Abstract
Ca2+-binding proteins of the S100 family participate in intracellular Ca2+ signaling by binding to and regulating specific cellular targets in their Ca2+-loaded conformation. Because the information on specific cellular targets of different S100 proteins is still limited, we developed an affinity approach that selects for protein targets only binding to the physiologically active dimer of an S100 protein. Using this approach, we here identify IQGAP1 as a novel and dimer-specific target of S100P, a member of the S100 family enriched in the cortical cytoskeleton. The interaction between S100P and IQGAP1 is strictly Ca2+-dependent and characterized by a dissociation constant of 0.2 μm. Binding occurs primarily through the IQ domain of IQGAP1 and the first EF hand loop of S100P, thus representing a novel structural principle of S100-target protein interactions. Upon cell stimulation, S100P and IQGAP1 co-localize at or in close proximity to the plasma membrane, and complex formation can be linked to altered signal transduction properties of IQGAP1. Specifically, the EGF-induced tyrosine phosphorylation of IQGAP1 that is thought to function in assembling signaling intermediates at IQGAP1 scaffolds in the subplasmalemmal region is markedly reduced in cells overexpressing S100P but not in cells expressing an S100P mutant deficient in IQGAP1 binding. Furthermore, B-Raf binding to IQGAP1 and MEK1/2 activation occurring downstream of IQGAP1 in EGF-triggered signaling cascades are compromised at elevated S100P levels. Thus, S100P is a novel Ca2+-dependent regulator of IQGAP1 that can down-regulate the function of IQGAP1 as a signaling intermediate by direct interaction.
Keywords: Calcium-binding Proteins, Calmodulin, Cytoskeleton, MAPKs, Signal Transduction, Protein-Protein Interaction
Introduction
Intracellular Ca2+ signaling requires a network of different effectors that couple the signal, a transient and often locally restricted elevation in Ca2+ levels, to cellular responses. Among these effectors are families of Ca2+-binding proteins that function by interacting with and thereby modulating target proteins in their Ca2+-bound conformation. S100 proteins are the largest family of these effector proteins consisting of 21 different genes in humans (for reviews on S100 proteins, see Refs.1–4). They are characterized by two EF hand-type Ca2+-binding sites, helix-loop-helix structures that coordinate Ca2+ through carbonyl, hydroxyl, and carboxyl oxygens located within the intrahelical loop. S100 proteins form homo- and heterodimers, and mutagenesis experiments as well as high resolution structures of S100 dimers complexed with target peptides have revealed that these dimers represent the physiologically relevant entity (for reviews see Refs. 3, 5, 6). A number of S100 target proteins have been identified, including cytoskeletal elements, different enzymes, and receptors (1–4). However, the search for specific targets has been hampered by the fact that S100 proteins expose hydrophobic surfaces in their Ca2+-bound active conformation and thus tend to show unspecific interactions in this conformation.
To circumvent this problem of unspecific hydrophobic interactions, we designed a strategy to probe for S100 protein interactions that are Ca2+-dependent but only occur with the biologically active dimer. The basis for this approach was the identification of a single residue in the S100P protein, Phe-15, that proved to be crucial for dimerization. Mutation of phenylalanine to alanine yielded an S100P variant that failed to dimerize but retained the capability to bind Ca2+ and to respond to Ca2+ binding with the exposure of hydrophobic residues (7). Comparing the interaction partners of this S100P F15A derivative that we considered unspecific with those binding to the wild-type dimeric S100P led to the identification of the membrane-cytoskeleton cross-linker ezrin as a dimer-specific and physiologically important target of S100P (8). Extending this approach, we now show that IQGAP1 is another target protein of dimeric S100P that does not show an unspecific interaction with the monomeric variant of the S100 protein.
IQGAP1 is a ubiquitously expressed member of the IQGAP family, which also includes IQGAP2 and -3 in humans. It is a multidomain protein that can recruit several interaction partners and that has been shown to function in signal transduction pathways and regulation of actin cytoskeleton and microtubule dynamics (for reviews see Refs. 9–11). The N- to C-terminal IQGAP1 contains the following: an F-actin-binding calponin homology domain (CHD)2; a proline-rich WW domain known to bind ERK1 and -2; an IQ domain that has been shown to interact with calmodulin, S100B, B-Raf, and MEK1 and -2; a RasGAP-related domain (GRD) capable of binding Cdc42 and Rac1; and a RasGAP C terminus (RGCT) that can interact with cell contact proteins such as E-cadherin and β-catenin and also with cytoskeleton regulators such as CLIP-170, Dia1, and adenomatous polyposis coli. The two C-terminal domains, GRD and RGCT, can interact with one another resulting in an autoinhibited conformation that can be relieved by phosphorylation at a serine residue in the GRD (12). Activation of IQGAP1 occurs downstream of different types of cell surface receptors, specifically receptor tyrosine kinases, G protein-coupled receptors, and integrins and, at least in some cases, is accompanied by tyrosine phosphorylation of IQGAP1 (13, 14). Once activated, IQGAP1 can serve as a signaling intermediate in a number of pathways. Through direct interaction it can inhibit the GTPase activity of Cdc42 and Rac1 thereby stabilizing their active, GTP-bound forms and increasing neural Wiskott-Aldrich syndrome protein-Arp2/3-dependent actin polymerization that affects cell shape dynamics and motility (for review see Ref. 15). Interaction with E-cadherin enables IQGAP1 to regulate cell-cell contacts (16), whereas the ability to bind actin and microtubule regulators, e.g. Dia1 and CLIP170, respectively, allows IQGAP1 to bridge the actin and microtubule networks, a function particularly relevant in the leading edge of migrating cells. Finally, through functioning as a scaffolding protein for signaling complexes, IQGAP1 is involved in different signaling pathways. Importantly, it can associate with B-Raf, MEK, and ERK isoforms, and it participates in the MAPK cascade possibly by facilitating the spatial coupling of the different kinases (for review see Ref. 17).
The activity of IQGAP1 itself is also regulated by direct protein interactions. Apart from the activating Cdc42 and Rac1 associations, the effect of calmodulin binding is best studied. Calmodulin can bind to the IQ domain of IQGAP1, and when bound in its Ca2+-loaded conformation, it interferes with many of the other protein interactions of IQGAP1 thereby silencing IQGAP1 activity (18). For example, Ca2+/calmodulin binding abrogates the activating Cdc42 interaction and also inhibits the interaction of IQGAP1 with B-Raf, thereby affecting other kinases of the MAPK pathway and down-regulating MAPK signaling, e.g. downstream of the EGF receptor (19). Here, we obtained further evidence for a role of IQGAP1 in linking Ca2+ and MAPK signaling. We identified Ca2+-bound S100P as a novel interaction partner of IQGAP1 and could show that S100P binding down-regulates both EGF-triggered tyrosine phosphorylation of IQGAP1 and EGF-induced MEK activation, however, without affecting the IQGAP1 interactions with Cdc42 and Rac1. This indicates that S100P is a novel and selective Ca2+-dependent regulator of IQGAP1 functions.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS), 2 mm l-glutamine, and antibiotics in a 7% CO2 incubator at 37 °C. HeLa cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Spodoptera frugiperda 9 (SF9) insect cells were cultured in TC-100 medium with 10% FCS and 0.26% tryptose phosphate broth (Sigma) at 27 °C in air atmosphere.
Plasmid Construction
The generation of pET-32a+ and pcDNA3.1 plasmids encoding the IQGAP1 full-length protein and different domains of IQGAP1 has been described (20). The constructs encoding tagless, His-, or GFP-tagged versions of human S100P in pKK223-3-, pET-28a+-, or pEGFP-C2 as well as the constructs encoding YFP-tagged human S100A10 in pYFP-C1 and GST-tagged human N-ERMAD in pGEX-4T-1 have been described previously (8, 21, 22). cDNAs encoding His-tagged S100P truncation and deletion mutants were generated by PCR. PCR was performed using Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA), the pET-28a+ plasmid containing S100Pwt cDNA as a template, and oligonucleotide primers containing the respective deletions and restriction enzyme sites. For the expression of S100PΔ21–25 fused to an N-terminal GFP tag, the coding sequence of S100PΔ21–25 was cloned into the pEGFP-C2 vector using EcoRI and SalI restriction sites (Clontech).
Recombinant Protein Expression
Expression of full-length GST-tagged IQGAP1 in SF9 insect cells employed Autographa californica nuclear polyhedrosis baculoviruses encoding GST-IQGAP1. After 72 h of infection, cells were scraped off and harvested by centrifugation (500 × g, 15 min). After washing with phosphate-buffered saline (PBS), cells were resuspended in 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, 1 mm DTT, 10 μm PMSF, and 10 μg/μl leupeptin and subjected to sonication (duty cycle 30%, output control 3). The lysed cells were centrifuged (100,000 × g, 1 h), and the supernatant was applied to a glutathione-Sepharose 4B (GE Healthcare) column equilibrated with 20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA, and 1 mm DTT. After washing with the same buffer, the IQGAP1-containing matrix was used for in vitro binding assays.
His-tagged or tagless S100Pwt and S100P deletion mutants as well as GST-tagged N-ERMAD and the His-tagged IQ domain of IQGAP1 were expressed in Escherichia coli cells (strain BL21(DE3)pLysS). Transformed bacteria were grown to an A600 of 0.6 at 37 °C, and recombinant protein expression was then induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm. After incubation for 3 h at 37 °C, cells were harvested by centrifugation (2500 × g; 10 min) and resuspended in lysis buffer (40 mm Hepes, pH 7.4, 20 mm imidazole, pH 7.4, 300 mm NaCl, 1 mm EDTA, 10 mm β-mercaptoethanol, 1 mm PMSF). Cells were lysed by three freeze/thaw cycles and sonication (duty cycle 50%, output control 5), and the remaining cellular debris was removed by centrifugation for 1 h at 100,000 × g.
For the preparation of tagless S100Pwt and S100PΔ21–25 proteins, the resulting supernatants were dialyzed against 40 mm Hepes, pH 7.4, 30 mm NaCl, 10 mm β-mercaptoethanol, and 1 mm PMSF and applied to a DEAE-Sephacel (Sigma) column equilibrated in dialysis buffer. After washing with increasing concentrations of NaCl (30 and 40 mm), tagless S100P proteins were eluted with dialysis buffer containing 200 mm NaCl. The pooled fractions were then dialyzed against 40 mm Hepes, pH 7.4, 80 mm NaCl, 10 mm β-mercaptoethanol, and 2 mm CaCl2. The dialyzed proteins were adjusted to a CaCl2 concentration of 5 mm and applied to a phenyl-Sepharose (GE Healthcare) column equilibrated in dialysis buffer. After washing with the same buffer, bound S100P protein was eluted with dialysis buffer containing 2 mm EGTA.
His-tagged S100Pwt or S100P deletion mutants were obtained by adjusting the supernatants after the 100,000 × g centrifugation step to a CaCl2 concentration of 5 mm and applying them to a phenyl-Sepharose column equilibrated in lysis buffer containing 0.5 mm CaCl2 and no EDTA. After washing with the same buffer, bound S100P proteins were eluted with lysis buffer containing 1 mm EGTA. The eluted S100P-containing fractions were pooled and dialyzed against 20 mm imidazole, pH 7.4, 150 mm NaCl, 10 mm β-mercaptoethanol, and 1 mm PMSF. Subsequently, the proteins were applied to a Ni-NTA-agarose (Qiagen, Hilden, Germany) column equilibrated in dialysis buffer. After washing with increasing concentrations of imidazole (25 and 35 mm), His-tagged S100P proteins were eluted with elution buffer (250 mm imidazole, pH 7.4, 150 mm NaCl, 10 mm β-mercaptoethanol, 1 mm PMSF).
The His-tagged IQ domain of IQGAP1 was prepared by applying the supernatant obtained after the ultracentrifugation step to a Ni-NTA-agarose column equilibrated with 40 mm Hepes, pH 7.4, 20 mm imidazole, pH 7.4, 300 mm NaCl, 20 mm β-mercaptoethanol, and 1 mm PMSF. After washing with the same buffer containing increasing concentrations of imidazole (25, 35, and 100 mm), His-tagged IQ domain was eluted with this buffer supplemented with 250 mm imidazole.
GST-tagged N-ERMAD was obtained by applying the soluble bacterial proteins to a glutathione-Sepharose 4B column equilibrated with PBS. After washing with PBS, bound GST-tagged N-ERMAD was eluted with a buffer containing 100 mm glutathione and 50 mm Tris-HCl, pH 8.0.
In Vitro Transcription/Translation
The TnT® Quick-coupled transcription/translation system (Promega, Madison, WI) was used to produce [35S]methionine-labeled IQGAP1 domains or the full-length protein. Therefore, 1 μg of pcDNA3.1 plasmid containing the respective cDNA was incubated with 12 μl of TnT® Quick Master Mix and 9 μCi of Redivue® l-[35S]methionine (GE-Healthcare) at 30 °C for 60 min. The success of the reaction was analyzed by SDS-PAGE and subsequent autoradiography, and the TnT® products were then further used for in vitro binding assays.
Affinity Chromatography of Placental Proteins on Immobilized S100P
His-tagged S100Pwt and S100P F15A were purified as described previously (8) and used as affinity tools for probing placental protein extracts. Extracts were prepared by homogenizing frozen placenta in homogenization buffer (30 mm Hepes, pH 7.2, 140 mm NaCl, 2 mm MgCl2, 1% Triton X-100, 1 mm DTT, 1 mm EDTA, 10 μg/ml pepstatin, 35 μg/ml aprotinin, 3 μm leupeptin, 0.5 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 1.5 mm PMSF) using a Waring blender. After centrifugation at 22,000 × g for 30 min at 4 °C, the resulting supernatant was further centrifuged at 100,000 × g for 1 h at 4 °C. The lysate was then dialyzed against 30 mm Hepes, pH 7.2, 20 mm imidazole, pH 7.2, 300 mm NaCl, 2 mm MgCl2, 10 mm β-mercaptoethanol, 10 μg/ml pepstatin, 35 μg/ml aprotinin, 3 μm leupeptin, 0.5 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 1.5 mm PMSF, and 2 mm NaN3. To screen for Ca2+-dependent interactions, CaCl2 was added to the placental protein extract to a final concentration of 0.7 mm. 30 mg of Ca2+-containing placental protein extract were then loaded onto Ni-NTA-agarose affinity columns containing 5 mg of purified His-tagged S100Pwt or S100P F15A, respectively. After washing the columns with dialysis buffer containing 0.7 mm CaCl2, Ca2+-dependently bound proteins were eluted with dialysis buffer containing 0.7 mm EGTA. The collected fractions were analyzed in gradient (7–15%) SDS-polyacrylamide gels stained with Coomassie Brilliant Blue.
In Vitro Binding Assay
In vitro binding assays were performed using different column-based affinity chromatography approaches. To analyze the interaction of IQGAP1 and S100P, glutathione-Sepharose 4B beads containing 3 μg of immobilized GST-IQGAP1 were equilibrated in buffer A (40 mm Hepes, pH 7.4, 20 mm imidazole, pH 7.4, 150 mm NaCl, 1 mm DTT, 1.5 mm PMSF) containing either 3 mm CaCl2 or 2 mm EGTA. 30 μg of tagless S100P protein were dialyzed against the respective equilibration buffer and added in the fluid phase. After incubation at 4 °C for 1 h and washing with the same buffers, bound S100P protein was eluted with buffer A containing 7 mm EGTA. Fractions of the flow-through and of washing and elution steps were collected and analyzed by SDS-PAGE and Western blots employing a monoclonal mouse anti-S100P antibody3 and a goat anti-mouse POX-labeled secondary antibody (Dianova, Hamburg, Germany).
For precise mapping of the interaction motif in IQGAP1, His-tagged S100Pwt was immobilized on Ni-NTA-agarose beads in buffer A containing 0.5 mm CaCl2 or 2 mm EGTA. To confirm that equivalent amounts of S100P protein were immobilized, small aliquots of the resin were subjected to SDS-PAGE and stained with Coomassie Brilliant Blue or probed by immunoblotting prior to the experiment. In vitro translated [35S]methionine-labeled IQGAP1 domains were added to the immobilized His-S100Pwt and incubated for 2 h at 4 °C. As a control, equal amounts of the in vitro translation products were incubated with Ni-NTA-agarose alone. After washing with the respective binding buffers, bound proteins were eluted with boiling SDS sample buffer and analyzed by SDS-PAGE and autoradiography.
For mapping of the IQGAP1-binding site in S100P, the recombinantly expressed His-tagged S100P truncation or deletion mutants was immobilized on Ni-NTA-agarose as described above, and in vitro translated full-length IQGAP1 protein was added. Analysis of IQGAP1 bound to the different S100P derivatives was performed as described above.
To compare S100Pwt and S100PΔ21–25 in terms of their ability to bind to the N-ERMAD of ezrin, 30 μg of the respective His tagged-S100P protein were immobilized on Ni-NTA-agarose beads equilibrated in buffer A containing 0.5 mm CaCl2. After incubation with 100 μg of GST-N-ERMAD for 2 h and extensive washing with the same buffer, bound GST-N-ERMAD was eluted with buffer A containing 5 mm EGTA. The collected fractions were analyzed by SDS-PAGE and Western blot using a monoclonal mouse anti-GST antibody (B14, Santa Cruz Biotechnology, Heidelberg, Germany) and a goat anti-mouse POX-labeled secondary antibody.
Real Time Binding Measured by Surface Plasmon Resonance (SPR)
SPR experiments were performed using a Biacore 3000 system (GE Healthcare) employing CM5 sensor chips. The flow buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 0.005% Tween 20) was filtered through 0.22-μm filters (Millipore) and degassed before use. About 3000 response units of purified S100Pwt or S100PΔ21–25 were immobilized on the sensor chip using the amine coupling method according to the manufacturer's instructions. Briefly, using a flow rate of 5 μl/min, the chip surface was activated with a 7-min injection of a freshly prepared 1:1 mixture of 0.2 m N-ethyl-N(3-diethylaminopropyl) carbodiimide and 0.05 m N-hydroxysuccinimide solution, followed by injection of S100Pwt or S100PΔ21-25 at a concentration of 30 μg/ml in acetate buffer, pH 4.0. When the desired level of immobilization was achieved, unreacted N-hydroxysuccinimide-ester groups were blocked with a 7-min injection of 1 m ethanolamine hydrochloride.
Sensorgrams (response units versus time) were recorded at 25 °C with a typical flow rate of 60 μl/min and injection times of 3 min. Controls for the contribution of the change in bulk refractive index were performed in parallel with blank flow cells that were activated with N-ethyl-N-(3-diethylaminopropyl) carbodiimide/N-hydroxysuccinimide and deactivated with ethanolamine in the absence of protein to be coupled and subtracted from all binding sensorgrams. Blank injections of the running buffer were also performed and subtracted from all the kinetic measurements.
To analyze the binding affinity of the IQ domain to immobilized S100Pwt or S100PΔ21–25, different concentrations of the IQ domain covering the concentration range between 50 nm and 1.5 μm diluted in 10 mm Hepes, pH 7.4, 150 mm NaCl, 0.005% Tween 20, 500 μm CaCl2 and 450 μm EGTA were injected into the flow cells. Each injection was repeated at least 2-fold for reproducibility. Regeneration of the surface was performed using one injection of 10 mm EGTA followed by a pulse injection of 50 mm NaOH. Approximate equilibrium dissociation constants (KD) were obtained by measuring the equilibrium resonance (Req) units at several ligand concentrations at equilibrium. The affinity of the interaction, i.e. the equilibrium dissociation constant (KD), was determined from the level of binding at equilibrium as a function of the sample concentrations by BIAEvaluation software version 4.1 (Biacore, Inc.). The steady state binding level is related to the concentration according to the Scatchard equation, Req/C = KARmax − ReqKA, where Rmax is the resonance signal at saturation; C is the concentration of free analyte, and KA is the equilibrium association constant.
The Ca2+ dependence of IQ-S100P interaction was verified by performing IQ injections on the immobilized S100Pwt in flow buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 0.005% Tween 20) containing either 2 mm HEDTA or 0.77 mm CaCl2 plus 2 mm HEDTA. The latter corresponded to a free Ca2+ concentration of 2.5 μm as determined by MaxChelator software.
Binding of the IQ domain of IQGAP1 to S100Pwt and S100PΔ21–25 was directly compared by reaction of similar concentrations of this domain with the immobilized S100P proteins. Therefore, the two S100P proteins were immobilized to the same level of response units (flow cells 2 and 4 were used for S100P and S100PΔ21–25, respectively, and flow cells 1 and 3 were used as blank references). Injections of the IQ domain in running buffer (10 mm Hepes, pH 7.4, 150 mm NaCl, 0.005% Tween 20, 500 μm CaCl2, and 450 μm EGTA) were then performed on all flow cells allowing the direct comparison between the response of IQ interaction with S100Pwt and S100PΔ21–25.
Co-immunoprecipitation
HeLa cells were transiently transfected with the pEGFP-S100Pwt expression construct. After 24 h, cells were washed once with PBS and scraped into 0.5 ml of lysis buffer containing 2.5 μm free Ca2+ (10 mm Hepes, pH 7.4, 150 mm NaCl, 1% Triton X-100, 2 mm HEDTA, 0.77 mm CaCl2, and Complete protease inhibitor mixture (Roche Diagnostics)). Cell lysis was performed on an overhead shaker for 30 min at 4 °C followed by centrifugation (300 × g, 30 min, 4 °C). The resulting post-nuclear supernatant (PNS) was applied to protein A-Sepharose CL-4B (GE Healthcare) equilibrated in lysis buffer either together with a polyclonal rabbit anti-IQGAP1 antibody (H-108, Santa Cruz Biotechnology) or nonspecific anti-mouse IgG polyclonal rabbit antibodies (DAKO, Glostrup, Denmark). After incubation for 2 h at 4 °C and washing with lysis buffer, bound proteins were eluted with boiling SDS sample buffer. Samples of the PNS and the eluted proteins were analyzed by SDS-PAGE and Western blot using a monoclonal mouse anti-IQGAP1 antibody (BD Biosciences) and a monoclonal mouse anti-GFP antibody (Clontech) followed by a goat anti-mouse POX-labeled secondary antibody.
Immunofluorescence Analysis
HeLa cells were grown on coverslips and serum-starved for 16 h. To stimulate cells, EGF was added to a final concentration of 50 ng/ml for 5 min at 37 °C. Control cells were maintained in FCS-free medium. Cells were then fixed with 3% formaldehyde in PBS for 10 min at room temperature. Following quenching in 50 mm NH4Cl for 7 min and permeabilization with 0.2% Triton X-100 in PBS for 2 min, cells were treated with 2% BSA in PBS for 30 min. Cells were then incubated with a mouse monoclonal anti-S100P antibody (MAb 18-9, a gift from A. Gibadulinova, Slovak Academy of Sciences, Bratislava, Slovakia) and a rabbit polyclonal anti-IQGAP1 antibody (raised against amino acids 1–863) each for 45 min and washed with PBS. Primary antibodies were detected with Texas Red-conjugated goat anti-mouse (Dianova) or Alexa Fluor 488 goat anti-rabbit (Invitrogen) secondary antibodies, respectively. Stained cells were mounted in Mowiol and analyzed using a confocal laser scanning microscope (LSM 510; Zeiss, Jena, Germany).
IQGAP1 Phosphorylation and B-Raf Binding Assay
HeLa cells were transiently transfected with pGFP-S100Pwt, pGFP-S100PΔ21–25, or pYFP-S100A10. After 16 h of serum starvation, cells were stimulated with EGF (E9644-.2MG, Sigma) at a final concentration of 50 ng/ml for 5 min at 37 °C.
Tyrosine phosphorylation of IQGAP1 was determined by immunoprecipitation. Therefore, cells were lysed in lysis buffer (50 mm Hepes, pH 7.4, 5 mm EDTA, pH 8.0, 1% Triton X-100, 50 mm NaCl, 1 mm PMSF, PhosStop phosphatase inhibitor mixture, and Complete protease inhibitor mixture (both from Roche Diagnostics)), collected by scraping, and sonicated for 15 s. The lysate was cleared by centrifugation (4000 × g, 5 min, 4 °C), and equal amounts of total soluble protein were incubated with a polyclonal rabbit anti-IQGAP1 antibody coupled to magnetic sheep anti-rabbit Dynabeads® (Invitrogen) for 2 h at 4 °C. After washing with lysis buffer, bound proteins were eluted with SDS sample buffer. The tyrosine phosphorylation of precipitated IQGAP1 was analyzed by SDS-PAGE and Western blot using a mouse monoclonal anti-phosphotyrosine antibody (4G10® Platinum, Millipore, Schwalbach, Germany) and a goat anti-mouse POX-labeled secondary antibody. Reprobing of the blot for total IQGAP1 using a monoclonal mouse anti-IQGAP1 antibody followed by an appropriate secondary antibody was used as internal loading control. IQGAP1-B-Raf complex formation was analyzed by probing the IQGAP1 immunoprecipitates with a monoclonal mouse anti-B-Raf antibody (F-3, Santa Cruz Biotechnology).
MEK1/2 Activation Assay
MEK1/2 activation was monitored by assessment of MEK1/2 phosphorylation on serine residue 221. Transfected HeLa cells were treated as described for the IQGAP1 phosphorylation assay. After removal of the medium, cells were washed, scraped into PBS, and pelleted by centrifugation (12,000 × g, 1 min, 4 °C). The cells were then lysed in lysis buffer (10 mm Tris-HCl, pH 7.4, 100 mm NaCl, 1% Triton X-100, 10% glycerol, 0.1% SDS, 1 mm EDTA, 1 mm EGTA, PhosStop phosphatase inhibitor mixture, and Complete protease inhibitor mixture) and incubated on ice for 30 min with vortexing every 10 min. The total lysates were analyzed by SDS-PAGE and Western blot using an anti-phospho-MEK1/2 antibody (166F8, Cell Signaling Technology, Beverly, MA) and a goat anti-rabbit POX-labeled secondary antibody (Dianova). The amount of total MEK1/2 in the lysates was assessed by reprobing of the blot with an anti-MEK1/2 antibody (47E6, Cell Signaling Technology) followed by a goat anti-rabbit POX-labeled secondary antibody.
Miscellaneous
ECL signal intensities were quantified by densitometric analysis using ImageJ software (National Institutes of Health). Statistical significance of the results was evaluated by Student's t test using GraphPad Prism version 4.00 (GraphPad Software, San Diego). All values were expressed as mean ± S.E., and a p value of less than 0.05 was considered significant.
RESULTS
Identification of IQGAP1 as a Novel S100P Target Protein
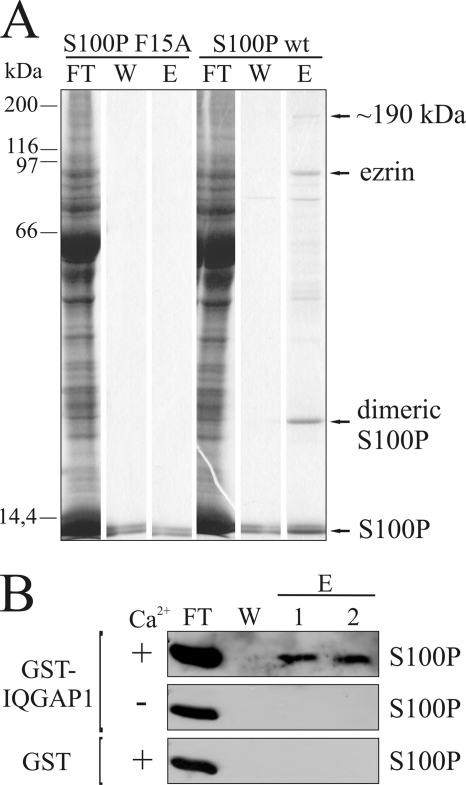
Ca2+-dependent affinity chromatography utilizing immobilized S100P was performed to identify new S100P binding partners in a total placental protein lysate. To increase the specificity of this approach, we probed for those proteins that bind only to the dimeric form of S100P, which is considered to be the biologically active entity (3). This was achieved by using the constitutively monomeric S100P F15A mutant (7) as a control in a parallel experimental setup. The affinity approach yielded a number of proteins that only bound to dimeric and not to monomeric S100P in a Ca2+-dependent manner (Fig. 1A). One of these proteins was identified previously as ezrin, a membrane-F-actin cross-linker (8). A second high molecular weight band only present in the EGTA eluate of the S100Pwt (dimeric) column was detected at ∼190 kDa. MALDI-MS analysis and data base searches identified this 190-kDa protein as human IQGAP1.
FIGURE 1.
Identification of IQGAP1 as a novel S100P target protein. A, affinity chromatography of placental proteins on columns containing immobilized S100Pwt or the monomeric F15A mutant. Equal amounts of placental protein extract were loaded onto the columns in a Ca2+-containing buffer, and flow-through fractions were collected (FT). Columns were washed, (W) and Ca2+-dependently bound proteins were eluted with an EGTA-containing buffer (E). The collected fractions were subjected to SDS-PAGE and stained with Coomassie. MALDI-TOF analysis identified the protein migrating at an apparent molecular mass of 190 kDa as human IQGAP1. This protein was only present in the eluate of the S100Pwt column. B, affinity chromatography with S100P and IQGAP1 reveals that the interaction is direct and Ca2+-dependent. S100Pwt was loaded onto columns containing immobilized GST-IQGAP1 or GST alone in a Ca2+- or EGTA-containing buffer, and flow-through fractions were collected (FT). Columns were washed (W) and Ca2+-dependently bound S100Pwt was eluted with an EGTA-containing buffer (E). The collected fractions were analyzed by immunoblotting for S100P.
To rule out the possibility that the observed interaction is indirect, i.e. mediated by other proteins present in the placental lysate, an affinity chromatography approach was performed using purified proteins. GST-tagged IQGAP1 expressed in SF9 insect cells was immobilized on glutathione-Sepharose, followed by addition of bacterially expressed S100P. As shown in Fig. 1B, S100P bound specifically to IQGAP1 and not to GST alone, and this binding was not observed in the absence of Ca2+. Thus, the S100P-IQGAP1 interaction is direct and strictly Ca2+-dependent.
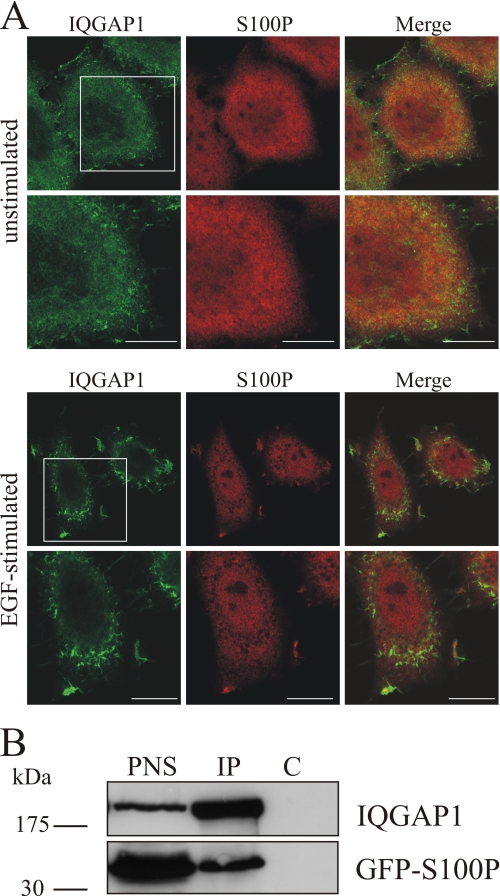
Next we analyzed whether S100P and IQGAP1 could also form a complex within cells. Therefore, we first visualized the intracellular localization of the two proteins using immunofluorescence microscopy (Fig. 2A). In serum-starved HeLa cells IQGAP1 was present throughout the cytoplasm showing some enrichment in plasma membrane protrusions, whereas S100P was evenly distributed throughout the cells. EGF stimulation, known to increase intracellular Ca2+ levels, triggered a co-recruitment of both proteins to membrane ruffles indicative of intracellular complex formation. More directly, an in vivo association of S100P and IQGAP1 was analyzed by co-immunoprecipitation approaches. These experiments employed HeLa cells expressing a GFP-S100P fusion protein to circumvent the limited reactivity of our anti-S100P antibodies in Western blots. Fig. 2B reveals that the anti-IQGAP1 antibody co-immunoprecipitated GFP-S100P at a free Ca2+ concentration of 2.5 μm, thereby providing additional evidence for an intracellular complex formation between S100P and IQGAP1.
FIGURE 2.
Co-localization and co-immunoprecipitation of S100P and IQGAP1. A, EGF-induced co-localization of endogenous S100P and IQGAP1 to membrane ruffles. HeLa cells were serum-starved for 16 h and either kept unstimulated or were stimulated with 50 ng/ml EGF for 5 min. After fixation and permeabilization, cells were stained with a polyclonal antibody against human IQGAP1 (green channel) and a monoclonal antibody against human S100P (red channel) followed by appropriate fluorescently labeled secondary antibodies. The lower panels show magnifications of the boxed areas. Scale bars, 10 μm. B, co-immunoprecipitation of GFP-S100Pwt with IQGAP1. HeLa cells were transiently transfected with pEGFP-S100Pwt. After 24 h, cells were lysed, and equal amounts of the PNS were subjected to immunoprecipitation (IP) with a polyclonal anti-IQGAP1 antibody or nonspecific anti-mouse IgG polyclonal antibodies (control, C) in the presence of 2.5 μm free Ca2+. The PNS and the precipitated proteins were analyzed by Western blot using a monoclonal anti-IQGAP1 and a monoclonal anti-GFP antibody.
S100P-IQGAP1 Complex Defines a Novel Type of S100-Target Protein Interaction
The interaction of S100P and IQGAP1 was analyzed in more detail by mapping the respective binding sites. Therefore, a series of affinity chromatography approaches was performed employing mutant forms of the two proteins.
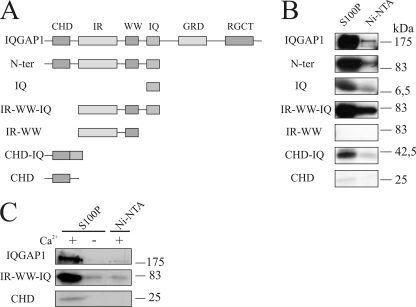
To identify the S100P binding domain in IQGAP1, [35S]methionine-labeled IQGAP1 mutants were used (Fig. 3A). In a first set of experiments, we could rule out that the C-terminal half of IQGAP1 contains the binding motif (data not shown). Therefore, we next analyzed mutants encompassing the N-terminal IQGAP1 domains. The in vitro translated mutants as well as full-length IQGAP1 were probed for their binding to bacterially expressed His-S100Pwt immobilized on Ni-NTA-agarose using Ni-NTA-agarose beads as a negative control. Fig. 3B reveals that all derivatives containing the IQ domain showed significant binding to S100P. A considerably weaker interaction was seen for the isolated CHD, whereas the IR-WW mutant showed no binding. As compared with all other mutants tested, IR-WW is characterized by the lack of both the CHD and the IQ domain, indicating that the latter two contain binding sites for S100P.
FIGURE 3.
Mapping of the S100P-binding site in IQGAP1. A, schematic overview of IQGAP1 mutants that were tested for S100P binding. Abbreviations used are as follows: IR, domain with six IQGAP-specific internal repeats; WW, domain with two conserved Trp (W) residues; IQ, domain harboring four IQ-motifs. B, binding to immobilized S100P. The indicated IQGAP1 mutants comprising the different domains as well as the full-length protein were translated and [35S]methionine-labeled in vitro and subsequently loaded onto Ni-NTA-agarose beads alone or beads containing immobilized His-S100Pwt in the presence of Ca2+. After washing with a Ca2+-containing buffer, bound proteins were eluted and detected by autoradiography. Note that all IQGAP1 domains except the IR-WW mutant bound to His-S100Pwt, although the interaction of CHD was rather weak. N-ter, N terminus. C, analysis of the Ca2+-dependent interaction of the CHD and the IQ domain with His-S100P. [35S]Methionine-labeled full-length IQGAP1, CHD, or IR-WW-IQ was loaded onto Ni-NTA-agarose beads containing His-S100Pwt or Ni-NTA-agarose beads alone, and beads were then processed as described above. The Ca2+ dependence of the interaction was analyzed by using either Ca2+- or EGTA-containing buffers.
The CHD and IQ domains had previously been identified as sites of interaction for other Ca2+-binding EF hand proteins. IQ harbors the binding site for calmodulin, S100B as well as for myosin essential light chain (20, 23, 24), whereas the CHD was shown to contain an additional low affinity binding site for calmodulin (25). This scenario is reminiscent of our findings concerning the S100P interaction with IQGAP1, which also occurs with high affinity in the IQ domain and to a weaker degree in the CHD (see also below).
Having identified the S100P binding domains in IQGAP1, we next analyzed if the interaction with these domains is still Ca2+-dependent. Therefore, the affinity approaches using immobilized S100P were repeated including EGTA controls. Fig. 3C reveals that the interaction of S100P with the respective IQGAP1 domains remained strictly Ca2+-dependent.
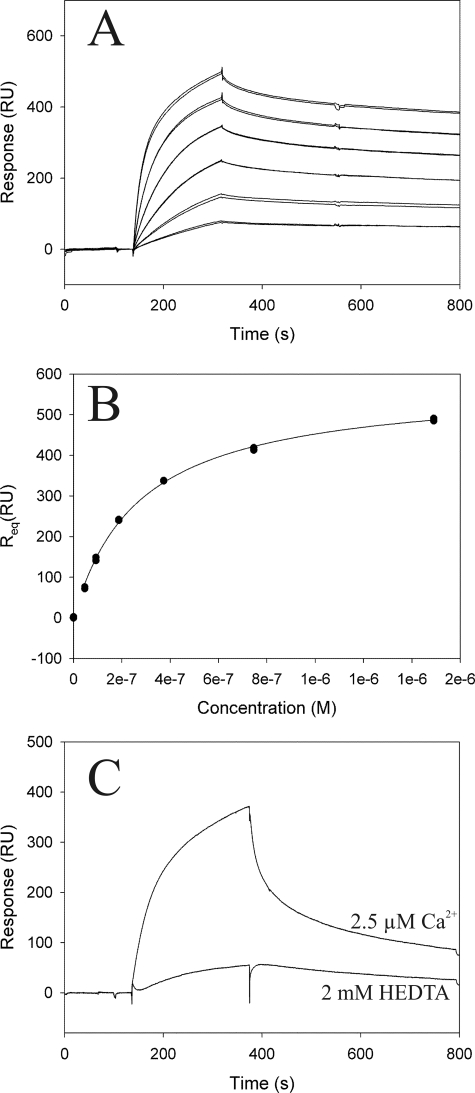
Because the IQ domain of IQGAP1 represented the main binding site for S100P, we employed SPR to obtain quantitative information on the affinity of this interaction (Fig. 4). Purified S100Pwt protein was immobilized on a CM5 sensor chip, and increasing concentrations of purified His-tagged IQ domain were injected in the presence of 50 μm Ca2+. This Ca2+ concentration was chosen because it falls between the KD of the N-terminal (800 μm) and C-terminal EF hand (1.6 μm) of S100P (26). Analysis of the data using a solid state affinity (Scatchard) approach revealed a KD of ∼0.2 μm for the IQ-S100P interaction (Fig. 4, A and B). A comparison of the SPR signal intensities of IQ domain injections on immobilized S100P in the presence or absence of Ca2+ verified the Ca2+ dependence of this interaction (Fig. 4C). This analysis revealed that interaction already occurs at free Ca2+ concentrations of 2.5 μm, which are observed in stimulated cells. The affinity of the IQ-S100P interaction (KD of 0.2 μm) proved to be identical at 50 (Fig. 4, A and B) and 2.5 μm Ca2+ (titrations not shown) indicating that Ca2+ binding to the high affinity C-terminal EF hand is solely responsible for inducing the IQ-S100P interaction.
FIGURE 4.
Interaction of S100Pwt with the IQ domain of IQGAP1 analyzed by SPR. Tagless S100Pwt was immobilized on a CM5 sensor chip and used for analysis of interaction kinetics with the purified IQ domain. A, IQ domain was injected at concentrations of 0.05–1.5 μm and a flow rate of 60 μl/min, and the average binding level at the end of injection (Req) was used for calculation of the steady state affinity. B, steady state affinity determined from the level of binding at equilibrium (Req) as a function of the sample concentration. Calculations were carried out using the BIAevaluation software version 4.1. An approximate dissociation constant of 2e−7 m was calculated for the binding of the IQ domain to S100P. C, Ca2+ dependence of the interaction between the IQ domain and S100Pwt was analyzed by comparison of the signal intensity in the presence or absence of 2.5 μm free Ca2+. RU, response units.
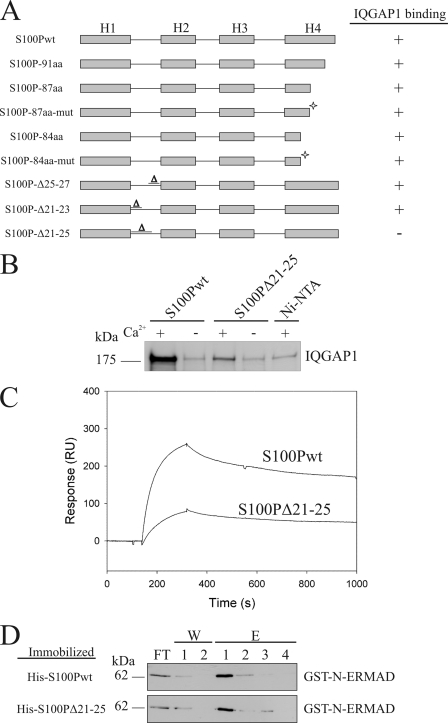
The IQGAP1-binding site in S100P was mapped by employing the S100P mutants shown in Fig. 5A. We first concentrated on the C-terminal extension of helix 4 because this C-terminal sequence displays the highest sequence variations among the S100 proteins and was also shown to contain the ezrin-binding site (21). However, and in contrast to most other interactions of S100 proteins with their targets, the variable C-terminal extension in S100P is not involved in IQGAP1 binding. [35S]Methionine-labeled full-length IQGAP1 still associated with all C-terminally truncated S100P mutants in the presence of Ca2+ (Fig. 5A). Therefore, we next focused on other variable regions in S100P as potential binding sites for IQGAP1. To identify such regions, we compared the three-dimensional structure of Ca2+-loaded S100P with that of S100A10, an S100 protein incapable of binding IQGAP1 (data not shown). The comparison revealed that, in addition to the C-terminal extension, an area within the first EF hand loop showed major structural differences between the two S100 proteins. Specifically, S100A10 has suffered a three amino acid deletion within this loop that could potentially contribute to IQGAP1 binding in S100P. Thus, we introduced similar deletions in S100P and analyzed their effect on IQGAP1 binding. Of these deletion mutants, S100PΔ21–25 showed a strongly reduced interaction with IQGAP1, which could also be confirmed by SPR measurements (Fig. 5, B and C). A 10-fold lower KD of ∼2 μm could be determined in titration experiments, thus identifying amino acids 21–25 of the first EF hand of S100P as important for IQGAP1 binding.
FIGURE 5.
Mapping of the IQGAP1-binding site in S100P. A, overview of the different S100P mutants that were tested for IQGAP1 binding. Binding or no binding is indicated by + or −. C-terminally truncated mutants, some containing additional mutations in helix 4 as indicated by an asterisk (S100P-87aa-mut: T82A, S83L, H86N; S100P-84aa-mut: T82A, S83L), as well as mutants with deletions in the first EF hand loop were analyzed in comparison with the full-length protein. B, amino acids (aa) 21–25 are essential for IQGAP1 binding. [35S]Methionine-labeled full-length IQGAP1 was added to immobilized His-S100Pwt or His-S100PΔ21–25 in the presence or absence of Ca2+. Bound IQGAP1 was detected by autoradiography. Note the strongly reduced amount of IQGAP1 bound to immobilized S100PΔ21–25 as compared with S100Pwt. C, SPR analysis comparing the binding of IQ to S100Pwt and S100PΔ21–25. 3000 response units (RU) of S100Pwt or S100PΔ21–25 were immobilized on a CM5 sensorchip. Sensorgrams were recorded following the injection of purified IQ domain on the immobilized S100P proteins in the presence of 50 μm Ca2+. D, analysis of the interaction of S100PΔ21–25 and S100Pwt with ezrin's N-ERMAD. GST-N-ERMAD was added to the immobilized His-tagged S100P proteins in the presence of Ca2+, and flow-through fractions were collected (FT). After washing with a Ca2+-containing buffer (W), bound GST-N-ERMAD was eluted with EGTA (E). The collected fractions were analyzed by immunoblotting using a monoclonal anti-GST antibody.
Given the significant IQGAP1-binding defect in S100PΔ21-25, we next aimed to establish whether other biological properties of S100P are affected by the Δ21–25 deletion. In these studies, we concentrated on the well documented interaction of S100P with the N-terminal domain of ezrin (N-ERMAD), which is Ca2+-dependent and dimer-specific (8). An affinity chromatography employing human N-ERMAD revealed that this binding was not compromised in S100PΔ21–25 (Fig. 5C). Furthermore, the interaction remained strictly Ca2+-dependent because no binding of N-ERMAD to immobilized S100PΔ21–25 was seen under EGTA conditions (data not shown). Thus, we conclude that only the Ca2+-dependent IQGAP1 binding is affected in the S100PΔ21–25 mutant, identifying this mutant as an ideal control for studying cellular effects of S100P that are mediated by binding to IQGAP1.
S100P Affects EGF-dependent MEK1/2 Activation via Interaction with IQGAP1
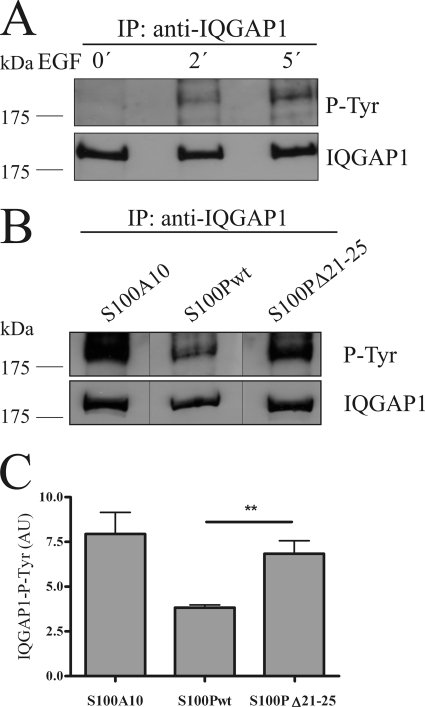
To study the effect of the S100P-IQGAP1 complex formation on the regulatory functions of IQGAP1, we analyzed whether S100P binding affects the interaction with the actin regulators Cdc42 or Rac1. However, no alteration in the binding of Cdc42 and Rac1 to IQGAP1 could be observed in the presence of S100P (for Cdc42 see supplemental Fig. 1), and the expression of S100P had no significant effect on Cdc42-mediated actin rearrangements in migrating cells (data not shown). Therefore, we focused on other regulatory functions of IQGAP1, specifically those associated with tyrosine phosphorylation of IQGAP1 that had been implicated in the activation of its function in signaling cascades (13, 14, 27, 28). We first established that EGF stimulation, which is known to elicit Ca2+ transients and also triggers MAPK signaling, induces a time-dependent tyrosine phosphorylation of IQGAP1 in HeLa cells (Fig. 6A). Next, we analyzed whether this tyrosine phosphorylation of IQGAP1 was affected by overexpression of S100P. To verify that this overexpression approach specifically targeted IQGAP1, we overexpressed closely related but IQGAP1 binding-deficient S100 derivatives in control experiments. Two controls were employed as follows: S100PΔ21–25 that shows the same properties as S100Pwt except IQGAP1 binding and S100A10. Like S100Pwt both were expressed as GFP- or YFP-tagged fusion proteins showing comparable expression rates as verified by fluorescence microscopy (data not shown). Interestingly, overexpression of GFP-S100Pwt led to a significantly reduced EGF-triggered tyrosine phosphorylation of IQGAP1 as compared with that observed in cells expressing the IQGAP1-binding deficient mutant GFP-S100PΔ21–25 (Fig. 6, B and C). The latter cells showed a similar rate of IQGAP1 tyrosine phosphorylation as cells overexpressing YFP-S100A10, i.e. an S100 protein incapable of interacting with IQGAP1.
FIGURE 6.
S100P interferes with EGF-induced tyrosine phosphorylation of IQGAP1. A, HeLa cells were serum-starved and either kept unstimulated or were stimulated with 50 ng/ml EGF for 2 or 5 min. Cells were then lysed, and the cleared cell lysates were subjected to immunoprecipitation (IP) using a polyclonal anti-IQGAP1 antibody. Time-dependent tyrosine phosphorylation of the precipitated IQGAP1 was analyzed by immunoblotting using a monoclonal anti-phosphotyrosine (P-Tyr) antibody (upper panel). Reprobing of the blot with a monoclonal anti-IQGAP1 antibody was carried out to verify that equal amounts of protein were precipitated (lower panel). B, HeLa cells expressing GFP-S100Pwt, GFP-S100PΔ21–25, or YFP-S100A10 (an S100 protein incapable of binding IQGAP1 used as control) were serum-starved and stimulated with 50 ng/ml EGF for 5 min. The tyrosine phosphorylation status of the precipitated IQGAP1 was analyzed as in A. Black lines indicate that the gel was cut between the lanes. C, statistical evaluation of EGF-induced IQGAP1 tyrosine phosphorylation in the presence of different S100 derivatives. ECL signal intensities of precipitated tyrosine-phosphorylated IQGAP1 protein bands from five independent experiments (a typical example is shown in B) were quantified by densitometry, and results were corrected for the amount of IQGAP1 in the corresponding sample, as revealed by blotting for total IQGAP1. Data represent means ±S.E. Statistical significance was calculated using Student's t test (**, p < 0.01). AU, arbitrary units.
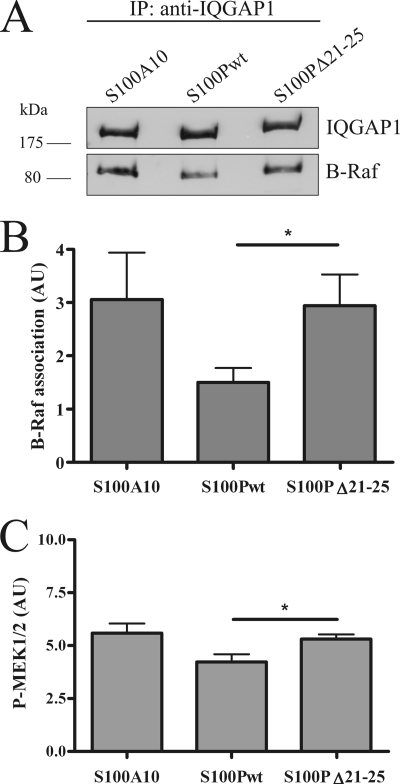
IQGAP1 can serve as a scaffolding protein in MAPK signaling cascades. It directly interacts with B-Raf, MEK1/2, and Erk1/2, and cellular IQGAP1 is required for efficient signal transduction following EGF stimulation that involves tyrosine phosphorylation (14, 29–31). Because S100P interfered with EGF-triggered tyrosine phosphorylation of IQGAP1, we next examined whether this has an impact on the scaffolding function of IQGAP1 in EGF-dependent MAPK signaling. Therefore, we analyzed IQGAP1-B-Raf complex formation and MEK1/2 activation in HeLa cells overexpressing S100Pwt, S100PΔ21–25, or S100A10. Fig. 7, A and B, reveals that B-Raf binding to IQGAP1 is significantly impaired in the presence of elevated S100Pwt. Underscoring the specificity of this effect, no reduction in IQGAP1-B-Raf interaction was observed in cells overexpressing the S100PΔ21–25 mutant. In line with reduced B-Raf binding to IQGAP1, overexpression of S100Pwt also reduced MEK1/2 activation as compared with overexpression of S100PΔ21–25 or S100A10 (Fig. 7C). Although this effect is statistically significant, it is less pronounced than the S100P-induced reduction in EGF-triggered tyrosine phosphorylation of IQGAP1. Most likely, this reflects the fact that MEK1/2 are intermediates of the MAPK signaling cascades that are activated downstream of tyrosine-phosphorylated IQGAP1 but also receive activation signals from EGF-controlled upstream mediators other than IQGAP1.
FIGURE 7.
S100P affects IQGAP1-B-Raf complex formation and reduces EGF-dependent MEK1/2 activation. A, HeLa cells expressing GFP-S100Pwt, GFP-S100PΔ21–25, or YFP-S100A10 were serum-starved and stimulated with 50 ng/ml EGF for 5 min. Cells were lysed, and the cleared cell lysates were subjected to immunoprecipitation (IP) using a polyclonal anti-IQGAP1 antibody. IQGAP1 and co-precipitated B-Raf were identified by immunoblotting using a monoclonal anti-IQGAP1 antibody and a monoclonal anti-B-Raf antibody. B, statistical evaluation of EGF-induced B-Raf-IQGAP1 complex formation in cells overexpressing different S100 derivatives. ECL signal intensities of co-precipitated B-Raf protein bands from seven independent experiments were quantified by densitometry, and results were corrected for the amount of precipitated IQGAP1 in the corresponding sample. Data represent means ± S.E. Statistical significance was calculated using Student's t test (*, p < 0.05). C, HeLa cells expressing different S100 proteins were subjected to analysis of EGF-induced MEK1/2 activation. Cells were serum-starved and stimulated with 50 ng/ml EGF for 5 min. Whole cell lysates were then subjected to SDS-PAGE and Western blot analysis using an anti-phospho-MEK1/2 antibody and, following stripping, an antibody detecting total MEK1/2. The amount of phospho-MEK1/2 was quantified by densitometric analysis of blots from six independent experiments, and in each case was corrected for the amount of total MEK1/2 in the corresponding lysate. Data of the densitometric scans are expressed as means ± S.E. Statistical significance was calculated using Student's t test (*, p < 0.05). AU, arbitrary units.
DISCUSSION
The multidomain structure of IQGAP1 enables the protein to function as an important scaffold in the cortical cytoskeleton, among other things activating actin polymerization and dynamics through Cdc42 and Rac1 and also assembling signaling intermediates of the MAPK pathway to achieve spatial proximity required for more efficient signal transduction. These diverse functions are controlled by regulation of IQGAP1 itself. One important feature of this regulatory circuit is the autoinhibition of IQGAP1 by intramolecular association of the GRD and RGCT domains that affects the IQGAP1 interaction with other effectors (12). Protein kinase C-mediated phosphorylation can relieve this intramolecular masking of binding sites resulting in an interaction-competent conformation known to bind both regulators of cytoskeleton and cell adhesion as well as signaling molecules of the MAPK pathway. Once the open conformation is established, the activity of IQGAP1 as an assembly factor can still be controlled by regulatory protein interactions. A prominent example is the interaction with calmodulin that can occur in the presence and absence of Ca2+ albeit with different consequences (32, 33). Although Ca2+-free calmodulin, which binds to the third and fourth IQ motif of IQGAP1, does not affect other protein interactions, Ca2+-loaded calmodulin, which can bind to all four IQ motifs, essentially abrogates the interactions with other IQGAP1 targets. Thus, by inhibiting the association of IQGAP1 with Cdc42 and F-actin as well as with proteins of the MAPK module, Ca2+/calmodulin silences the stimulatory action of IQGAP1 in both Cdc42-induced cortical cytoskeleton dynamics and MAPK signaling. Here, we identify S100P as another Ca2+-regulated EF hand protein that can represent a link between Ca2+ signaling and IQGAP1 activity. However, in contrast to Ca2+/calmodulin, Ca2+/S100P selectively interferes only with the IQGAP1-dependent MAPK activation, which occurs following EGF stimulation but not with the IQGAP1-Cdc42 interaction. Thus S100P appears to regulate only a set of IQGAP1-target protein interactions in response to intracellular Ca2+ transients.
Like calmodulin, Ca2+/S100P binds to the IQ domain of IQGAP1. The interaction is of considerable affinity with a KD of 0.2 μm and occurs within stimulated cells, as revealed here by EGF treatment that is known to elevate intracellular Ca2+. Structural information on the S100-target protein complexes available to date has identified amphipathic helices as binding sites in the target proteins. The hydrophobic face of these helices binds to a hydrophobic cleft in the S100 protein that in part is formed by hydrophobic side chains of the C-terminal extension of the second F helix (3). Mutations within or deletions of the C-terminal extension typically affect this type of target protein binding and, in the case of S100P, abrogate the interaction with ezrin (21). This scenario is different for the binding of Ca2+/S100P to IQGAP1, which does not require the C-terminal extension of the S100 protein. Rather, a sequence in the first EF hand loop appears to constitute at least part of the binding site. This first EF hand loop of S100 proteins is considered atypical as it contains 14 instead of 12 residues (1, 2) and in S100P has a greatly reduced affinity toward Ca2+ as compared with the canonical C-terminal EF hand (26). Thus, the N-terminal EF hand loop is expected to remain Ca2+-free even at the elevated Ca2+ levels occurring in stimulated cells and hence is available for protein interactions. The S100P-IQGAP1 interaction therefore appears to constitute a novel type of target binding executed by S100 proteins. The structural principle of this interaction is probably shared with S100B because the C-terminal extension in S100B is also dispensable for IQGAP1 binding (20).
The Ca2+-driven binding of S100P to IQGAP1 described here appears to selectively participate in modulating signaling functions of IQGAP1 that at least in part involve EGF-stimulated tyrosine phosphorylation and cross-talk with the MAPK pathway. Tyrosine phosphorylation of IQGAP1 has been shown to occur downstream of the activation of several receptor tyrosine kinases, including VEGF and EGF receptors with c-Src probably functioning as an intermediate in the signaling cascade (13, 14, 27). Within this pathway, tyrosine-phosphorylated IQGAP1 signals to B-Raf and downstream MAPKs, and in endothelial cells this signaling has been shown to be required for efficient cell proliferation (14). IQGAP1 directly binds B-Raf and other downstream kinases of the MAPK/ERK module thereby activating the signaling cascade, and tyrosine phosphorylation of IQGAP1 has been implicated in positively regulating this activation (14, 29, 31). Overexpression of S100P significantly reduces the EGF-dependent tyrosine phosphorylation of IQGAP1 indicating that S100P can function as a negative feedback regulator responding to Ca2+ elevations, which are also initiated upon EGF stimulation. This negative regulatory role also reflects itself in the S100P-induced reduction of MEK1/2 activation that is seen in EGF-treated HeLa cells. It is most likely due to a reduced binding of B-Raf to IQGAP1 that could be caused by reduced tyrosine phosphorylation in IQGAP1. Importantly, the phenotypes seen upon overexpression of S100P specifically reflect the consequence of the S100P-IQGAP1 complex formation because they are not observed when the binding-deficient mutant S100PΔ21–25 is employed instead of wild-type S100P.
Our data indicate that Ca2+ signaling in response to EGF or other stimuli can induce at least two responses with respect to IQGAP1 activity. On one hand, and as shown before, it can trigger the formation of a Ca2+/calmodulin-IQGAP1 complex that appears to reside in an inhibited conformation unable to interact with and thereby activate mediators of MAPK signaling and regulators of the F-actin and microtubule dynamics (18). On the other hand, Ca2+ transients can lead to the binding of Ca2+/S100P to IQGAP1, which will interfere with MAPK signaling but not with the Cdc42-mediated activation of actin rearrangements, for example those that are required for efficient cell migration. In line with this assumption, overexpression of S100P, although negatively affecting tyrosine phosphorylation on IQGAP1 and MEK1/2 activation, has no significant effect on actin rearrangements known to be triggered by Cdc42.4 How these two effects, i.e. partial and full inactivation of IQGAP1 by Ca2+/S100P and Ca2+/calmodulin binding, respectively, are regulated themselves is not known. Differences in the respective Ca2+ transient required for activating S100P and calmodulin binding, respectively, or tissue- and cell type-specific differences in the relative expression levels of S100P and calmodulin could be relevant for an initiation of only one of the two scenarios. Future experiments have to reveal this, for example by determining the intracellular Ca2+ threshold for binding of S100P versus calmodulin to IQGAP1.
Supplementary Material
Acknowledgment
We thank A. Gibadulinova (Slovak Academy of Sciences, Bratislava, Slovakia) for monoclonal anti-S100P antibodies.
This work was supported by Grant Ge2/016/10 from the Interdisciplinary Centre for Clinical Research of the Muenster University Medical School (to V. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
V. Gerke, unpublished data.
A. Heil and V. Gerke, unpublished data.
- CHD
- calponin homology domain
- GRD
- GAP related domain
- RGCT
- Ras GAP C-terminal domain
- N-ERMAD
- N-terminal ERM association domain
- PNS
- post-nuclear supernatant
- SPR
- surface plasmon resonance
- Ni-NTA
- nickel-nitrilotriacetic acid
- HEDTA
- N-(2-hydroxyethyl)ethylenediaminetriacetic acid.
REFERENCES
- 1. Donato R. (1999) Biochim. Biophys. Acta 1450, 191–231 [DOI] [PubMed] [Google Scholar]
- 2. Heizmann C. W., Fritz G., Schäfer B. W. (2002) Front. Biosci. 7, d1356–d1368 [DOI] [PubMed] [Google Scholar]
- 3. Santamaria-Kisiel L., Rintala-Dempsey A. C., Shaw G. S. (2006) Biochem. J. 396, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leclerc E., Fritz G., Vetter S. W., Heizmann C. W. (2009) Biochim. Biophys. Acta 1793, 993–1007 [DOI] [PubMed] [Google Scholar]
- 5. Zimmer D. B., Wright Sadosky P., Weber D. J. (2003) Microsc. Res. Tech. 60, 552–559 [DOI] [PubMed] [Google Scholar]
- 6. Bhattacharya S., Bunick C. G., Chazin W. J. (2004) Biochim. Biophys. Acta 1742, 69–79 [DOI] [PubMed] [Google Scholar]
- 7. Koltzscher M., Gerke V. (2000) Biochemistry 39, 9533–9539 [DOI] [PubMed] [Google Scholar]
- 8. Koltzscher M., Neumann C., König S., Gerke V. (2003) Mol. Biol. Cell 14, 2372–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown M. D., Sacks D. B. (2006) Trends Cell Biol. 16, 242–249 [DOI] [PubMed] [Google Scholar]
- 10. Brandt D. T., Grosse R. (2007) EMBO Rep. 8, 1019–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnson M., Sharma M., Henderson B. R. (2009) Cell. Signal. 21, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 12. Grohmanova K., Schlaepfer D., Hess D., Gutierrez P., Beck M., Kroschewski R. (2004) J. Biol. Chem. 279, 48495–48504 [DOI] [PubMed] [Google Scholar]
- 13. Yamaoka-Tojo M., Ushio-Fukai M., Hilenski L., Dikalov S. I., Chen Y. E., Tojo T., Fukai T., Fujimoto M., Patrushev N. A., Wang N., Kontos C. D., Bloom G. S., Alexander R. W. (2004) Circ. Res. 95, 276–283 [DOI] [PubMed] [Google Scholar]
- 14. Meyer R. D., Sacks D. B., Rahimi N. (2008) PLoS One 3, e3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Noritake J., Watanabe T., Sato K., Wang S., Kaibuchi K. (2005) J. Cell Sci. 118, 2085–2092 [DOI] [PubMed] [Google Scholar]
- 16. Kuroda S., Fukata M., Nakagawa M., Fujii K., Nakamura T., Ookubo T., Izawa I., Nagase T., Nomura N., Tani H., Shoji I., Matsuura Y., Yonehara S., Kaibuchi K. (1998) Science 281, 832–835 [DOI] [PubMed] [Google Scholar]
- 17. Sacks D. B. (2006) Biochem. Soc. Trans. 34, 833–836 [DOI] [PubMed] [Google Scholar]
- 18. Briggs M. W., Sacks D. B. (2003) FEBS Lett. 542, 7–11 [DOI] [PubMed] [Google Scholar]
- 19. Ren J. G., Li Z., Sacks D. B. (2008) J. Biol. Chem. 283, 22972–22982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mbele G. O., Deloulme J. C., Gentil B. J., Delphin C., Ferro M., Garin J., Takahashi M., Baudier J. (2002) J. Biol. Chem. 277, 49998–50007 [DOI] [PubMed] [Google Scholar]
- 21. Austermann J., Nazmi A. R., Müller-Tidow C., Gerke V. (2008) J. Biol. Chem. 283, 29331–29340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zobiack N., Gerke V., Rescher U. (2001) FEBS Lett. 500, 137–140 [DOI] [PubMed] [Google Scholar]
- 23. Hart M. J., Callow M. G., Souza B., Polakis P. (1996) EMBO J. 15, 2997–3005 [PMC free article] [PubMed] [Google Scholar]
- 24. Weissbach L., Bernards A., Herion D. W. (1998) Biochem. Biophys. Res. Commun. 251, 269–276 [DOI] [PubMed] [Google Scholar]
- 25. Ho Y. D., Joyal J. L., Li Z., Sacks D. B. (1999) J. Biol. Chem. 274, 464–470 [DOI] [PubMed] [Google Scholar]
- 26. Becker T., Gerke V., Kube E., Weber K. (1992) Eur. J. Biochem. 207, 541–547 [DOI] [PubMed] [Google Scholar]
- 27. Yamaoka-Tojo M., Tojo T., Kim H. W., Hilenski L., Patrushev N. A., Zhang L., Fukai T., Ushio-Fukai M. (2006) Arterioscler. Thromb. Vasc. Biol. 26, 1991–1997 [DOI] [PubMed] [Google Scholar]
- 28. Usatyuk P. V., Gorshkova I. A., He D., Zhao Y., Kalari S. K., Garcia J. G., Natarajan V. (2009) J. Biol. Chem. 284, 15339–15352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren J. G., Li Z., Sacks D. B. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10465–10469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roy M., Li Z., Sacks D. B. (2004) J. Biol. Chem. 279, 17329–17337 [DOI] [PubMed] [Google Scholar]
- 31. Roy M., Li Z., Sacks D. B. (2005) Mol. Cell. Biol. 25, 7940–7952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li Z., Sacks D. B. (2003) J. Biol. Chem. 278, 4347–4352 [DOI] [PubMed] [Google Scholar]
- 33. Briggs M. W., Sacks D. B. (2003) EMBO Rep. 4, 571–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.