Abstract
The ability of interferons (IFNs) to inhibit viral replication and cellular proliferation is well established, but the specific contribution of each IFN-stimulated gene (ISG) to these biological responses remains to be completely understood. In this report we demonstrate that ISG54, also known as IFN-induced protein with tetratricopeptide repeats 2 (IFIT2), is a mediator of apoptosis. Expression of ISG54, independent of IFN stimulation, elicits apoptotic cell death. Cell death and apoptosis were quantified by propidium iodide uptake and annexin-V staining, respectively. The activation of caspase-3, a key mediator of the execution phase of apoptosis, was clearly apparent in cells expressing ISG54. The anti-apoptotic B cell lymphoma-xl (Bcl-xl) protein inhibited the apoptotic effects of ISG54 as did the anti-apoptotic adenoviral E1B-19K protein. In addition, ISG54 was not able to promote cell death in the absence of pro-apoptotic Bcl family members, Bax and Bak. Analyses of binding partners of ISG54 revealed association with two homologous proteins, ISG56/IFIT1 and ISG60/IFIT3. In addition, ISG60 binding negatively regulates the apoptotic effects of ISG54. The results reveal a previously unidentified role of ISG54 in the induction of apoptosis via a mitochondrial pathway and shed new light on the mechanism by which IFN elicits anti-viral and anti-cancer effects.
Keywords: Apoptosis, Cell Death, Cytokine, Innate Immunity, Interferon, Mitochondrial Apoptosis, shRNA
Introduction
Interferons (IFNs) are well recognized as essential mediators of innate immunity by their ability to inhibit viral replication and stimulate immune effector cells (1, 2). They are also known for their ability to inhibit cellular proliferation, and for this reason they have been used clinically as anti-neoplastic agents for decades (3–7). There are three main types of IFN, classified by their ability to bind distinct receptors (8). IFNs elicit these biological responses by binding to cell surface receptors that activate a Janus kinase signal transducer and activator of transcription (JAK-STAT) pathway and other signal pathways to regulate gene expression (9–13). Genes that are induced transcriptionally by IFNs, often referred to as IFN-stimulated genes (ISGs),4 can play roles that affect both the inhibition of viral replication and the inhibition of cellular proliferation. For example, genes that promote apoptosis can impact viral replication by sacrificing the cell for the survival of the host and impact the progression of cancer by reducing transformed cell survival.
Type I and type III IFNs confer their biological effects by stimulating the activation of a transcription factor complex called IFN-stimulated gene factor 3 (ISGF3)(14, 15). ISGF3 contains tyrosine-phosphorylated dimers of STAT1 and STAT2, and the STAT2-associated factor IFN regulatory factor 9 (IRF-9) (9, 16, 17). ISGF3 is responsible for the transcriptional induction of a set of ISGs, one of which is ISG54 (18–21). Notably, the ISG54 gene is also induced in response to viral infection, independent of IFN but dependent on activation of the IRF-3 transcription factor, a documented stimulator of apoptosis (22–26).
The response of cells to IFN is complex and can depend on tissue type and state of cellular transformation. Although IFN has been found to promote apoptosis in cancerous cells, conversely it has been shown to stimulate the proliferation of primary normal human cells (21). The biological response to IFN is a concerted effect of multiple induced genes, both antiproliferative and proliferative, and has been the subject of many investigations (21, 27, 28). Some of the IFN-induced genes that have been described for their roles in apoptosis include promyelocytic leukemia (PML), protein kinase R (PKR), IRF-1, 2′-5′ oligoadenylate synthase (OAS), and TNF-related apoptosis inducing ligand (TRAIL) among others (3, 29–32). IFN-mediated apoptosis also was found to be associated with activation of caspases by way of a mitochondrial cell death pathway. Treatment of cancer cells with IFN caused a loss of mitochondrial membrane potential and release of cytochrome c (6). Still, a direct link of ISGs to mitochondrial-mediated cell death remains to be characterized. In this report we identify ISG54 as a mediator of mitochondrial cell death.
The ISG54 gene codes for a protein of ∼54 kDa (472 aa) with tetratricopeptide repeats (TPR) and has also been designated IFN-induced protein with tetratricopeptide repeats 2 (IFIT2). It is one of four related human ISGs with characteristic TPR motifs (33). This motif is a sequence of 34 moderately conserved amino acids that form a structure composed of two antiparallel helices and is involved in protein-protein interactions (34). The function of the TPR motifs in ISG54 remains to be ascertained. However, a TPR motif of a related family member, ISG56, has been reported to bind to the human Papillomavirus E1 replication protein and inhibit its function (35). In addition, studies addressing the function of ISG54 and ISG56 have indicated a negative effect on translation by interacting with the eukaryotic initiation factor 3 (36–38). In this study we have determined that ISG54 forms a multiprotein complex with the related proteins ISG56 and ISG60 and stimulates cell death via a mitochondrial pathway. The results indicate that ISG54 plays a significant role in the mediation of cellular apoptosis in response to viral infection or IFN signaling.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cell lines were obtained from American Type Culture Collection and cultured in DMEM with 8% FBS. Wild type baby mouse kidney (BMK) cells and bax−/−, bak−/− double knock-out BMK cells (39) were kind gifts of Dr. Wei-Xing Zong (Stony Brook University).
Plasmids and Transfections
Full-length human ISG54 cDNA was cloned using PCR into the following vectors: pcDNA3 (Invitrogen) with an N-terminal T7 tag, pCGN with an N-terminal HA tag (Addgene), pEF-1V5-HisB with an C-terminal V5 tag (Invitrogen), and pEGFP-N1 with a C-terminal monomeric GFP tag (Clontech). The pEGFP-N1 plasmid was modified to introduce mutations A206K, L221K, and F223R to ensure a monomeric GFP (mGFP) (40). ISG54-mGFP was subcloned into the tetracycline-inducible vector pREvTRE (Clontech). pRev-ISG54-mGFP expression was induced with pLib-rtTAm2-iresTRSID-iresPuro, a gift of Dr. Michael J. Ausserlechner (Medical University Innsbruck) (41) Truncated forms of ISG54 encompassing 1–4TPR domains (aa 1–208), 2–9TPR domains (aa 94–472), 3–9TPR domains (aa 138–472), and 4–9TPR domains (aa 172–472) were cloned into pEF-1V5-HisB. Human ISG56, ISG58, and ISG60 genes were similarly cloned. pcDNA3-Bcl-xL plasmid was a gift from Dr. Colin Duckett (University of Michigan). pSPLuc-poly(A) plasmid containing firefly luciferase gene with 60 adenine bases at 3′ terminus was a gift from Dr. Philip Marsden (University of Toronto) (42). The pSPLuc-poly(A) luciferase gene was cloned into pcDNA3 to generate pcDNA3-luc-poly(A) with a T7 promoter site. IRES-luc-poly(A) plasmid was constructed by cloning hepatitis C virus internal ribosome entry site (IRES) from H4325Wt (a gift from Dr. Eckard Wimmer, Stony Brook University) into pcDNA3-lucPolyA. The dominant negative p53 plasmid (aa 320–393) was a kind gift from Dr. Ute Moll (Stony Brook University) (43). Plasmid encoding adenoviral E1B-19K was a kind gift of Dr. Lynne Vales (University of Medicine and Dentistry, New Jersey) (44). Plasmids encoding adenoviral E1B-55K and E4-Orf6 were kind gifts of Dr. Patrick Hearing (Stony Brook University). TransIT LT1 reagent (Mirus) was used for DNA transfections. TransMessenger Transfection Reagent (Qiagen) was used for RNA transfections.
Western Blot, Immunoprecipitation, and Antibodies
For Western blots, cell lysates were prepared in 0.5% Nonidet P-40 buffer, and proteins were separated by SDS-PAGE (45). Proteins were transferred to nitrocellulose membranes (Thermo Scientific), and reactive signals were detected with the Odyssey Imager (Li-COR Biosciences). For immunoprecipitation, 400–500 μg of protein were incubated with antibody overnight at 4 °C, and immunocomplexes were collected on protein G-agarose beads (Invitrogen). Polyclonal rabbit anti-ISG54 antibodies were generated against GST-ISG54 (381–473 aa). Commercial antibodies used included monoclonal anti-V5 (Invitrogen), polyclonal rabbit anti-HA (Santa Cruz Biotechnology), monoclonal anti-FLAG (Sigma), anti-active caspase-3 (Cell Signaling), monoclonal anti-calnexin (Santa Cruz Biotechnology), secondary antibody conjugated to TRITC (The Jackson Laboratory), normal IgG (Santa Cruz Biotechnology), and secondary antibodies for Odyssey Imager anti-mouse (Rockland) and anti-rabbit (Invitrogen). Images were analyzed using Image J software (NIH).
Cell Death
Cells were trypsinized from plates, washed with PBS, and stained with propidium iodide according to the manufacturer's instructions (Invitrogen). For annexin V staining cells were resuspended in staining buffer, and allophycocyanin-conjugated annexin V was added according to manufacturer instructions (BD Pharmingen). Cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences). The gate was set for GFP expression, and 10,000 cells in each population were analyzed with BD CellQuest software. Cells positive for GFP expression were isolated by fluorescence-activated cell sorting (FACS) with a BD Biosciences FACS Vantage cell sorter. Salubrinal was used at the concentrations indicated (Tocris Bioscience) (46).
Microscopy
Immunofluorescence was performed after cell fixation with 4% paraformaldehyde and permeabilization in 0.2% Triton X-100. Cells were incubated with primary antibody followed by secondary antibody conjugated to TRITC. Cells were visualized with Zeiss Axiovert 200M and Axiovision Version 4.5. Live cell imaging was performed with cells seeded on glass-bottom plates (Mattek Corp.). Mitochondria were visualized by incubation with 500 nm MitoTracker Orange CMTMRos before imaging (Invitrogen). The Zeiss Tempcontrol 37–2 Digital and CTI Controller 3700 was used with the Zeiss LSM 510 laser scanning microscope system (Zeiss) and an alpha Plan-FLUAR 100×/1.45 objective. Live cell images were captured using Zeiss LSM 5 Pascal imaging software.
Translation Analysis
mRNAs encoding the luciferase gene were transfected into cells expressing ISG54 and evaluated for translation. In vitro transcription was performed with Ambion mMessage mMachine T7 kit. Luciferase RNA was synthesized from a T7 promoter in pcDNA3-luc-poly(A), and a 5′-7-methyl guanosine (m7G) cap was added to the mRNA in vitro. IRES-luc-poly(A) was used to prepare 5′-IRES-regulated luciferase mRNA. After reactions, DNA templates were degraded with DNase, and mRNA was precipitated with LiCl. Integrity and the amount of transcribed RNA were evaluated with electrophoresis and spectrophotometry. Cells were transfected with ISG54-mGFP or mGFP, and the following day they were transfected with the 5′-cap luciferase mRNA or IRES luciferase mRNA. Five hours after RNA transfection the cells were sorted for GFP fluorescence (BD Biosciences FACS Vantage cell sorter) and then tested for luciferase activity with Promega Dual-Luciferase Reporter Assay and Lumat LB9507 Luminometer (EG G Berthold).
Mass Spectrometry Analysis
HeLa cells were transfected with plasmids T7-ISG54 pcDNA3 or T7-pcDNA3 and were subsequently treated with 1000 units/ml IFN-α (gift from Roche Applied Science, Nutley, NJ) overnight. Cell lysates were prepared and incubated with anti-T7 antibodies conjugated to agarose beads (Novagen). Proteins were eluted with Novagen elution buffer, neutralized, and treated with iodoacetamide. Proteins were separated by SDS-PAGE and stained with SilverQuest (Invitrogen). Protein samples were analyzed by mass spectrometry by ProtTech Inc. (Norristown, PA).
Glycerol Gradient Sedimentation
Cell lysates were prepared with isotonic buffer (140 mm NaCl, 5 mm Tris, pH 8.2, 5 mm EDTA, 0.5% Nonidet P-40) and clarified by centrifugation at 18,000 × g for 15 min. Samples were concentrated with Amicon Ultra4 Filter columns, and 700 μg of protein were applied to the top of 25–40% glycerol gradients (47). One gradient was prepared with molecular mass references corresponding to 50 μg of bovine serum albumin (66 kDa), alcohol dehydrogenase (150 kDa), catalase (250 kDa), and apoferritin (448 kDa). Samples were centrifuged in Beckman SW60Ti rotor for 40 h at 40,000 rpm at 4 °C, and 150-μl fractions were collected from the top of each gradient for analysis. Mass marker references were visualized with Coomassie R250 staining.
shRNA Knockdown
Four double-stranded oligonucleotides targeting human ISG54 cDNA were designed for use in the Ambion pSilencerTM system and were cloned into pSilencer 2.1-U6-puro vector. The oligonucleotides corresponded to nucleotides (nt) 136 (5′-GATCCGCTTCATAAGATGCGTGAATTCAAGAGATTCACGCATCTTATGAAGCTTTTTTGGAAA-3′), nt 652 (5′-GATCCGGAATTCAGTAAAGAGCTTCTCAAGAGAAAGCTCTTTACTGAATTCCTTTTTTGGAAA-3′), nt 1075 (5′-GATCCGGAATTCAGTAAAGAGCTTCTCAAGAGAAAGCTCTTTACGAATTCCTTTTTTGGAAA-3′), and nt 1203 (5′-GATCCACCAGAAATCAAGGGAGAATTCAAGAGATTCTCCCTTGATTTCTGGTTTTTTTGGAAA-3′). HeLa cells were transfected with one of the pSilencer ISG54 shRNA plasmids or with a pSilencer control containing a random shRNA sequence (Ambion). Stable cell lines were selected for resistance to 660 ng/ml puromycin. ISG54 knockdown efficiency was evaluated by Western-blot and ImageJ software (NIH).
RESULTS
ISG54 Expression Promotes Cell Death
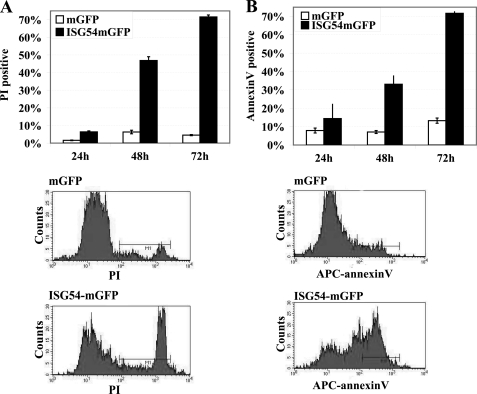
The negative effect of ISG54 on cellular proliferation was first detected by monitoring the expression of ISG54-mGFP in transfected HeLa cell cultures. The percentage of cells expressing ISG54-mGFP was low and decreased dramatically with time in comparison to cells expressing mGFP (supplemental Fig. 1A). To evaluate the viability of cells expressing ISG54, we quantified the percent of dead cells in the culture by staining with propidium iodide (PI). PI is excluded from viable cells but enters cells with disrupted membranes and intercalates the DNA. HeLa cells were transfected with plasmids encoding ISG54-mGFP or mGFP, and at 24, 48, and 72 h post-transfection the cells were harvested and incubated with PI. Flow cytometry was used to quantify cells positive for both GFP and PI fluorescence. At 24 h post-transfection the cells expressing ISG54-GFP showed a slight increase in cell death compared with the mGFP control, and at 48 and 72 h there was a dramatic increase in cell death (Fig. 1A). Analysis of total cell cultures transfected with ISG54-mGFP clearly indicated cell death was apparent in the ISG54-GFP-positive cells and not in GFP-negative cells (supplemental Fig. 1B). Because expression of ISG54-mGFP promoted cell death, it was challenging to establish a stable inducible cell line due to basal expression of the repressed gene. However, transient inducible expression of ISG54 regulated by a tetracycline-responsive promoter showed a similar lethal effect of ISG54 in cell cultures (supplemental Fig. 1C).
FIGURE 1.
Expression of ISG54 promotes cell death by apoptosis. A, HeLa cells were transfected with plasmids encoding ISG54-mGFP or mGFP, stained with PI at 24, 48, and 72 h posttransfection, and analyzed with flow cytometry. The percent of cells positive for both GFP and PI was quantified. Lower panels, the single dimension histogram of flow cytometry data of cells expressing mGFP or ISG54-mGFP after 48 h transfection is shown. B, cells were transfected with mGFP or ISG54-mGFP for 24, 48, or 72 h, stained with allophycocyanin-annexin V, and analyzed with flow cytometry. A gate was set for GFP expression, and the distribution of annexin V signal was analyzed. The percent of cells positive for GFP and annexin V staining is shown. Lower panels, single dimension histograms of flow cytometry data are shown for the 48-h time point. APC, allophycocyanin.
Apoptosis in Response to ISG54
To evaluate whether ISG54-expressing cells die by way of apoptosis, we measured staining with annexin V. Annexin V binds to the phosphatidylserine groups that are exposed on the outer leaflet of the plasma membrane in apoptotic cells (48). HeLa cells were transfected with plasmids encoding ISG54-mGFP or mGFP, and at 24, 48, and 72 h they were stained with allophycocyanin-annexin V. Cells expressing ISG54-mGFP were found to display a high percent of annexin V staining in comparison with the population of cells expressing mGFP (Fig. 1B). This result is consistent with the loss of membrane phospholipid asymmetry, a molecular change indicative of apoptosis. ISG54 expression promoted apoptosis in various human cell lines including HT1080 and HEC1B (supplemental Fig. 2, A and B).
To ensure the expression level of ISG54 was at par with that expressed during a response to IFN, we compared protein levels by Western blot (supplemental Fig. 2, C and D). Endogenous ISG54 is clearly evident in the cells as early as 6 h after IFN stimulation (supplemental Fig. 2C). To compare the levels of ISG54-GFP with endogenous ISG54 protein, GFP-positive cells expressing ISG54-mGFP were isolated by FACS, and the levels of ISG54 were compared with cells treated with IFNα. A Western blot comparison with antibody to ISG54 clearly showed the protein expression of ISG54-mGFP in cells was similar to that of endogenous ISG54 induced by IFN, indicating physiologically relevant levels.
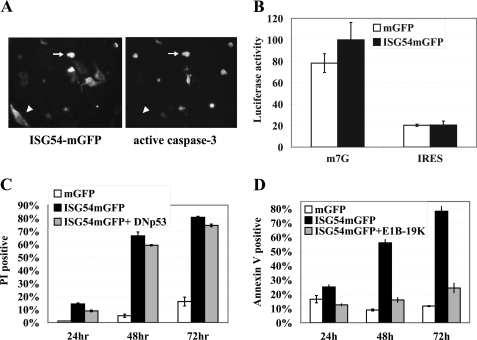
Activation of the family of caspase proteases is essential for the characteristic changes associated with apoptosis (49–52). The caspases are synthesized as zymogens, and proteolytic processing is usually required for their activation. Caspase-3 is a primary executioner enzyme responsible for cleavage of many cellular substrates, and for this reason we evaluated the activation of caspase-3 in cells expressing ISG54. Cells were transfected with ISG54-mGFP and evaluated for the presence of the activated form of caspase-3 by immunofluorescence (Fig. 2A). There was a significant correlation between cells expressing ISG54-mGFP and activated caspase-3 (>60%). This was particularly evident in cells displaying morphological changes of apoptosis. In healthy cells, ISG54-mGFP resided in the cytoplasm and did not appear to have active caspase-3. Caspase-3 activation became evident with cell shrinkage associated with apoptosis. To evaluate the effect of caspase inhibition on ISG54-induced apoptosis, a pan-caspase inhibitor (benzyloxycarbonyl-VAD-fluoromethyl keto) was tested. The results indicated this peptide inhibitor reduced cell death, although not completely (supplemental Fig. 3A). Together the results suggest that ISG54 promotes cell death via apoptosis.
FIGURE 2.
Mechanistic evaluation of ISG54-mediated apoptosis. A, shown is an image of fluorescent cells transfected with ISG54-GFP (left) and an image of immunofluorescence staining with anti-active caspase-3 at 48 h after transfection (right). The arrows indicate apoptotic cells positive for ISG54 and caspase-3, and arrowheads indicate healthy cell positive for ISG54 and negative for caspase-3. B, shown is the effect of ISG54 on translation. HeLa cells were transfected with plasmids expressing mGFP or ISG54-mGFP for 16 h and subsequently were transfected with m7G-capped and -polyadenylated mRNA encoding luciferase (m7G) or with polyadenylated mRNA encoding luciferase regulated by a 5′ IRES (IRES). GFP-positive cells were isolated by FACS 5 h after mRNA transfection, and luciferase activity was measured. C, the effect of dominant negative p53 on ISG54-mediated cell death is shown. HeLa cells were co-transfected with plasmids encoding mGFP, ISG54-mGFP, or ISG54-mGFP and a FLAG-tagged truncation of p53 (DNp53) (aa 320–393). Expression of dominant negative FLAG-p53 was confirmed by the immunofluorescence (not shown). Cells were harvested 24, 48, or 72 h after transfection, and cell death was quantified by PI staining of GFP-positive cells with flow cytometry. D, the effect of adenoviral E1B-19K is shown. HeLa cells were transfected with plasmids encoding mGFP, ISG54-mGFP, or ISG54-mGFP and adenoviral E1B-19K. Cells were harvested at 24, 48, or 72 h post-transfection, and apoptosis was quantified by annexin V staining of GFP-positive cells with flow cytometry.
Because previous reports suggested that ISG54 and ISG56 inhibited translation and this effect might trigger apoptosis, we assessed the in vivo effect of ISG54 on translation (36–38). Cells expressing either ISG54-mGFP or mGFP were transfected with 5′-m7G cap luciferase mRNA or 5′- IRES luciferase mRNA that was synthesized in vitro. Cells expressing green fluorescence were isolated by FACS 5 h after RNA transfection. The cells were lysed, and luciferase activity was measured as a read-out of translation. ISG54 did not demonstrate any negative effects on the translation of either capped or IRES mRNA in this assay (Fig. 2B). Therefore, we propose that ISG54-induced apoptosis is not the sole result of translation inhibition.
To evaluate whether transcriptional regulation by the p53 tumor suppressor is involved in the initiation of apoptosis by ISG54, we co-expressed ISG54-mGFP with a dominant negative p53 mutant. The p53 mutant encodes the carboxyl domain of p53 and oligomerizes with wt p53 to inhibit formation of functional p53 tetramers, thereby disrupting its nuclear function as a transcription factor (43, 53). Expression of the dominant negative p53 mutant did not have a significant effect on ISG54 induced cell death (Fig. 2C). Adenoviral proteins that have been characterized as inhibitors of p53, E1B-55K, and E4 orf6 also did not influence the ability of ISG54 to induce cell death (supplemental Fig. 3B) (54, 55). These results indicate the apoptotic effect of ISG54 is independent of p53. Adenovirus does express another viral protein that inhibits cellular apoptosis, E1B-19K (56). We tested the effect of E1B-19K expression and found that it abolished the ability of ISG54 to promote cellular apoptosis (Fig. 2D).
ISG54-induced Apoptosis Is Executed via a Mitochondrial Pathway
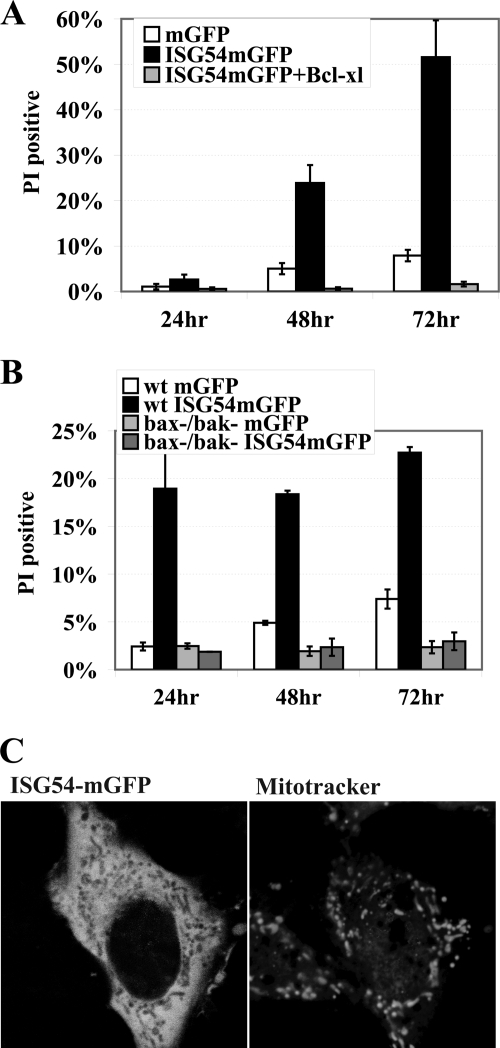
Apoptosis triggered by a mitochondrial pathway is mediated primarily by the B cell lymphoma-2 (Bcl-2) family of proteins (57, 58). The Bcl-2 proteins characteristically have either pro-apoptotic or anti-apoptotic activities by regulating mitochondrial outer membrane permeability. Because the E1B-19K adenoviral protein is an anti-apoptotic Bcl2 homologue and it blocked apoptosis by ISG54, we investigated the effects of cellular Bcl-2 proteins. The effect of Bcl-xl, a potent anti-apoptotic inhibitor of mitochondrial permeability, was evaluated by co-transfection with ISG54-mGFP (59). Overexpression of Bcl-xL was found to abolish ISG54-induced cell death, indicating ISG54 promoted a mitochondrial pathway of apoptosis (Fig. 3A).
FIGURE 3.
ISG54-induced apoptosis is executed via a mitochondrial pathway. A, Bcl-xL blocks cell death induced by ISG54. HeLa cells were transfected with plasmids encoding mGFP, ISG54-mGFP, or ISG54-mGFP co-transfected with Bcl-xL, and GFP-positive cells were analyzed by flow cytometry for PI staining at 24, 48, or 72 h post transfection. B, cells lacking Bax and Bak genes are resistant to ISG54-induced apoptosis. Wild type BMK cells or bax−/− bak−/− double knock-out BMK cells were transfected with mGFP or ISG54-mGFP for the times indicated, and GFP-positive cells were analyzed by flow cytometry after PI staining. C, live-cell imaging of cells expressing ISG54-mGFP (left) and stained with MitoTracker Orange (right) reveals the ISG54mGFP localizes in the cytoplasm but is excluded from mitochondria.
In a converse approach we evaluated the response of cells that lack the pro-apoptotic Bax and Bak genes (39, 60). Bax and Bak proteins induce mitochondrial outer membrane permeability resulting in caspase activation (57, 58). The bax−/− bak−/− double knock-out cells are resistant to diverse stimuli that trigger mitochondrial apoptosis. These cells and the corresponding wild type cells were transfected with ISG54-mGFP. There was no increase in cell death in bax−/− bak−/− cells even after 72 h of ISG54 expression, indicating that Bax and Bak activity is required for apoptosis in response to ISG54 (Fig. 3B). Because the results suggested that ISG54 promoted apoptosis via a mitochondrial pathway, we evaluated whether ISG54 localized to mitochondria. Cells expressing ISG54-mGFP were stained with MitoTracker orange, a fluorescent probe that accumulates in metabolically active mitochondria. Live cell imaging was performed and showed ISG54-mGFP expression diffuse throughout the cytoplasm but excluded from mitochondria (Fig. 3C). The results indicate that although ISG54 triggers a mitochondrial pathway of apoptosis, it does not enter the mitochondria.
The Bcl proteins are also known to localize to the endoplasmic reticulum (ER) and to be involved in ER stress as it affects mitochondrial apoptosis (61–63). To determine whether ISG54-GFP is present in the ER, we evaluated co-localization by immunofluorescence with antibodies to calnexin, an integral ER protein. Results indicated ISG54 is present in the ER but not at higher levels than in the cytoplasm (supplemental Fig. 4, A and B). In addition, an inhibitor of ER stress, salubrinal, did not block the ability of ISG54 to promote cell death (supplemental Fig. 4C) (46).
ISG54 Forms Oligomers with Itself and ISG56 and ISG60
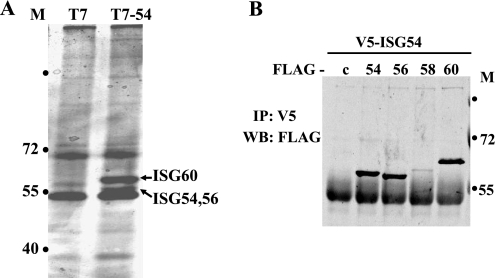
Preliminary studies with a yeast two-hybrid screen indicated ISG54 could bind to ISG56 and ISG60 (data not shown). To pursue this observation we evaluated protein-protein interactions in mammalian cells. Cells were transfected with T7-ISG54 expression plasmid or T7 empty vector, and immunocomplexes were collected with anti-T7 antibody. Eluted proteins were separated by SDS-PAGE and stained with silver (Fig. 4A). Distinct protein bands were apparent in the immunocomplexes from cells expressing T7-ISG54. These proteins were excised from the gel and identified by mass spectrometry as ISG60, ISG56, and ISG54.
FIGURE 4.
ISG54 forms oligomers with itself and ISG56 and ISG60. A, HeLa cells were transfected with T7-ISG54 or T7-empty vector for 24 h, and immunocomplexes were collected from cell lysates with anti-T7 antibodies. Proteins were separated by SDS-PAGE and silver-stained, and proteins specific to ISG54 expression were identified by mass spectrometry as ISG60 (upper band) and ISG56 and ISG54 (lower band). Molecular mass standards are indicated (M) in kDa. B, cells were co-transfected with V5-ISG54 and FLAG-empty vector (c) or FLAG-ISG54 (54), FLAG-ISG56 (56), FLAG-ISG58 (58), or FLAG-ISG60 (60). ISG54 protein was immunoprecipitated (IP) with anti-V5 antibodies, and associated proteins were detected by Western blot (WB) with anti-FLAG antibodies.
To confirm these protein interactions, co-immunoprecipitation studies were performed with cells co-transfected with plasmids encoding ISG54-V5 and either FLAG-ISG60, FLAG-ISG58, FLAG-ISG56, FLAG-ISG54, or FLAG-empty vector (c). ISG54 was immunoprecipitated with anti-V5, and the associated proteins were detected by Western blot with anti-FLAG antibody (Fig. 4B). Input analyses demonstrated efficient expression of all proteins (supplemental Fig. 5A). The co-immunoprecipitations indicated ISG54 was able to bind to itself, ISG60, and ISG56 but not efficiently to ISG58.
ISG54 TPR Binding Domains
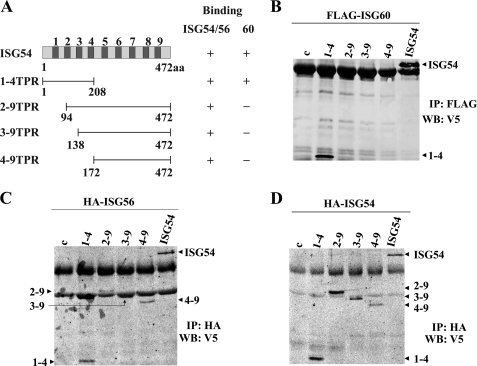
The structure of the ISG54 protein is predicted to contain nine TPRs distributed uniformly along the molecule (Fig. 5A). To determine the regions of ISG54 that are critical for binding to its partners, we created V5-tagged truncations of the protein. The 1–4TPR construct encodes the first four TPR domains at the N terminus, whereas 2–9TPR, 3–9TPR, and 4–9TPR lack the N terminus but contain noted TPR domains and the complete C terminus. These V5-tagged truncations of ISG54 were expressed with either FLAG-ISG60, HA-ISG56, or HA-ISG54 and evaluated for their ability to bind the proteins by co-immunoprecipitation and Western blot. The ISG54 proteins were efficiently expressed (supplemental Fig. 5, B–D). Analysis of ISG60 interactions with the ISG54 truncations demonstrated a binding requirement of the first TPR domain of ISG54. Only full-length ISG54 or the 1–4TPR protein could co-immunoprecipitate with ISG60, whereas the 2–9TPR, 3–9TPR, and 4–9TPR did not bind ISG60 (Fig. 5B). The co-immunoprecipitation results were markedly different with ISG56 or ISG54. All of the ISG54 truncations could bind to the ISG56 and ISG54 proteins (Fig. 5, C and D). The results indicate that the interface of ISG54 association with itself or with ISG56 is distinct from the interface of ISG54 with ISG60.
FIGURE 5.
Domains of ISG54 that recognize binding partners. A, a schematic diagram of ISG54-V5 noting the location of the nine TPR motifs (78) is shown. Deletion mutations of ISG54-V5 are shown with amino acid positions and TPR content. The column to the right indicates the ability of ISG54, ISG56, or ISG60 to bind to ISG54-V5 proteins (+). B, HeLa cells were co-transfected with FLAG-ISG60 and either empty vector (c), one of the truncations of ISG54-V5 (noted by TPR content), or full-length ISG54-V5. Immunoprecipitation (IP) of ISG60 was performed with anti-FLAG antibody, and Western blot (WB) was performed with anti-V5 antibody to detect associated ISG54 fragments. C, cells were co-transfected with HA-ISG56 and versions of ISG54-V5 as in B. Immunoprecipitation of ISG56 was performed with anti-HA antibody, and associated ISG54 fragments were detected by Western blot with anti-V5 antibody. D, cells were co-transfected with HA-ISG54 and versions of ISG54-V5 as in B. Immunoprecipitation of ISG54 was performed with anti-HA antibody, and associated ISG54 fragments were identified by Western blot with anti-V5 antibody.
ISG54, ISG56, and ISG60 Exist as 150–200-kDa Complexes in the Cytoplasm
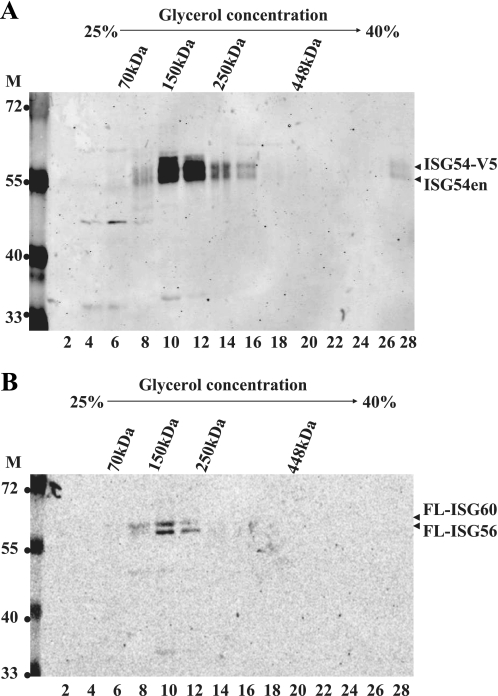
Considering the ability of ISG54 to bind to itself and to ISG54 and ISG60, we investigated the oligomeric nature of the complexes. The sedimentation rate of the proteins through glycerol gradients was used to estimate the molecular mass of the complexes. Cells were co-transfected with plasmids ISG54-V5, FLAG-ISG56, and FLAG-ISG60 and stimulated with IFN-α overnight. Detergent lysates were sedimented through 25–40% glycerol gradients, and fractions containing the proteins were identified by Western blot. The position of molecular mass markers in gradients was used to approximate the mass of the protein complexes. ISG54-V5 sedimented primarily in fractions corresponding to 150–200 kDa, and the endogenous ISG54 protein was found to sediment in the same fractions (Fig. 6A). Western blots detecting the ISG56 and ISG60 proteins demonstrated that they sedimented in similar fractions consistent with a molecular mass of 150–200 kDa (Fig. 6B). Comparable experiments were performed without IFN treatment and provided similar results (data not shown). Notably, monomeric forms of the ISGs were never detected.
FIGURE 6.
Glycerol gradient sedimentation of ISG protein complexes. HeLa cells were co-transfected with ISG54-V5, FLAG-ISG56, and FLAG-ISG60 plasmids and treated with IFN-α for 18 h. Lysates were sedimented through 25–40% glycerol gradients, and fractions were collected. A, shown is a Western blot with anti-ISG54 antibody detected ISG54-V5 and endogenous ISG54 (ISG54en). B, A Western blot with anti-FLAG (FL) antibody detected both FLAG-ISG56 and FLAG-ISG60. The sedimentation positions of molecular mass markers in the gradients are indicated at the top. Molecular mass standards for Western blot are indicated (M) in kDa.
ISG54 Knockdown Decreases Sensitivity to IFN-α-induced Cell Death
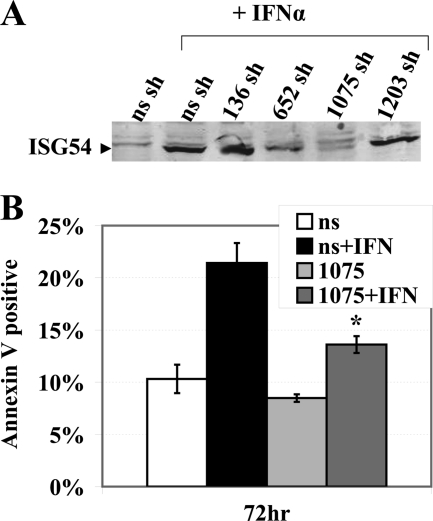
IFN induces the transcription of a number of genes that can have concerted effects on apoptosis. To determine whether a decrease in one gene product, ISG54, would have a significant impact on the ability of IFN to stimulate apoptosis, we evaluated shRNA knockdown of ISG54. The Ambion pSilencer shRNA system was used to design and generate four plasmids expected to produce shRNA corresponding to different regions of ISG54 mRNA (supplemental Fig. 6A). These plasmids and a nonspecific shRNA control plasmid were used to generate stable HeLa cell lines. The cells were evaluated by Western blot for the level of ISG54 protein produced in response to IFN-α stimulation (Fig. 7A). One of the shRNA cell lines, 1075, showed reduced protein levels of ISG54 by more than 60% as assayed with ImageJ software. Therefore, we evaluated the ability of IFNα to stimulate apoptosis in the ISG54 shRNA 1075 knock-down cell line in comparison with the nonspecific shRNA cell line (Fig. 7B). Cells were treated with IFNα for 72 h and were evaluated by flow cytometry for annexin V staining. The cells expressing ISG54 1075 shRNA displayed significantly less apoptosis. PI staining confirmed reduced cell death in the 1075 shRNA population with effective knockdown of ISG54 (data not shown). Reduced apoptosis in the 1075 shRNA cells was also evident in comparison to other ISG54 shRNA cell lines like 136 that did not have reduced ISG54 protein levels (supplemental Fig. 6B). These results indicate that ISG54 plays a major role in apoptosis in response to IFNα.
FIGURE 7.
Knockdown of ISG54 by shRNA decreases IFN-α-induced apoptosis. A, stable cell lines were established expressing shRNA plasmids directed at four different fragments of ISG54 cDNA or nonspecific (ns) shRNA plasmid. IFN-stimulated cells were analyzed for knockdown efficiency of endogenous ISG54 protein by Western blot with anti-ISG54 antibody. B, cells expressing stable knockdown of ISG54 with 1075 shRNA or nonspecific (ns) shRNA were evaluated for their apoptotic response to IFNα by annexin V staining and flow cytometry. *, calculated p value comparing effects of IFN in cells with 1075 shRNA or nonspecific shRNA was <0.03.
ISG60 Modulates ISG54-induced Apoptosis
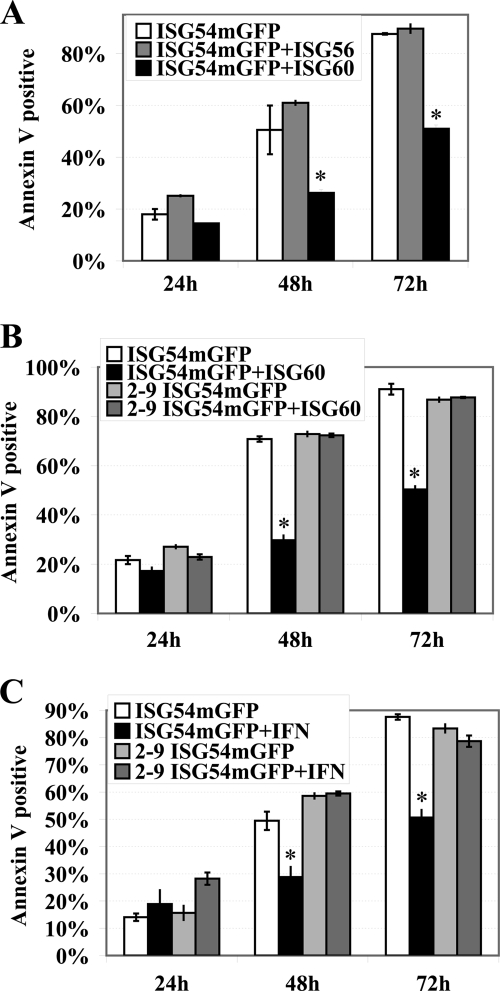
IFN induces transcription of multiple genes that regulate cell survival and apoptosis, and the concerted effects of these ISGs result in specific biological responses. For this reason we determined whether the ISG54 binding partners, ISG56 and ISG60, could modulate the pro-apoptotic effects of ISG54. Apoptosis was evaluated by annexin V staining of cells expressing ISG54-mGFP or ISG54-mGFP with V5-tagged ISG56 or ISG60 (Fig. 8A). ISG56 did not appear to influence the apoptotic effects of ISG54; however, co-expression with ISG60 significantly reduced apoptosis. The modulatory effect of ISG60 was not due to reduced ISG54-mGFP expression; in fact, the number of cells expressing ISG54-mGFP increased with co-expression of ISG60 (supplemental Fig. 7A). Western blot analyses also indicated that ISG60 expression significantly increased ISG54 protein levels (supplemental Fig. 7B). To determine if direct binding of ISG60 to ISG54 was required for its inhibitory effects, we evaluated the truncation of ISG54 that lacks the first TPR and cannot bind ISG60 (Fig. 8B). ISG54-mGFP with TPR domains 2–9 was able to promote apoptosis similar to wt ISG54; however, this response was not suppressed by ISG60. Although ISG60 could inhibit apoptosis induced by wt ISG54, it could not inhibit apoptosis by ISG54 lacking the first TPR. This result indicates that ISG60 must bind to ISG54 to have a suppressive effect.
FIGURE 8.
ISG60 inhibits the proapoptotic effects of ISG54. A, HeLa cells were co-transfected with plasmids encoding either ISG54-mGFP and empty vector or ISG54-mGFP and ISG56-V5 or ISG54-mGFP and ISG60-V5. GFP-positive cells were analyzed for annexin V staining by flow cytometry after 24, 48, or 72 h post-transfection. *, calculated p values for the inhibitory effect of ISG60 were <0.02 and <0.007 at 48 and 72 h, respectively. B, cells were transfected with either ISG54-mGFP and empty vector, ISG54-mGFP and ISG60-V5, 2–9TPR-ISG54-mGFP and empty vector, or 2–9TPR-ISG54-mGFP and ISG60-V5. GFP-positive cells were analyzed for annexin V staining by flow cytometry after 24, 48, or 72 h post-transfection. *, calculated p values for the inhibitory effect of ISG60 were <0.003 and < 0.01 at 24 and 48 h, respectively. C, IFN suppresses the apoptotic effects of ISG54 dependent on the first TPR. HeLa cells were transfected with ISG54-mGFP or 2–9TPR-ISG54-mGFP, and 12-h after transfection the cells were either untreated or treated with 2000 units/ml IFNα. GFP-positive cells were quantified for apoptosis by annexin V staining with flow cytometry at 24, 48, and 72 h post-transfection. *, calculated p values for the inhibitory effects of IFN were <0.01 and <0.008 at 48 and 72 h, respectively.
Expression of ISG54 effectively promoted apoptosis outside the context of IFN stimulation. The ability of ISG60 to suppress apoptosis by ISG54 suggested that IFN stimulation could induce genes that influenced the biological effects of ISG54. To evaluate the effect of IFNα, cells were transfected with ISG54-mGFP or the deletion 2–9TPR-ISG54-mGFP and subsequently were treated with IFNα (Fig. 8C). Analyses with annexin V staining clearly indicated that IFNα treatment reduced the level of apoptosis elicited by full-length ISG54. The pro-survival effects were also evident with an increased number of live cells expressing ISG54-mGFP (supplemental Fig. 7C). In contrast, IFNα had no effect on apoptosis induced by the ISG54 deletion that cannot bind ISG60, 2–9TPR-ISG54-mGFP. Both ISG60 and IFNα treatment suppressed the apoptotic effects of ISG54, and this effect required the first TPR domain of ISG54. The results suggest that ISG60 may be the primary modulator of ISG54 during the IFNα response.
DISCUSSION
IFNs are unique among the cytokines for their ability to confer cellular resistance to viral infections and for their ability to inhibit the growth of cancer cells. Several IFN-induced genes have been recognized for their role in viral defense (64, 65), but a clear understanding of the mechanisms by which IFNs are effective in the treatment of cancer remains to be determined. Type I IFN has been reported to induce gene products that can lead to growth arrest, apoptotic signaling, or alternatively, proliferation (3, 20, 21, 66, 67). The biological response to IFN is complex, which is evident in its ability to stimulate immunosurveillance and cancer immuno-editing as part of its anti-tumor effects (68). In this report we focused our attention on ISG54 in view of the fact that this gene is robustly induced by type I IFNs (18, 19), viral infection (22), or DNA damage (25) and that apoptosis can occur coordinately in reaction to each stimulus.
Our study demonstrates that expression of ISG54 promotes cell death via apoptosis. Flow cytometry was used to quantify cell viability by PI staining and cellular apoptosis by annexin V staining. Because caspase activation is central to the execution phase of apoptosis, we evaluated the activation of caspase-3 by immunofluorescence. Cells expressing ISG54 developed morphological changes that accompany apoptosis, and caspase-3 activity became apparent with these changes. This is the first description of the pro-apoptotic function of ISG54. Previous studies linked ISG54 and ISG56 to the inhibition of translation, particularly translation initiated via an IRES (36–38, 69, 70). However, in our system ISG54 did not decrease cap-dependent or IRES-dependent translation. Therefore although ISG54-induced cell death may be connected to inhibition of protein synthesis, it appears to be triggered by other cellular pathways.
Pathways that lead to caspase activation and apoptosis are often designated as either extrinsic or intrinsic (52, 57, 58, 71). The extrinsic pathway initiates outside the cell by transmembrane death receptors and the subsequent activation of caspases. The intrinsic pathway, also called the mitochondrial pathway, is dependent on pro-apoptotic proteins such as Bax or Bak that induce mitochondrial outer membrane permeability, release of apoptogenic molecules, and activation of caspases. The designations are not completely accurate as the receptor-mediated extrinsic pathway can also trigger the intrinsic pathway. We have found that overexpression of the anti-apoptotic Bcl-xl protein dramatically decreases cell death induced by ISG54 (59). In addition, cells that lack expression of pro-apoptotic Bax and Bak proteins are resistant to the apoptotic effects of ISG54. These data indicate that ISG54 functions to directly or indirectly trigger a mitochondrial pathway of apoptosis. Because the p53 tumor suppressor induces the transcription of pro-apoptotic Bcl-2 proteins NOXA and PUMA, we evaluated the possible contribution of p53 using a dominant negative mutant (43, 53, 72–75). ISG54-induced apoptosis was found to continue unabated in the presence of the p53 interfering mutant. These effects also were apparent with anti-apoptotic viral genes. The adenoviral Bcl-2 homologue, E1B-19K, was able to block the apoptotic effects of ISG54. However, the adenoviral E1B-55K and E4 orf6 proteins that inhibit the function of p53 did not influence the ability of ISG54 to promote apoptosis.
ISG54 triggers cell death via a mitochondrial response that appears to initiate in the cytoplasm. To investigate the means by which ISG54 stimulates this biological response, we explored protein-protein interactions by affinity tag purification and mass spectrometry. This approach identified interactions with two other TPR-containing family members, ISG56 and ISG60, but not with the fourth member of this family, ISG58. The interactions were verified with co-immunoprecipitation methods and revealed that ISG54 also forms complexes with itself. The protein-protein interaction between ISG54 and ISG60 is distinct from that of ISG54 and ISG56 or with ISG54 and itself. The first TPR domain of ISG54 is essential for binding to ISG60, but it is not required for binding ISG54 or ISG56. We have also determined that ISG56 can bind to ISG60 and that both ISG54 and ISG60 are able to form homomeric complexes (data not shown).
The numerous possibilities for protein-protein interactions among these three ISGs led us to estimate the mass of the complex. Glycerol gradient sedimentation indicated the presence of ISG54, ISG56, and ISG60 in multimeric complexes of ∼150–200 kDa. Considering the molecular mass of these ISG proteins, they may form trimeric or tetrameric complexes. The exact stoichiometry and composition of the complex remain to be established, and the range of possible protein interactions evokes many theoretical combinations. Considering the fact that TPR motifs are known to play a role in formation of multiprotein complexes, the concept that these ISGs create a scaffold for docking target proteins is intriguing.
Expression of ISG54 clearly destines the cell for apoptotic death. Notably, however, IFNα is not a strong apoptotic agent, although it induces the transcription of ISG54. In fact IFNα can stimulate cellular proliferation of normal primary cells (21) The biological responses to IFN are cell context-specific and the result of induced expression of many diverse genes. Because ISG54 physically interacts with two other IFN-induced genes, ISG56 and ISG60, we evaluated the effect of these genes on ISG54-induced apoptosis. ISG60 was found to significantly reduce the ability of ISG54 to promote apoptosis. This effect required the first TPR domain of ISG54, which is also required for ISG60 binding to ISG54. Treatment with IFNα elicited a similar pro-survival response, suggesting that ISG60 induced by IFNα is a primary regulator of ISG54-induced cell death.
Results with the shRNA knockdown of ISG54 suggest that it is one of the factors responsible for IFN-stimulated cell death. By promoting apoptosis, ISG54 may also play a critical role in defense of viral infections. The sacrifice of an infected cell can ensure survival of the host, evident in the fact that many viruses have evolved mechanisms to block the cellular apoptotic defense (76, 77). Deciphering the mechanisms by which IFN can promote apoptosis in cancer cells or infected cells can define new therapeutic targets of intervention. Our findings with ISG54 and its interactive partners have identified new possibilities for directed therapy development.
Supplementary Material
Acknowledgments
We extend our appreciation to Dr. Velasco Cimica for sharing his expertise with imaging, Dr. Janaki Iyer for generating the monomeric GFP, Dr. Wei-Xing Zong for advice with apoptosis studies, and to everyone in the laboratory for continual support.
This work was supported, in whole or in part, by National Institutes of Health Grant R21AI067885 (to N. C. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–7.
- ISG
- interferon-stimulated gene
- IRF
- interferon regulatory factor
- Bcl
- B cell lymphoma
- PI
- propidium iodide
- TPR
- tetratricopeptide repeat
- BMK
- baby mouse kidney
- mGFP
- monomeric GFP
- IRES
- internal ribosome entry site
- TRITC
- tetramethylrhodamine isothiocyanate
- aa
- amino acids
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Stark G. R., Kerr I. M., Williams B. R., Silverman R. H., Schreiber R. D. (1998) Annu. Rev. Biochem. 67, 227–264 [DOI] [PubMed] [Google Scholar]
- 2. Borden E. C., Sen G. C., Uze G., Silverman R. H., Ransohoff R. M., Foster G. R., Stark G. R. (2007) Nat. Rev. Drug Discov. 6, 975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clemens M. J. (2003) J. Interferon Cytokine Res. 23, 277–292 [DOI] [PubMed] [Google Scholar]
- 4. Chawla-Sarkar M., Lindner D. J., Liu Y. F., Williams B. R., Sen G. C., Silverman R. H., Borden E. C. (2003) Apoptosis 8, 237–249 [DOI] [PubMed] [Google Scholar]
- 5. Miller C. H., Maher S. G., Young H. A. (2009) Ann. N.Y. Acad. Sci. 1182, 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Thyrell L., Erickson S., Zhivotovsky B., Pokrovskaja K., Sangfelt O., Castro J., Einhorn S., Grandér D. (2002) Oncogene 21, 1251–1262 [DOI] [PubMed] [Google Scholar]
- 7. Li W., Lewis-Antes A., Huang J., Balan M., Kotenko S. V. (2008) Cell Prolif. 41, 960–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fensterl V., Sen G. C. (2009) Biofactors 35, 14–20 [DOI] [PubMed] [Google Scholar]
- 9. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 10. Schindler C., Levy D. E., Decker T. (2007) J. Biol. Chem. 282, 20059–20063 [DOI] [PubMed] [Google Scholar]
- 11. Reich N. C., Liu L. (2006) Nat. Rev. Immunol. 6, 602–612 [DOI] [PubMed] [Google Scholar]
- 12. Platanias L. C. (2005) Nat. Rev. Immunol. 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 13. Pokrovskaja K., Panaretakis T., Grandér D. (2005) J. Interferon Cytokine Res. 25, 799–810 [DOI] [PubMed] [Google Scholar]
- 14. Levy D. E., Kessler D. S., Pine R., Darnell J. E., Jr. (1989) Genes Dev. 3, 1362–1371 [DOI] [PubMed] [Google Scholar]
- 15. Fu X. Y., Kessler D. S., Veals S. A., Levy D. E., Darnell J. E., Jr. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 8555–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martinez-Moczygemba M., Gutch M. J., French D. L., Reich N. C. (1997) J. Biol. Chem. 272, 20070–20076 [DOI] [PubMed] [Google Scholar]
- 17. Gutch M. J., Daly C., Reich N. C. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 11411–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Levy D., Larner A., Chaudhuri A., Babiss L. E., Darnell J. E., Jr. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 8929–8933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lafage M., Clauss I., Couez D., Simonetti J., Wathelet M. G., Huez G. (1992) Genomics 13, 458–460 [DOI] [PubMed] [Google Scholar]
- 20. de Veer M. J., Holko M., Frevel M., Walker E., Der S., Paranjape J. M., Silverman R. H., Williams B. R. (2001) J. Leukoc. Biol. 69, 912–920 [PubMed] [Google Scholar]
- 21. Gomez D., Reich N. C. (2003) J. Immunol. 170, 5373–5381 [DOI] [PubMed] [Google Scholar]
- 22. Andersen J., VanScoy S., Cheng T. F., Gomez D., Reich N. C. (2008) Genes Immun. 9, 168–175 [DOI] [PubMed] [Google Scholar]
- 23. Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. (2002) J. Virol. 76, 5532–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. (1998) EMBO J. 17, 1087–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Weaver B. K., Ando O., Kumar K. P., Reich N. C. (2001) FASEB J. 15, 501–515 [DOI] [PubMed] [Google Scholar]
- 26. Weaver B. K., Kumar K. P., Reich N. C. (1998) Mol. Cell Biol. 18, 1359–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Samarajiwa S. A., Forster S., Auchettl K., Hertzog P. J. (2009) Nucleic Acids Res. 37, D852–D857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Der S. D., Zhou A., Williams B. R., Silverman R. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Z. G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P. P. (1998) Nat. Genet. 20, 266–272 [DOI] [PubMed] [Google Scholar]
- 30. Jagus R., Joshi B., Barber G. N. (1999) Int. J. Biochem. Cell Biol. 31, 123–138 [DOI] [PubMed] [Google Scholar]
- 31. Tamura T., Ishihara M., Lamphier M. S., Tanaka N., Oishi I., Aizawa S., Matsuyama T., Mak T. W., Taki S., Taniguchi T. (1995) Nature 376, 596–599 [DOI] [PubMed] [Google Scholar]
- 32. Zhou A., Paranjape J., Brown T. L., Nie H., Naik S., Dong B., Chang A., Trapp B., Fairchild R., Colmenares C., Silverman R. H. (1997) EMBO J. 16, 6355–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fensterl V., White C. L., Yamashita M., Sen G. C. (2008) J. Virol. 82, 11045–11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Blatch G. L., Lässle M. (1999) Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 35. Terenzi F., Saikia P., Sen G. C. (2008) EMBO J. 27, 3311–3321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hui D. J., Bhasker C. R., Merrick W. C., Sen G. C. (2003) J. Biol. Chem. 278, 39477–39482 [DOI] [PubMed] [Google Scholar]
- 37. Wang C., Pflugheber J., Sumpter R., Jr., Sodora D. L., Hui D., Sen G. C., Gale M., Jr. (2003) J. Virol. 77, 3898–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Terenzi F., Pal S., Sen G. C. (2005) Virology 340, 116–124 [DOI] [PubMed] [Google Scholar]
- 39. Degenhardt K., Sundararajan R., Lindsten T., Thompson C., White E. (2002) J. Biol. Chem. 277, 14127–14134 [DOI] [PubMed] [Google Scholar]
- 40. Shaner N. C., Steinbach P. A., Tsien R. Y. (2005) Nat. Methods 2, 905–909 [DOI] [PubMed] [Google Scholar]
- 41. Ausserlechner M. J., Obexer P., Deutschmann A., Geiger K., Kofler R. (2006) Mol. Cancer Ther. 5, 1927–1934 [DOI] [PubMed] [Google Scholar]
- 42. Mawji I. A., Marsden P. A. (2006) Exp. Biol. Med. 231, 704–708 [PubMed] [Google Scholar]
- 43. Stommel J. M., Marchenko N. D., Jimenez G. S., Moll U. M., Hope T. J., Wahl G. M. (1999) EMBO J. 18, 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vales L. D., Darnell J. E., Jr. (1989) Genes Dev. 3, 49–59 [DOI] [PubMed] [Google Scholar]
- 45. Kumar K. P., McBride K. M., Weaver B. K., Dingwall C., Reich N. C. (2000) Mol. Cell Biol. 20, 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boyce M., Bryant K. F., Jousse C., Long K., Harding H. P., Scheuner D., Kaufman R. J., Ma D., Coen D. M., Ron D., Yuan J. (2005) Science 307, 935–939 [DOI] [PubMed] [Google Scholar]
- 47. Kotanides H., Moczygemba M., White M. F., Reich N. C. (1995) J. Biol. Chem. 270, 19481–19486 [DOI] [PubMed] [Google Scholar]
- 48. Koopman G., Reutelingsperger C. P., Kuijten G. A., Keehnen R. M., Pals S. T., van Oers M. H. (1994) Blood 84, 1415–1420 [PubMed] [Google Scholar]
- 49. Porter A. G., Jänicke R. U. (1999) Cell Death Differ 6, 99–104 [DOI] [PubMed] [Google Scholar]
- 50. Earnshaw W. C., Martins L. M., Kaufmann S. H. (1999) Annu. Rev. Biochem. 68, 383–424 [DOI] [PubMed] [Google Scholar]
- 51. Kumar S. (2007) Cell Death Differ. 14, 32–43 [DOI] [PubMed] [Google Scholar]
- 52. Strasser A., O'Connor L., Dixit V. M. (2000) Annu. Rev. Biochem. 69, 217–245 [DOI] [PubMed] [Google Scholar]
- 53. Shaulian E., Haviv I., Shaul Y., Oren M. (1995) Oncogene 10, 671–680 [PubMed] [Google Scholar]
- 54. Querido E., Blanchette P., Yan Q., Kamura T., Morrison M., Boivin D., Kaelin W. G., Conaway R. C., Conaway J. W., Branton P. E. (2001) Genes Dev. 15, 3104–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dobner T., Horikoshi N., Rubenwolf S., Shenk T. (1996) Science 272, 1470–1473 [DOI] [PubMed] [Google Scholar]
- 56. Han J., Sabbatini P., Perez D., Rao L., Modha D., White E. (1996) Genes Dev. 10, 461–477 [DOI] [PubMed] [Google Scholar]
- 57. Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 58. Adams J. M., Cory S. (2007) Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 60. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Krajewski S., Tanaka S., Takayama S., Schibler M. J., Fenton W., Reed J. C. (1993) Cancer Res. 53, 4701–4714 [PubMed] [Google Scholar]
- 62. Zong W. X., Li C., Hatzivassiliou G., Lindsten T., Yu Q. C., Yuan J., Thompson C. B. (2003) J. Cell Biol. 162, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Szegezdi E., Macdonald D. C., Ní Chonghaile T., Gupta S., Samali A. (2009) Am. J. Physiol. Cell Physiol. 296, C941–C953 [DOI] [PubMed] [Google Scholar]
- 64. Sadler A. J., Williams B. R. (2008) Nat. Rev. Immunol. 8, 559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. George C. X., Li Z., Okonski K. M., Toth A. M., Wang Y., Samuel C. E. (2009) J. Interferon Cytokine Res. 29, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Katsoulidis E., Carayol N., Woodard J., Konieczna I., Majchrzak-Kita B., Jordan A., Sassano A., Eklund E. A., Fish E. N., Platanias L. C. (2009) J. Biol. Chem. 284, 25051–25064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kayagaki N., Yamaguchi N., Nakayama M., Eto H., Okumura K., Yagita H. (1999) J. Exp. Med. 189, 1451–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dunn G. P., Koebel C. M., Schreiber R. D. (2006) Nat. Rev. Immunol. 6, 836–848 [DOI] [PubMed] [Google Scholar]
- 69. Terenzi F., Hui D. J., Merrick W. C., Sen G. C. (2006) J. Biol. Chem. 281, 34064–34071 [DOI] [PubMed] [Google Scholar]
- 70. Hui D. J., Terenzi F., Merrick W. C., Sen G. C. (2005) J. Biol. Chem. 280, 3433–3440 [DOI] [PubMed] [Google Scholar]
- 71. Wang C., Youle R. J. (2009) Annu. Rev. Genet. 43, 95–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000) Science 288, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 73. Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 74. Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 75. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 76. Tschopp J., Thome M., Hofmann K., Meinl E. (1998) Curr. Opin. Genet. Dev. 8, 82–87 [DOI] [PubMed] [Google Scholar]
- 77. Best S. M. (2008) Annu. Rev. Microbiol. 62, 171–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Karpenahalli M. R., Lupas A. N., Söding J. (2007) BMC Bioinformatics 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.