identity of these polypeptides, gel slices containing these bands were excised, trypsin-digested, and subjected to LTQ FT-ICR mass spectrometry identification. Mass spectrometry analysis revealed that these two polypeptides are ubiquitin-specific protease 12 (USP12, accession number 32698815) and ubiquitin-specific protease 46 (USP46, accession number 31377709), both containing the catalytic cysteine triad characteristic of ubiquitin-specific proteases. Although there are no other visible bands exhibiting similar eluting profile (intensity: fractions 64 > 61 > 67), we still determined whether there are other deubiquitinases that could also contribute to the observed deubiquitination activity. For this purpose, we identified by mass spectrometry all proteins in fraction 64, including gel slices without observable silver-stained bands. No other putative deubiquitinases were identified in fraction 64 (data not shown).
FIGURE 1.
Purification and identification of the Ubp-M-independent histone H2A deubiquitinase. A, HeLa cells contain a Ubp-M-independent H2A deubiquitination activity. Top panel, Western blot analysis of Ubp-M in HeLa nuclear proteins fractionated on DE52 and P11 columns. Middle and bottom panels, histone deubiquitination assay using HeLa nuclear proteins fractionated on DE52 and P11 columns with ubH2A-containing mononucleosomes as substrates. Numbers on the top of the panels indicate the salt concentration (M) for step elution. NE and NP represent nuclear extracts and nuclear pellet, respectively. Nonspecific signals from protein fractions (middle panel) are indicated by asterisks. Antibodies used are indicated on the left side of the panels. B, schematic representation of the steps used to purify the Ubp-M-independent H2A deubiquitination activity. Numbers represent the salt concentrations (mm) at which the H2A deubiquitination activity elutes from the columns. C, identification of USP12 and USP46 as the deubiquitinase(s) responsible for the Ubp-M-independent H2A deubiquitination activity. Silver staining of an SDS-PAGE (top panel), H2A deubiquitination assay (2nd and 3rd panels), and Western blot analysis (bottom two panels) of fractions derived from the Superose 6 column. The elution profile of the protein markers is indicated on the top of the panels. Antibodies used are indicated on the left side of the panels. D, an enlarged image showing bands from silver-stained gel correlated with H2A deubiquitination activity. The candidate proteins that co-fractionated with the deubiquitinase activity are indicated by dots. E, USP46 and/or USP12 are responsible for the Ubp-M-independent H2A deubiquitination activity. Western blot analysis (top panel) and H2A deubiquitination assay (bottom two panels) of samples derived from input (In), flow-through (Ft), and bound (B) using purified rabbit IgG and USP46 antibodies. Antibodies used for Western blot assay are indicated on the left side of the panels.
To further confirm that USP12 and/or USP46 are responsible for the Ubp-M-independent H2A deubiquitination activity, antibodies were generated against full-length USP12 and USP46. Because of the similarity of USP12 and USP46 (88% amino acid identity), cross-reactions were found between these two antibodies (supplemental Fig. S1). Western blot analysis with these antibodies of fractions derived from the Superose 6 column as well as the hydroxyapatite, Mono S, and Mono Q columns revealed that USP12 and USP46 are indeed co-purified with the H2A deubiquitination activity (Fig. 1C, bottom two panels; supplemental Fig. S2, and data not shown). To further confirm this result, we performed an immunodepletion assay. As shown in Fig. 1E, when USP46 was immunodepleted from an aliquot of the Mono Q fraction (top panel, compare lanes 2–4 with 5–7), the H2A deubiquitination activity was also depleted (bottom two panels, compare lanes 2–4 with 5–7). Because of antibody cross-reaction, USP12 was also immunodepleted (data not shown). Nonetheless, the immunodepletion assay confirmed that USP12 and/or USP46 are responsible for the Ubp-M-independent H2A deubiquitination activity.
WDR48 Interacts with USP12 and USP46 and Is Required for the Deubiquitination Activity
To further determine that USP12 and/or USP46 are indeed responsible for the H2A deubiquitination activity, we purified recombinant USP12 and USP46 from Sf9 cells (supplemental Fig. S3A). As shown in supplemental Fig. S3, B and C, both recombinant USP12 and USP46 eluted out between fractions 70 and 76 on the Superose 6 column with a molecular weight smaller than that of the native complex. These data suggest that USP12 and USP46 interact with other protein(s) in vivo. Histone H2A deubiquitination assay revealed that recombinant USP12, USP46, or USP12 plus USP46 could not deubiquitinate histone H2A (supplemental Fig. S3D, bottom two panels). These results indicate that protein(s) interacting with USP12 and/or USP46 are required for the H2A deubiquitination activity.
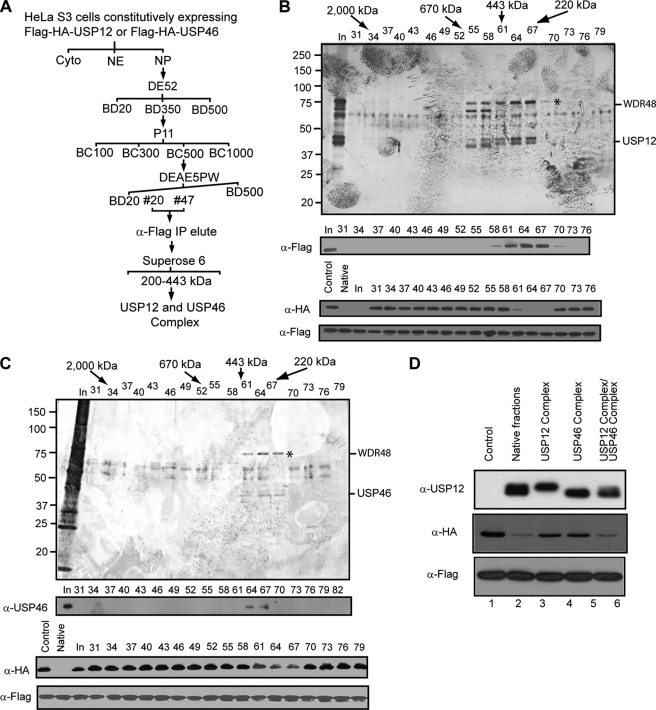
To identify the USP12- and USP46-interacting protein(s), we established HeLa cell lines that stably express FLAG-HA-USP12 or FLAG-HA-USP46 with the retrovirus-mediated protein expression system. The expression levels of epitope-tagged USP12 and USP46 within total cell lysate are beyond the detection limit of Western blot assay, and protein fractionation experiments revealed that both epitope-tagged USP12 and USP46 are exclusively localized in nuclear pellets, similar to the respective endogenous protein (data not shown). With these cell lines, employing a combination of conventional and affinity purification approaches (Fig. 2A), we purified the USP12 and USP46 complex to homogeneity (Fig. 2, B and C). Both USP12 and USP46 complexes were eluted between fractions 61 and 67 on the Superose 6 column (Fig. 2, B and C, top two panels), with the exact profile as the native complex (Fig. 1C). Histone deubiquitination assay revealed that both USP12 and USP46 complex can deubiquitinate histone H2A and that the H2A deubiquitination activity is correlated with the elution profile of USP12 and USP46 (Fig. 2, B and C, bottom two panels). Silver staining of an SDS-PAGE containing the same fractions revealed that USP12 and USP46 (marked with “*”, as confirmed by Western blot) co-purified with one other dominant polypeptide with a molecular mass of 80 kDa. Mass spectrometry analysis revealed the protein band as WDR48 (Fig. 2, B and C). Spectrometry analysis also identified lower abundance proteins DMWD, WDR20, WDR26, and WDR77. Relative abundance of the identified proteins based on the number of observed peptides in the mass spectrometry analysis are provided in supplemental Table S1. Based on these comparisons, we concluded that WDR48 was the primary interacting protein for the two proteases. Interestingly, Western blotting assay revealed that USP12 and USP46 likely exist in mutually exclusive complexes (Fig. 2D, top panel, compare lane 3 with 4). Because mass spectrometry analysis did not reveal other putative deubiquitinases in fraction 64, we estimated whether USP12 and USP46 could account for the observed H2A deubiquitination activity in native fractions. For this purpose, we compared the H2A deubiquitination activity of native complex, USP12 complex, USP46 complex, and a mixture of USP12 and USP46 complexes. As shown in Fig. 2D, a mixture of USP12 and USP46 complexes with ratios similar to that of the native fractions display a similar activity with the native fractions, indicating that the deubiquitinase activity can be attributed to USP12 and USP46 (compare lane 2 with 3–6).
FIGURE 2.
WDR48 interacts with USP12 and USP46 and is required for the histone deubiquitination activity. A, schematic representation of the steps used to purify the USP12 and USP46 complexes from HeLa S3 cells stably expressing FLAG-HA-USP12 and FLAG-HA-USP46. Numbers represent the salt concentrations (mm) at which USP12 or USP46 elutes from the columns. B, silver staining (top panel), Western blot (2nd panel), and histone H2A deubiquitination assay (bottom two panels) of the affinity-purified USP12 complex separated on a Superose 6 column. Protein identities are labeled on the right side of the top panel. Antibodies used are indicated on the left side of the panels. C, silver staining (top panel), Western blot (2nd panel), and histone H2A deubiquitination assay (bottom two panels) of the affinity-purified USP46 complex separated on a Superose 6 column. Protein identities are labeled on the right side of the top panel. Antibodies used are indicated on the left side of the panels. D, comparison of the H2A deubiquitination activity of affinity-purified USP12 and USP46 complexes with native fractions. Top panel, Western blot analysis of USP12 in affinity-purified USP12 and USP46 complexes as well as native fractions. Bottom two panels, histone H2A deubiquitination assay of affinity-purified USP12 and USP46 complexes and native fractions. Antibodies used are indicated on the left side of the panels.
USP12 and USP46 Prefer Nucleosome Substrates and Deubiquitinate Both H2A and H2B in Vitro and in Vivo
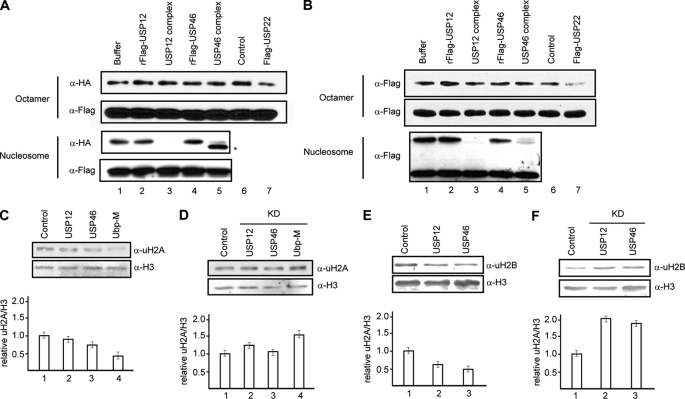
To gain insight into the function of USP12 and USP46-mediated histone deubiquitination, we determined the substrate preference and specificity of the USP12 and USP46 complexes. To determine whether WDR48 regulates USP12 and USP46 substrate preference and specificity, we also included USP12 and USP46 single subunits in these assays. To determine the substrate specificity, we prepared histone octamer and mononucleosomes, which contain equal amount of ubH2A as described in previous studies (14). When these substrates were used in histone deubiquitination assays, we found that USP12 and USP46 complexes, but not USP12 and USP46 single subunits, could deubiquitinate histone H2A and that this activity is specific to nucleosomes (Fig. 3A, compare lanes 2–5 with lane 1 in 1st and 3rd panels). We confirmed that the failure of USP12 and USP46 complexes to deubiquitinate H2A in octamer content is not due to the quality of substrates, as these substrates can be deubiquitinated by USP22 (Fig. 3A, top two panels, compare lane 6 with 7) (50). These results indicate that USP12 and USP46 complexes are specific for nucleosomal H2A.
FIGURE 3.
USP12 and USP46 prefer nucleosomal substrates and deubiquitinate both H2A and H2B in vitro and in vivo. A, histone H2A deubiquitination assay of USP12 and USP46 complexes and single subunits with histone octamers (top two panels) and mononucleosomes (bottom two panels) as substrates. Antibodies used are indicated on the left side of the panels. USP22 was used as a control in lane 7. Nonspecific signal from fraction is indicated by an asterisk. B, histone H2B deubiquitination assay of USP12 and USP46 complexes and single subunits with histone octamers (top two panels) and mononucleosomes (bottom two panels) as substrates. Antibodies used are indicated on the left side of the panels. USP22 was used as a control. C and E, overexpression of USP12 and USP46 reduced both H2A and H2B ubiquitination. HeLa cells transfected with plasmids as indicated on the top were analyzed by Western blot assay with antibodies indicated on the left side of the panels. Quantitations of three independent experiments are shown in the bottom panel. Error bars indicate standard deviation from the mean calculated from three independent samples. Ubp-M was used as a positive control. D and F, knockdown of USP12 and USP46 results in an increase of both H2A and H2B ubiquitination. HeLa cells transfected with siRNA as indicated on the top were analyzed by Western blot assay with antibodies indicated on the left side of the panels. Error bars indicate standard deviation from the mean calculated from four independent samples.
To determine whether USP12 and USP46 complexes are specific for histone H2A, we prepared histone octamer and mononucleosomes containing ubiquitinated H2B (39). When these substrates were used for histone deubiquitination assay, we found that USP12 and USP46 complexes, but not USP12 and USP46 single subunits, could deubiquitinate histone H2B (Fig. 3B, 3rd panel, compare lanes 2–5 with lane 1). Interestingly, USP12 and USP46 complexes deubiquitinate histone H2B as efficiently as H2A. The deubiquitination activity toward H2B is also specific for nucleosomes (Fig. 3B, compare lanes 2–5 with lane 1 in 1st and 3rd panels), although these substrates can also be deubiquitinated by USP22 (Fig. 3B, top panel, compare lane 6 with 7). Based on these results, we concluded that USP12 and USP46 complexes are nucleosome-specific histone deubiquitinases with dual substrate specificity for both histone H2A and H2B.
USP12 and USP46 Regulate H2A and H2B Deubiquitination in Vivo
To determine whether USP12 and USP46 regulate histone deubiquitination in vivo, we transfected USP12 and USP46 into HeLa cells and examined the effects on the levels of H2A and H2B ubiquitination. As a positive control, we also included Ubp-M in this assay. As shown in Fig. 3, C and E, transfection of USP12 and USP46 resulted in a decrease of both H2A and H2B ubiquitination levels (top panels, compare 2 and 3 with 1; see quantification in bottom panels). These data suggest that USP12 and USP46 regulate H2A and H2B ubiquitination in vivo. To further confirm this notion, we transfected control siRNA (scramble) and siRNA against USP12 and/or USP46 into HeLa cells. As shown in supplemental Fig. S4, transfection of siRNA against USP12 and USP46 reduced the levels of USP12 and USP46 mRNA significantly, compared with control knockdown (top three panels, compare lane 1 with 2–6). Interestingly, we noticed that when USP12 was knocked down, the levels of USP46 mRNA increased over 2-fold and vice versa (supplemental Fig. S4, top two panels, compare lane 1 with 2 and 3 and 4 and 5, numbers represent the relative fold change). This result suggests that the expression of USP12 and USP46 is coordinated in vivo.
To determine whether knockdown of USP12 and/or USP46 affects H2A and H2B ubiquitination in vivo, we employed the sensitive and quantitative LI-COR Odyssey Infrared Imaging System for Western blot assay. As shown in Fig. 3, D and F, when USP12 and/or USP46 was knocked down, we consistently observed an increase of both ubiquitinated H2A and H2B (top panels, compare lanes 2 and 3 with lane 1, see quantification in bottom panels). This result suggests that USP12 and USP46 regulate the levels of both H2A and H2B deubiquitination in vivo.
Previous studies indicated that both H2A and H2B ubiquitination regulate Hox gene expression (12, 22). To determine whether USP12- and USP46-mediated histone deubiquitination regulates Hox gene expression, we investigated whether knockdown of USP12 and/or USP46 affects the expression of Hox genes. Semi-quantitative RT-PCR assay revealed that knockdowns of USP12 and/or USP46 do not affect the expression of selected Hox genes (data not shown). However, we noticed that prolonged knockdown (96 h) of USP12 and/or USP46 resulted in a significant decrease of the expression of HOXB1 and HOXD10 genes (supplemental Fig. S4, 2nd to 6th panels, compare lane 1 with lanes 2–6). To determine whether this regulation involves USP12 and/or USP46, we used ChIP assay to detect the binding of FH-USP12 to HOXB1 and HOXD10 genes but failed to detect the binding of USP12 to HOXB1 and HOXD10 genes (data not shown). These results suggest that the effect of prolonged knockdown on Hox gene expression might be indirect.
USP12 Regulates Xenopus Early Embryonic Development
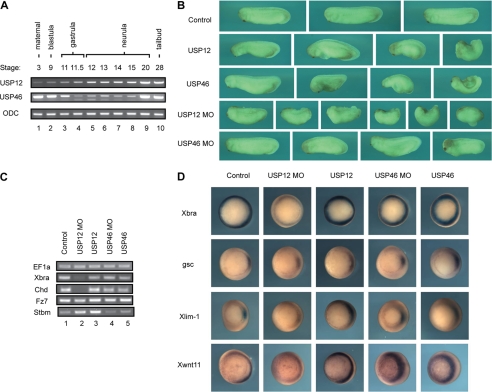
To determine the in vivo function of USP12 and USP46, we chose to investigate in a developmental context, using Xenopus laevis as our model. For this purpose, we first examined the expression of USP12 and USP46 during Xenopus early development. As shown in Fig. 4A, the mRNAs of USP12 and USP46 are expressed maternally, and the expression persists throughout all developmental stages tested (top two panels). Interestingly, an additional USP46 transcript was detected at stages 11–15, suggesting that there might be an alternative splice variant (Fig. 4A, middle panel, lanes 4–8).
FIGURE 4.
USP12 and USP46 regulate Xenopus development. A, semi-quantitative RT-PCR analysis of USP12 (top panel) and USP46 (middle panel) expression in X. laevis at different stages of embryonic development. Ornithine decarboxylase (ODC) was used as a loading control. B, USP12 regulates Xenopus gastrulation. Control uninjected Xenopus embryos (top row), embryos injected with USP12 and USP46 mRNA (2nd and 3rd row), or USP12 or USP46 MOs (4th and 5th row) are shown by the side view with the head to the left. Embryos injected with USP12 and USP46 mRNA had a smaller head and reduced body length, and many embryos did not go through gastrulation efficiently, so that they displayed an open blastopore phenotype. Embryos injected with USP12 MOs displayed severe reduction of both head and tail structures, and many embryos showed gastrulation defects with the open blastopore phenotype. Embryos injected with USP46 MOs developed relatively normally, with only minor defects in the head in certain embryos. C, semi-quantitative RT-PCR analysis of selected marker genes at gastrula stages in uninjected embryos (lane 1), embryos injected with USP12 mRNA and MOs (lanes 2 and 3), or USP46 mRNA and MOs (lanes 4 and 5). Translation elongation factor 1α (EF1a) was used as a control. D, In situ hybridization of selected markers in control embryos, embryos injected with USP12 mRNA and MOs, and USP46 mRNA and MOs.
To determine whether USP12 and USP46 regulate Xenopus development, we first used a gain-of-function approach. We injected mRNAs encoding USP12 and USP46 into the animal poles of two-cell stage embryos and allowed the embryos to develop to the tailbud to tadpole stages. As shown in Fig. 4B, enhanced expression of USP12 and USP46 led to severe defects in early Xenopus embryogenesis (2nd and 3rd rows). The heads of the injected embryos were reduced, and the body axes were shortened. In addition, many embryos did not go through gastrulation efficiently; they displayed an open blastopore phenotype with exposed endoderm. The defects were dose-dependent, so that at high doses of injected RNAs (2 ng), most embryos showed gastrulation defects and failed to close blastopore completely (data not shown). Control uninjected embryos or embryos injected with β-galactosidase RNA collected in parallel developed normally (Fig. 4B, top row, and data not shown).
To complement the gain-of-function approach, we further determined the role of USP12 and USP46 in Xenopus development with the loss-of-function strategy, i.e. by knocking down the endogenous protein level with specific antisense morpholino oligonucleotides (MOs). For this purpose, we injected 40 ng of USP12 or USP46 MOs into animal poles of two-cell stage embryos or dorsal marginal zones of four-cell stage embryos and incubated the embryos to the tailbud stages. As shown in Fig. 4B, depletion of USP12 resulted in severe reduction of both head and tail structures (4th row). Embryos lacked distinct somite development and often displayed split neural tube or open blastopore defects. In contrast, USP46 morphant embryos were largely normal, with only minor defects in head reduction in some embryos (Fig. 4B, bottom row). Injection of a control MO at the same or higher doses did not cause any embryonic defects (data not shown). Based on these data, we conclude that although enhanced expression of either USP12 or USP46 regulates gastrulation in Xenopus, an endogenous level of USP12, but not that of USP46, plays a more critical role in early Xenopus development.
To investigate the mechanism by which USP12 and USP46 regulate Xenopus development, we examined the expression of markers for mesodermal fate (Xbra for pan-mesoderm and Chd for dorsal mesoderm) as well as for gastrulation movements (Fz7 and Stbm are both required for convergent extension movements) by RT-PCR analysis. Although enhanced expression of USP12 and USP46 resulted in gastrulation defects, neither gene affected expression of any of these markers (Fig. 4C, compare lane 1 with lanes 3 and 5). The results indicate that overexpression of USP12/46 leads to direct regulation of cell movements without interfering with cell fate determination. In contrast, expression of the mesodermal marker Xbra and the organizer marker Chd was both decreased by depletion of USP12 (Fig. 4C, lane 2) but not by depletion of USP46 (lane 4), suggesting that USP12, but not USP46, modulates mesodermal cell fate determination. To further visualize how USP12 and USP46 regulate the spatial expression of the markers at gastrula stages, we performed whole mount in situ hybridization experiments. The transcription of Xbra was significantly reduced by the loss of USP12, and changes in Xlim-1 and Xwnt11 expression were also detected but to a lesser degree (Fig. 4D). In comparison, alteration of USP46 levels did not significantly change the expression levels of the markers assayed, but we did observe expansion of the expression domain of Xlim1 toward the lateral side when USP12 or USP46 was overexpressed. Taken together, these data reveal that endogenous USP12 regulates mesodermal cell fate during Xenopus embryogenesis, whereas ectopic expression of USP12 or USP46 modulates cell movements without affecting cell fate.
USP12 Regulates Histone Ubiquitination during Xenopus Embryonic Development
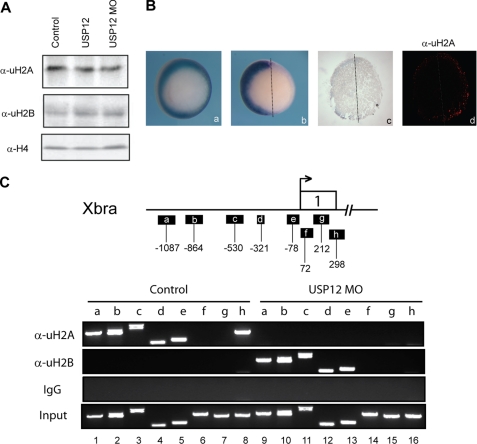
To determine whether the defects of Xenopus development we observed are linked to alternative histone ubiquitination, we measured the levels of ubiquitinated H2A and H2B when USP12 was overexpressed or knocked down. As shown in Fig. 5A, we failed to detect significant changes in the levels of global histone ubiquitination by Western blot. This could be due to the fact that USP12 only functions in certain cells in Xenopus embryos, and the changes could not be seen in bulk preparation of histones. To overcome this problem, we therefore examined the level of ubH2A by immunohistochemistry on embryonic sections. As shown in Fig. 5B, injection of USP12-MO in one of the two cells of early Xenopus embryos leads to reduction of Xbra on the injected side, and this correlates with increased ubH2A expression on this side (Fig. 5B, image c and d). The enhanced staining was found only in the mesodermal ring but not in the endodermal region, possible explaining why we did not detect global increase of ubH2A by Western blot analysis. These data suggest that USP12 can regulate histone H2A ubiquitination in Xenopus embryos. In the same assay, we did not detect positive staining for ubH2B (data not shown). This could be because the levels of ubH2B are low at this stage or the ubH2B antibody has weak affinity for Xenopus histones or that the antibody for ubH2B is not suitable for immunohistochemical staining.
FIGURE 5.
USP12 regulates H2A and H2B ubiquitination in Xenopus embryos. A, Western blot analysis of ubH2A and ubH2B level in control Xenopus embryos and embryos injected with USP12 mRNA and MOs. Histone H4 serves as a loading control. B, in situ hybridization for Xbra1 and immunohistochemistry staining for ubH2A on embryonic sections injected with USP12-MO in one of the two cells of two-cell stage Xenopus embryos. C, chromatin immunoprecipitation assay of uH2A and uH2B levels on Xba promoter and coding regions in control Xenopus embryos and embryos injected with USP12 MO. A diagram of Xba gene is shown on the top of the panels.
To determine whether the changes of mesodermal gene expression are linked to histone ubiquitination, we assayed for the occupancy of Xbra promoter and 5′-coding regions by ubH2A and ubH2B with chromatin immunoprecipitation assay, using anti-ubH2A and anti-ubH2B antibodies. As shown in Fig. 5C, ubH2A was enriched in Xbra promoter region (top panel, lanes 1–5) and coding region h (top panel, lane 8) but not in coding regions f and g (top panel, lanes 6 and 7). Surprisingly, knockdown of USP12 resulted in a decrease of H2A ubiquitination on the Xbra promoter and coding regions (Fig. 5C, top panel, compare lanes 1–8 with 9–16). In contrast, ubH2B was not detected in Xbra promoter and coding regions in control samples (Fig. 5C, 2nd panel, lanes 1–8). Knockdown of USP12 resulted in an increase of H2B ubiquitination in the Xbra promoter region (Fig. 5C, middle panel, compare lanes 1–5 with 9–13). These results are intriguing as ubiquitination of H2A and H2B has been generally linked to gene silencing and gene activation, respectively. However, recent studies indeed reveal that H2B ubiquitination has pleiotropic effects, with positive and negative influence on transcription (5, 34). Xbra gene may be a gene that is repressed by ubH2B at this developmental stage. These results are also intriguing as H2A ubiquitination was found to increase in the mesoderm upon knockdown of USP12 but not in the Xbra gene (Fig. 5B, image d). Addressing how USP12 coordinately regulates H2A and H2B ubiquitination to regulate Xbra gene expression and development will be a major undertaking. USP12 in Xenopus may provide a good experimental system to address the relation between H2A and H2B ubiquitination.
DISCUSSION
The human genome encodes 95 putative deubiquitinases; however, only a few have been functionally characterized, and limited numbers of substrate have been identified (51, 52). Using an unbiased biochemical approach, we purified from HeLa cells a histone deubiquitination activity as USP12 and USP46. Further characterization revealed that USP12 and USP46 regulate both H2A and H2B ubiquitination in vivo. Therefore, this study identifies histones (H2A and H2B) as the physiologically relevant substrates of USP12 and USP46. Although ubiquitination of H2A and H2B have both been implicated in Hox gene expression, USP12- and USP46-mediated histone deubiquitination is unlikely involved in this process. The identification of target genes of USP12 and USP46 will depend on applying global gene expression assay and sequencing genomic DNAs associated with USP12 and USP46 (ChIP-seq). It is intriguing that instead of regulating global histone ubiquitination nondiscriminatively, different histone ubiquitinases may be recruited to distinct sets of genes to regulate specific expression of these genes to affect different processes. Another possibility is that USP12- and USP46-mediated H2A and H2B deubiquitination may regulate nuclear events other than transcription, such as cell cycle progression. It is interesting to note that USP3, which also deubiquitinates both H2A and H2B, is required for cell cycle S phase progression and is involved in DNA damage response (33). Whether USP12 and USP46 play a role in these processes remains to be determined. USP12 and USP46 have been reported to interact with PHLPP and PHLPPL, two phosphatases that could dephosphorylate Akt (51, 53), a crucial enzyme involved in cell survival and cell cycle regulation. Consistent with the hypothesis that USP12 and USP46 must regulate important cellular processes, stable cell lines with high efficiency of USP12 or USP46 knockdowns could not be established (data not shown). Further investigations are needed to reveal cellular processes regulated by USP12 and USP46. In an attempt to address the function of these histone deubiquitinases in an organism, we studied X. laevis. Our results indicate that USP12 regulates Xenopus development during gastrula stages. USP12 is required for mesodermal cell fate determination and proper gastrulation movements. Xenopus may thus provide a good experimental system to further address the function of USP12 in a developmental context.
While our studies were underway, Sowa et al. (51) performed a global proteomic analysis of proteins interacting with human deubiquitination enzymes. Three WD-40 domain-containing proteins WDR48, WDR20, and DMWD and two protein phosphatases PHLPP and PHLPPL were reported to interact with USP12 and USP46, forming multiprotein complexes being part of the distributive networks (51). Of these interacting proteins, WDR48 was first identified as an activator of USP1 and has recently been shown to stimulate the deubiquitination activity of USP12 and USP46 on artificial substrates (36, 54). We reported in this study that WDR48 is definitely required for the deubiquitination activity of USP12 and USP46 toward its physiological substrate nucleosomal histones (51, 55). The critical control of USP12 and USP46 deubiquitinating activity by WDR48 is consistent with the hypothesis that most deubiquitination activities are cryptic to prevent inappropriate cleavage (52). Other than WDR48, WDR20 was also reported to stimulate the deubiquitination activity of USP12 and USP46 (51, 55). We initially purified USP12 and USP46 from HeLa nuclear pellet fractions as Ubp-M independent histone deubiquitinases (Fig. 1A), and our previous studies suggested that the H2A deubiquitination activity in nuclear extract fractions is attributed to Ubp-M (14). We therefore purified epitope-tagged USP12 and USP46 from nuclear pellet fractions of our stable cell lines. In the affinity purification, several WD repeat-containing proteins were identified, including WDR48, WDR20, DMWD, and two others in the nuclear extract. However, WDR48 was by far the most abundant WD repeat-containing protein in this fraction, suggesting that it was the dominant interacting protein in our complexes. We reasoned that this could be due to the approach we used to purify the USP12 and USP46 complexes. There could be USP12 and USP46 that interacts predominantly with WDR20 and/or other identified WD proteins in nuclear extract or cytoplasm fractions. This result suggests that proteins interacting with USP12 and USP46 may regulate the substrate specificity of deubiquitinating enzymes. In this scenario, USP12 and USP46 might deubiquitinate substrates other than histones when complexed with WDR20 in cytoplasm or nuclear extracts. Identifying these substrates will add our understanding of the physiological processes regulated by USP12 and USP46. Possibly due to same reasons, we also did not identify PHLPP and PHLPPL in USP12 and USP46 complexes purified from nuclear pellet fractions. Our results would suggest that future experiments that include quantitative assessment of associated proteins in each fraction will be vital in delineating the physiological processes that involve USP12 and USP46.
In summary, our studies here identified USP12 and USP46 as deubiquitinases that target histone H2A and H2B. Furthermore, our studies also reveal that USP12 regulates early embryonic development in Xenopus. Further identification of the cellular processes regulated by USP12- and USP46-mediated histone deubiquitination in different systems (cell lines and embryo development) will add significant information to our understanding of histone ubiquitination in higher eukaryotes. From this study and our previous studies on Ubp-M, we can envision that distinct histone deubiquitinases may be expressed at different developmental stages, targeting specific genes and regulating different cellular processes. Furthermore, our studies implicated that USP12 and USP46 might complex with different partners (WDR20 and WDR48), have different subcellular localization, and potentially target different substrates. The observation that ectopic expression of USP12 and USP46 leads to gastrulation defects without affecting marker gene expression brings up the intriguing possibility that these histone deubiquitinases may also function to remove ubiquitin in other molecules that are involved directly in controlling cell movements. This issue awaits further investigation.
Supplementary Material
Acknowledgments
We thank Dr. Moshe Oren for the anti-ubH2B antibody, Dr. Steven McMahon for FLAG-USP22 plasmid, and University of Alabama at Birmingham Fermentation Facility for mammalian cell culture.
This work was supported, in whole or in part, by National Institutes of Health Grant CA13148-35 from NCI (University of Alabama at Birmingham Comprehensive Cancer Center Collaborative Programmatic Development Grant Program to H. W. and C. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- LTQ
- linear trap quadrupole
- FT-ICR
- Fourier transform ion cyclotron resonance
- MO
- morpholino oligonucleotide.
REFERENCES
- 1. Orlando V. (2003) Cell 112, 599–606 [DOI] [PubMed] [Google Scholar]
- 2. Mohn F., Schübeler D. (2009) Trends Genet. 25, 129–136 [DOI] [PubMed] [Google Scholar]
- 3. Martin C., Zhang Y. (2005) Nat. Rev. Mol. Cell Biol. 6, 838–849 [DOI] [PubMed] [Google Scholar]
- 4. Campos E. I., Reinberg D. (2009) Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 5. Li B., Carey M., Workman J. L. (2007) Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 6. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 7. Ko M., Sohn D. H., Chung H., Seong R. H. (2008) Mutat. Res. 647, 59–67 [DOI] [PubMed] [Google Scholar]
- 8. Hirst M., Marra M. A. (2009) Int. J. Biochem. Cell Biol. 41, 136–146 [DOI] [PubMed] [Google Scholar]
- 9. Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osley M. A. (2006) Brief. Funct. Genomics Proteomics 5, 179–189 [DOI] [PubMed] [Google Scholar]
- 11. Jason L. J., Moore S. C., Lewis J. D., Lindsey G., Ausió J. (2002) BioEssays 24, 166–174 [DOI] [PubMed] [Google Scholar]
- 12. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 13. de Napoles M., Mermoud J. E., Wakao R., Tang Y. A., Endoh M., Appanah R., Nesterova T. B., Silva J., Otte A. P., Vidal M., Koseki H., Brockdorff N. (2004) Dev. Cell 7, 663–676 [DOI] [PubMed] [Google Scholar]
- 14. Joo H. Y., Zhai L., Yang C., Nie S., Erdjument-Bromage H., Tempst P., Chang C., Wang H. (2007) Nature 449, 1068–1072 [DOI] [PubMed] [Google Scholar]
- 15. Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010) Cell 141, 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu P., Zhou W., Wang J., Puc J., Ohgi K. A., Erdjument-Bromage H., Tempst P., Glass C. K., Rosenfeld M. G. (2007) Mol. Cell 27, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakagawa T., Kajitani T., Togo S., Masuko N., Ohdan H., Hishikawa Y., Koji T., Matsuyama T., Ikura T., Muramatsu M., Ito T. (2008) Genes Dev. 22, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Scheuermann J. C., de Ayala Alonso A. G., Oktaba K., Ly-Hartig N., McGinty R. K., Fraterman S., Wilm M., Muir T. W., Müller J. (2010) Nature 465, 243–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 20. Hwang W. W., Venkatasubrahmanyam S., Ianculescu A. G., Tong A., Boone C., Madhani H. D. (2003) Mol. Cell 11, 261–266 [DOI] [PubMed] [Google Scholar]
- 21. Wood A., Krogan N. J., Dover J., Schneider J., Heidt J., Boateng M. A., Dean K., Golshani A., Zhang Y., Greenblatt J. F., Johnston M., Shilatifard A. (2003) Mol. Cell 11, 267–274 [DOI] [PubMed] [Google Scholar]
- 22. Zhu B., Zheng Y., Pham A. D., Mandal S. S., Erdjument-Bromage H., Tempst P., Reinberg D. (2005) Mol. Cell 20, 601–611 [DOI] [PubMed] [Google Scholar]
- 23. Kim J., Hake S. B., Roeder R. G. (2005) Mol. Cell 20, 759–770 [DOI] [PubMed] [Google Scholar]
- 24. Kim J., Guermah M., McGinty R. K., Lee J. S., Tang Z., Milne T. A., Shilatifard A., Muir T. W., Roeder R. G. (2009) Cell 137, 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henry K. W., Wyce A., Lo W. S., Duggan L. J., Emre N. C., Kao C. F., Pillus L., Shilatifard A., Osley M. A., Berger S. L. (2003) Genes Dev. 17, 2648–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Daniel J. A., Torok M. S., Sun Z. W., Schieltz D., Allis C. D., Yates J. R., 3rd., Grant P. A. (2004) J. Biol. Chem. 279, 1867–1871 [DOI] [PubMed] [Google Scholar]
- 27. Emre N. C., Ingvarsdottir K., Wyce A., Wood A., Krogan N. J., Henry K. W., Li K., Marmorstein R., Greenblatt J. F., Shilatifard A., Berger S. L. (2005) Mol. Cell 17, 585–594 [DOI] [PubMed] [Google Scholar]
- 28. Gardner R. G., Nelson Z. W., Gottschling D. E. (2005) Mol. Cell. Biol. 25, 6123–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X. Y., Varthi M., Sykes S. M., Phillips C., Warzecha C., Zhu W., Wyce A., Thorne A. W., Berger S. L., McMahon S. B. (2008) Mol. Cell 29, 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao Y., Lang G., Ito S., Bonnet J., Metzger E., Sawatsubashi S., Suzuki E., Le Guezennec X., Stunnenberg H. G., Krasnov A., Georgieva S. G., Schüle R., Takeyama K., Kato S., Tora L., Devys D. (2008) Mol. Cell 29, 92–101 [DOI] [PubMed] [Google Scholar]
- 31. Weake V. M., Lee K. K., Guelman S., Lin C. H., Seidel C., Abmayr S. M., Workman J. L. (2008) EMBO J. 27, 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Knaap J. A., Kumar B. R., Moshkin Y. M., Langenberg K., Krijgsveld J., Heck A. J., Karch F., Verrijzer C. P. (2005) Mol. Cell 17, 695–707 [DOI] [PubMed] [Google Scholar]
- 33. Nicassio F., Corrado N., Vissers J. H., Areces L. B., Bergink S., Marteijn J. A., Geverts B., Houtsmuller A. B., Vermeulen W., Di Fiore P. P., Citterio E. (2007) Curr. Biol. 17, 1972–1977 [DOI] [PubMed] [Google Scholar]
- 34. Espinosa J. M. (2008) Genes Dev. 22, 2743–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Weake V. M., Workman J. L. (2008) Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 36. Cohn M. A., Kee Y., Haas W., Gygi S. P., D'Andrea A. D. (2009) J. Biol. Chem. 284, 5343–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sun Z. W., Allis C. D. (2002) Nature 418, 104–108 [DOI] [PubMed] [Google Scholar]
- 38. Chandrasekharan M. B., Huang F., Chen Y. C., Sun Z. W. (2010) Mol. Cell. Biol. 30, 3216–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Y., Reddy B., Thompson J., Wang H., Noma K., Yates J. R., 3rd., Jia S. (2009) Mol. Cell 33, 428–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. (1983) Methods Enzymol. 101, 582–598 [DOI] [PubMed] [Google Scholar]
- 41. Renfrow M. B., Mackay C. L., Chalmers M. J., Julian B. A., Mestecky J., Kilian M., Poulsen K., Emmett M. R., Marshall A. G., Novak J. (2007) Anal. Bioanal. Chem. 389, 1397–1407 [DOI] [PubMed] [Google Scholar]
- 42. Eng J. K., McCormack A. L., Yates Iii J. R. (1994) J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 43. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 44. Clauser K. R., Baker P., Burlingame A. L. (1999) Anal. Chem. 71, 2871–2882 [DOI] [PubMed] [Google Scholar]
- 45. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 46. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 47. Li P., Yao H., Zhang Z., Li M., Luo Y., Thompson P. R., Gilmour D. S., Wang Y. (2008) Mol. Cell. Biol. 28, 4745–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Blythe S. A., Reid C. D., Kessler D. S., Klein P. S. (2009) Dev. Dyn. 238, 1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chang C., Wilson P. A., Mathews L. S., Hemmati-Brivanlou A. (1997) Development 124, 827–837 [DOI] [PubMed] [Google Scholar]
- 50. Zhang X. Y., Pfeiffer H. K., Thorne A. W., McMahon S. B. (2008) Cell Cycle 7, 1522–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brognard J., Sierecki E., Gao T., Newton A. C. (2007) Mol. Cell 25, 917–931 [DOI] [PubMed] [Google Scholar]
- 54. Cohn M. A., Kowal P., Yang K., Haas W., Huang T. T., Gygi S. P., D'Andrea A. D. (2007) Mol. Cell 28, 786–797 [DOI] [PubMed] [Google Scholar]
- 55. Kee Y., Yang K., Cohn M. A., Haas W., Gygi S. P., D'Andrea A. D. (2010) J. Biol. Chem. 285, 11252–11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.